Abstract
cDNA clones for vitronectin, a cell adhesion-promoting plasma and tissue protein, were isolated from a lambda gt11 library containing cDNA inserts made from human liver mRNA. The library was screened with anti-vitronectin antibodies and the positive clones were further identified with synthetic oligonucleotide probes deduced from the partial amino acid sequence of vitronectin. Nucleotide sequence analysis showed that the largest insert was 1545 bp long and contained the whole sequence corresponding to plasma vitronectin. It showed that vitronectin contains the entire 44-amino acid somatomedin B peptide at its NH2 terminus and, near its COOH terminus, a 34-amino acid glycosaminoglycan binding site in which half of the amino acids are basic residues. Three potential carbohydrate attachment sites are present in the sequence. An Arg-Gly-Asp sequence, which has previously been shown to be the cell attachment site in fibronectin, was found in vitronectin immediately after the NH2-terminal somatomedin B sequence. No other homologies with fibronectin were found. The Arg-Gly-Asp sequence appears to constitute the cell attachment site of vitronectin, since it is in the region where we have previously localized the cell attachment site, its presence correlate with cell attachment activity among the insert-coded polypeptides, and because previous results have shown that synthetic peptides containing the Arg-Gly-Asp sequence inhibit the cell attachment function of vitronectin. The discovery of an Arg-Gly-Asp cell attachment site in a protein with a known cell attachment function emphasizes the general importance of this sequence in cell recognition.
Full text
PDF




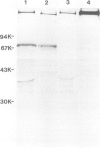
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes D. W., Silnutzer J. Isolation of human serum spreading factor. J Biol Chem. 1983 Oct 25;258(20):12548–12552. [PubMed] [Google Scholar]
- Barnes D. W., Silnutzer J., See C., Shaffer M. Characterization of human serum spreading factor with monoclonal antibody. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1362–1366. doi: 10.1073/pnas.80.5.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M. J., Oldberg A., Pierschbacher M. D., Ruoslahti E. Chondroitin/dermatan sulfate proteoglycan in human fetal membranes. Demonstration of an antigenically similar proteoglycan in fibroblasts. J Biol Chem. 1984 Nov 25;259(22):13742–13750. [PubMed] [Google Scholar]
- Charbit A., Clement J. M., Hofnung M. Further sequence analysis of the phage lambda receptor site. Possible implications for the organization of the lamB protein in Escherichia coli K12. J Mol Biol. 1984 May 25;175(3):395–401. doi: 10.1016/0022-2836(84)90355-3. [DOI] [PubMed] [Google Scholar]
- Fryklund L., Sievertsson H. Primary structure of somatomedin B: a growth hormone-dependent serum factor with protease inhibiting activity. FEBS Lett. 1978 Mar 1;87(1):55–60. doi: 10.1016/0014-5793(78)80132-x. [DOI] [PubMed] [Google Scholar]
- Ginsberg M., Pierschbacher M. D., Ruoslahti E., Marguerie G., Plow E. Inhibition of fibronectin binding to platelets by proteolytic fragments and synthetic peptides which support fibroblast adhesion. J Biol Chem. 1985 Apr 10;260(7):3931–3936. [PubMed] [Google Scholar]
- Hayman E. G., Engvall E., A'Hearn E., Barnes D., Pierschbacher M., Ruoslahti E. Cell attachment on replicas of SDS polyacrylamide gels reveals two adhesive plasma proteins. J Cell Biol. 1982 Oct;95(1):20–23. doi: 10.1083/jcb.95.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman E. G., Pierschbacher M. D., Ohgren Y., Ruoslahti E. Serum spreading factor (vitronectin) is present at the cell surface and in tissues. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4003–4007. doi: 10.1073/pnas.80.13.