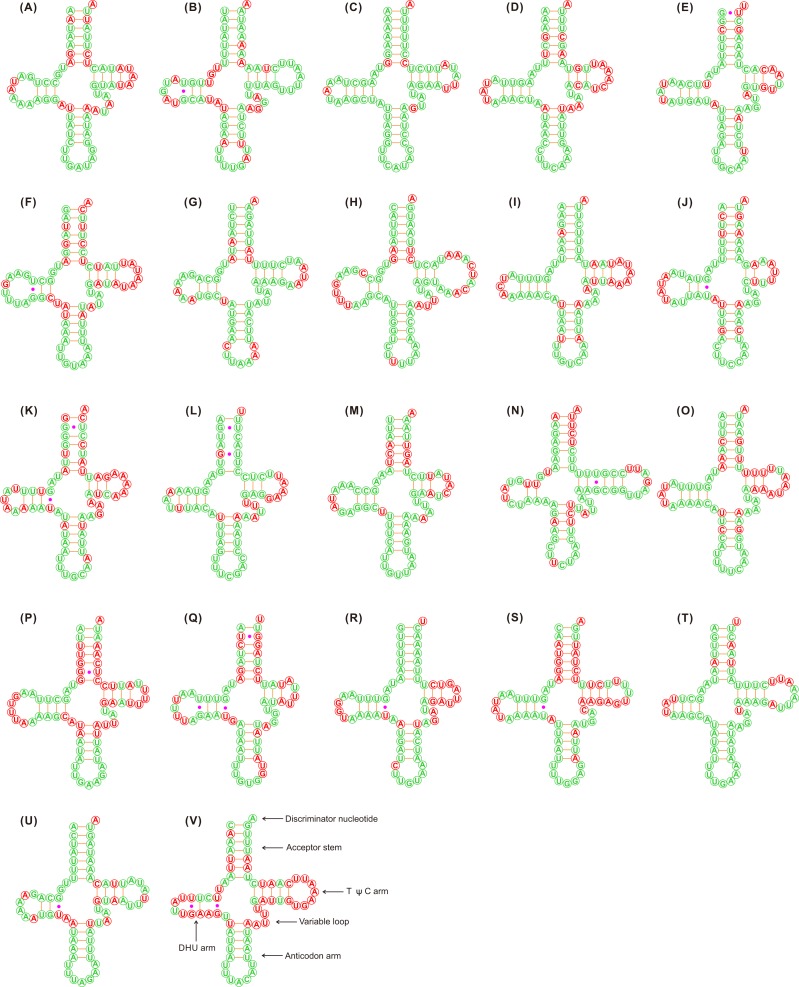
Figure 1. Putative secondary structures of the 22 tRNA genes identified in the mitochondrial genome of Adelphocoris lineolatus.
(A) trnI, (B) trnQ, (C) trnM, (D) trnW, (E) trnC, (F) trnY, (G) trnL2, (H) trnK, (I) trnD, (J) trnG, (K) trnA, (L) trnR, (M) trnN, (N) trnS1, (O) trnE, (P) trnF, (Q) trnH, (R) trnT, (S) trnP, (T) trnS2, (U) trnL1, (V) trnV. All tRNA genes are shown in the order of occurrence in the mitochondrial genome starting from trnI. The nucleotides showing 100% identity in the 15 mirid mitochondrial genomes are marked with green color, and the variable region are marked with red color. Bars indicate Watson-Crick base pairings, and dots between G and U pairs mark canonical base pairings in tRNA.