Abstract
In this study, we characterize the thermodynamics of hybridization, binding kinetics and conformations of four ribose-modified (2′-fluoro, 2′-O-propyl, 2′-O-methoxyethyl and 2′-O-aminopropyl) decameric mixed-sequence oligonucleotides. Hybridization to the complementary non-modified DNA or RNA decamer was probed by fluorescence and circular-dichroism spectroscopy and compared to the same duplex formed between two non-modified strands. The thermal melting points of DNA–DNA duplexes were increased by 1.8, 2.2, 0.3 and 1.3°C for each propyl, methoxyethyl, aminopropyl and fluoro modification, respectively. In the case of DNA–RNA duplexes, the melting points were increased by 3.1, 4.1 and 1.0°C for each propyl, methoxyethyl and aminopropyl modification, respectively. The high stability of the duplexes formed with propyl-, methoxyethyl- and fluoro-modified oligonucleotides correlated with high preorganization in these single-strands. Despite higher thermodynamic duplex stability, hybridization kinetics to complementary DNA or RNA was slower for propyl- and methoxyethyl-modified oligonucleotides than for the non-modified control. In contrast, the positively-charged aminopropyl-modified oligonucleotide showed rapid binding to the complementary DNA or RNA.
INTRODUCTION
Oligonucleotides and their mimics are becoming increasingly interesting for use as gene-targeted drugs and molecular biology tools (1,2). Several backbone-modified oligonucleotides have shown great potential as effective antisense therapeutics for various human diseases due to their promise of improved cell permeability, enhanced resistance to enzymatic degradation and tailored RNA binding properties (3,4). Upon oligonucleotide binding to target RNA, the hybrid duplexes that are formed are substrates of RNA nucleases, such as RNase H, resulting in RNA inactivation. Although a large number of modifications have been incorporated into oligonucleotides (5), only a limited few meet the stringent requirements for gene-targeting applications. To better design modifications, an understanding of not only the chemistry of new modifications but also their physical properties on a molecular level must be achieved. Physical characterization (thermodynamics and kinetics) of nucleic acid hybridization is important for predicting the behavior of designed oligonucleotides in both in vitro and in vivo applications.
Modifications have been made to the base, the ribose moiety and the phosphate backbone (4–6). To change the electrostatic properties of the negative phosphodiester backbone of natural DNA, backbone modifications such as phosphorothioate (one oxygen in the phosphate group is replaced with a sulfur) and phosphoroamidate modifications have been developed. Most is known about the analogs of the former class; several clinical trials are in progress with phosphorothioate-based drugs (4). Peptide nucleic acids (PNA) with polyamide backbones have also been well characterized (7). Several carbohydrate modifications at the 2′-position of the pentofuranose moiety have been studied as RNA mimics. 2′-O-methyl RNA, in which the 2′ position of the sugar moiety is replaced with a methoxy group, increases the chemical stability (stability of glycosidic linkage, resistance to alkali etc.) and biostability to a wide range of RNA- and DNA-specific nucleases (8,9). The alkyl group reduces the nucleophilic character of the 2′ oxygen and steric hindrance due to the bulky alkyl increases nuclease resistance of the adjacent phosphodiester bond relative to RNA. It is believed that 2′-modifications with electronegative substituents confer an RNA-like 3′-endo sugar conformation to oligonucleotides (10–13). Such preorganization will favor A-form duplex structures and should improve binding affinity to RNA.
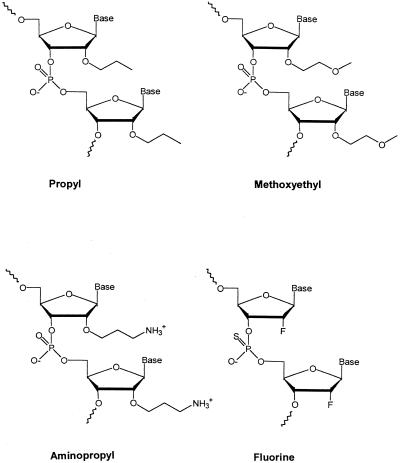
In addition to the 2′-O-methyl modification, the family of ribose-modified oligonucleotides includes structures with 2′-O-propyl (14), 2′-O-methoxyethyl (15), 2′-O-aminopropyl and 2′-fluoro modification (10,11,16). The crystal structure of fully modified 2′-O-methoxyethyl dodecamer shows that the sugars are locked in the 3′-endo conformation due to a Gauche effect between O4′-O2′. In addition, the side-chains themselves adopt organized structures due to Gauche effects in the -OCH2CH2O- linkages (13). In contrast, the 2′-O-aminopropyl-modification carries a positive charge in the amino group (pKa ∼9–10) at physiological pH (5). This modification incorporated into an oligonucleotide yields high nuclease resistance but only a modest stability increase (11,16). When the 2′ sugar position is directly linked to the electronegative substituent fluoro, a large increase in oligonucleotide binding affinity results (12). Oligonucleotides with 2′-O-alkyl, 2′-O-alkyl with glycerol–ether linkages, 2′-fluoro and 2′-O-aminoalkyl modifications have been studied for their pharmacokinetic, pharmacodynamic and pharmacological properties (10,11,17). These types of oligonucleotides all bind to complementary RNA with higher affinity than unmodified DNA and offer reasonably high nuclease resistance; thus, they have strong in vivo potency as gene-targeting agents.