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman E. G., Pierschbacher M. D., Ruoslahti E. Detachment of cells from culture substrate by soluble fibronectin peptides. J Cell Biol. 1985 Jun;100(6):1948–1954. doi: 10.1083/jcb.100.6.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O., Yamada K. M. Fibronectins: multifunctional modular glycoproteins. J Cell Biol. 1982 Nov;95(2 Pt 1):369–377. doi: 10.1083/jcb.95.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles B. B., Howe C. C., Aden D. P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980 Jul 25;209(4455):497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- Kwok S. C., Ledley F. D., DiLella A. G., Robson K. J., Woo S. L. Nucleotide sequence of a full-length complementary DNA clone and amino acid sequence of human phenylalanine hydroxylase. Biochemistry. 1985 Jan 29;24(3):556–561. doi: 10.1021/bi00324a002. [DOI] [PubMed] [Google Scholar]
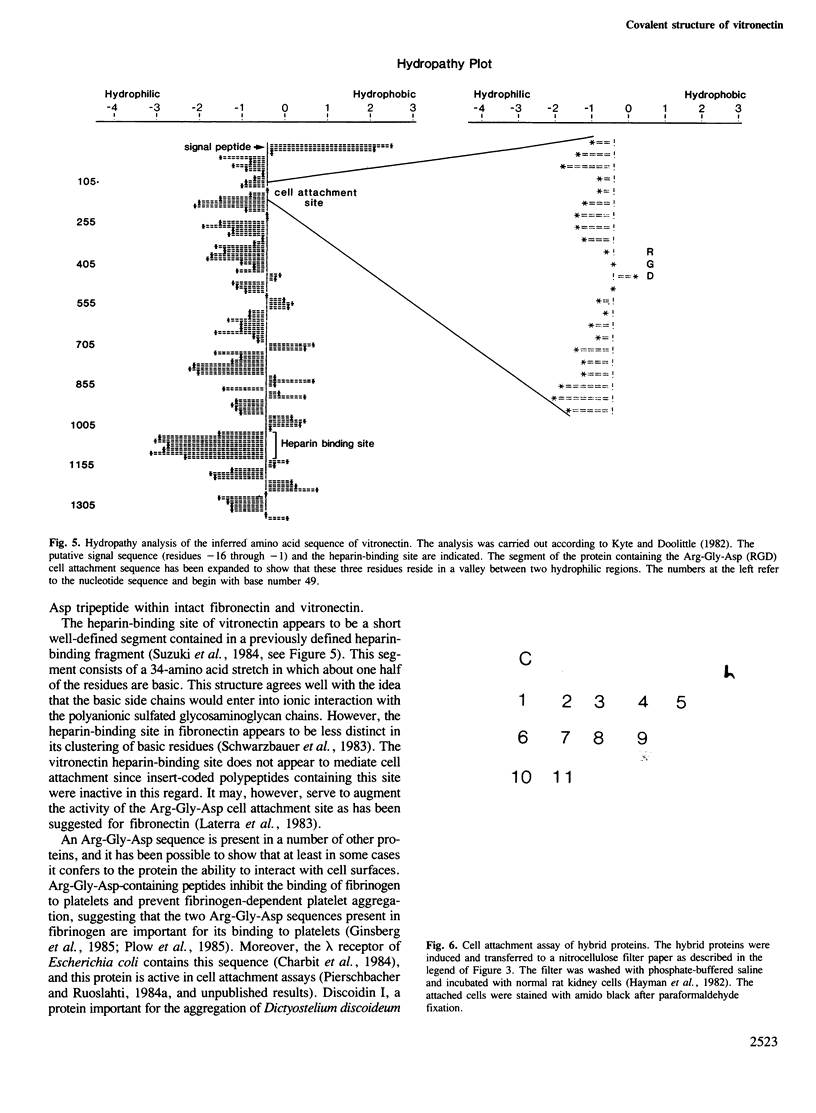
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laterra J., Norton E. K., Izzard C. S., Culp L. A. Contact formation by fibroblasts adhering to heparan sulfate-binding substrata (fibronectin or platelet factor 4). Exp Cell Res. 1983 Jun;146(1):15–27. doi: 10.1016/0014-4827(83)90320-8. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Miyake T., Hozumi T., Itakura K. Solid-phase synthesis of polynucleotides. II. Synthesis of polythymidylic acids by the block coupling phosphotriester method. Nucleic Acids Res. 1980 Nov 25;8(22):5473–5489. doi: 10.1093/nar/8.22.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldberg A., Linney E., Ruoslahti E. Molecular cloning and nucleotide sequence of a cDNA clone coding for the cell attachment domain in human fibronectin. J Biol Chem. 1983 Sep 10;258(17):10193–10196. [PubMed] [Google Scholar]
- Pierschbacher M. D., Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984 May 3;309(5963):30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- Pierschbacher M. D., Ruoslahti E. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5985–5988. doi: 10.1073/pnas.81.19.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pytela R., Pierschbacher M. D., Ruoslahti E. Identification and isolation of a 140 kd cell surface glycoprotein with properties expected of a fibronectin receptor. Cell. 1985 Jan;40(1):191–198. doi: 10.1016/0092-8674(85)90322-8. [DOI] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E., Engvall E., Hayman E. G. Fibronectin: current concepts of its structure and functions. Coll Relat Res. 1981;1(1):95–128. doi: 10.1016/s0174-173x(80)80011-2. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E., Hayman E. G., Pierschbacher M., Engvall E. Fibronectin: purification, immunochemical properties, and biological activities. Methods Enzymol. 1982;82(Pt A):803–831. doi: 10.1016/0076-6879(82)82103-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Tamkun J. W., Lemischka I. R., Hynes R. O. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell. 1983 Dec;35(2 Pt 1):421–431. doi: 10.1016/0092-8674(83)90175-7. [DOI] [PubMed] [Google Scholar]
- Springer W. R., Cooper D. N., Barondes S. H. Discoidin I is implicated in cell-substratum attachment and ordered cell migration of Dictyostelium discoideum and resembles fibronectin. Cell. 1984 Dec;39(3 Pt 2):557–564. doi: 10.1016/0092-8674(84)90462-8. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Pierschbacher M. D., Hayman E. G., Nguyen K., Ohgren Y., Ruoslahti E. Domain structure of vitronectin. Alignment of active sites. J Biol Chem. 1984 Dec 25;259(24):15307–15314. [PubMed] [Google Scholar]
- Tan Z. K., Ikuta S., Huang T., Dugaiczyk A., Itakura K. Solid-phase synthesis of polynucleotides. VIII: A simplified synthesis of oligodeoxyribonucleotides. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):383–391. doi: 10.1101/sqb.1983.047.01.045. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]