In this study, we have characterized the thermodynamics of hybridization, binding kinetics and the conformations of 2′-fluoro, 2′-O-propyl, 2′-O-methoxyethyl and 2′-O-aminopropyl backbone-modified decameric mixed-sequence oligonucleotides (Scheme 1) upon hybridization to the complementary non-modified DNA or RNA decamer. In accordance with a preference for RNA, we find the DNA–RNA duplex stability to be larger than the DNA–DNA duplex stability for the modified strands. The increases in Tm values of the DNA–RNA duplexes with the modified oligonucleotides correlate linearly with the amount of pre-organized structure in the single strands. Despite the high thermodynamic affinity, the hybridization rates of propyl- and methoxyethyl-modified oligonucleotides to complementary DNA and RNA are reduced when compared to the kinetics for non-modified, fluoro- and aminopropyl-modified oligonucleotides.
Scheme 1.
MATERIALS AND METHODS
Materials
The four 2′-ribose modified (see Scheme 1) oligonucleotides (DNA 1 sequence; note that these are not really DNAs but RNA mimics) were prepared at Isis Pharmaceuticals Inc. (Carlsbad, CA). The synthesis procedures are described in (12,18–20). Complementary non-labeled and fluorescein-labeled DNA/RNA decamers (DNA 2 and RNA 2 sequence) were purchased from Intergrated DNA technologies (Iowa). The oligonucleotides contain the following base sequences: DNA 1, 5′-GTAGATCACT-3′; DNA 2, 5′-AGTGATCTAC-3′; RNA 2, 5′-AGUGAUCUAC-3′.
DNA–DNA and DNA–RNA (combining DNA 1 with DNA 2 or RNA 2) duplexes for thermal experiments were prepared by initial heating to 90°C followed by slow cooling to room temperature. A 5 mM phosphate buffer with 100 mM NaCl pH 7.0, was used unless otherwise stated; normally the DNA concentrations were 5 µM duplexes (or 10 µM strands). Absorption measurements were made on a Cary-50 instrument; concentrations of DNA strands/duplexes were determined at 80°C. In case of DNA–RNA duplexes, DEPC treated buffer was used.
Circular dichroism (CD) melting curves
Thermal melting was monitored by far-UV CD on an OLIS spectropolarimeter connected to a digitally controlled water bath (Julabo). CD spectra of 5 µM duplex were recorded every five degrees (1 cm cell; 3 scans of 220–300 nm, averaged at each temperature; 1 data point per nm) from 10 to 95°C, with 5 min of equilibration time at each temperature (heating rate of ∼1°C per min). For all duplexes, thermal experiments were performed at least twice. The thermal transitions were analyzed using the Van’t Hoff equation (21). This two-state model assumes that single strands are in equilibrium with only base-paired duplexes, i.e. there are no partially base-paired structures in the melting process (22). Furthermore, the change in heat capacity (ΔCp) between the two states is assumed to be zero and the thermodynamic parameters ΔH and ΔS are considered temperature-independent (22).
Analysis requires converting the experimental CD versus temperature curve into a melted fraction versus temperature curve. The fraction of strands in duplex form (α) versus temperature (T) plots were obtained by fitting the melting profile to a two-state transition model with linearly sloping lower and upper baselines. The Tm values were obtained directly from the temperature at α = 0.5. Values of K, the equilibrium constant, were determined at each temperature using the equation (for a bimolecular reaction):
K = 2α/[(1 – α)2CT/C0]
where CT is the total single strand concentration and C0 is the unit concentration 1 M. For a two-state transition, if ΔH is independent of the temperature, then a plot of ln K versus 1/T is linear with –ΔH/R as the slope and ΔS/R as the intercept, where R is the gas constant (8.31 J/mol.K). For all the duplexes studied here, the ln K versus 1/T plots are linear, suggesting that for all duplexes, also the ones with modified strands, the thermal reactions occur with no or minimal ΔCP.
Fluorescence hybridization kinetics
Equilibrium fluorescein emission (500–650 nm, excitation 496 nm, 1 cm cell) was measured on a Varian Eclipse fluorometer. Kinetic association experiments (20°C) were made on an Applied Photophysics SX.18MV stopped-flow reaction analyzer, emission mode (496 nm excitation, 510 nm cut-off filter), 1:1 mixing, path length 2 mm. No amplitude changes occurred in the dead time (<2 ms) of the instrument. For each experiment, a minimum of eight kinetic traces was averaged. The fluorescence versus time data was analyzed by non-linear least-squares fits to an integrated second-order rate equation. Additional experiments, under pseudo first-order conditions, gave essentially the same rate constants as that of the 1:1 mixings. To minimize contributions from the back (dissociation) reactions, only the first 50% of the kinetic traces were used in the fits.
RESULTS
Thermodynamic stability of 2′-sugar modified DNA–DNA and DNA–RNA duplexes
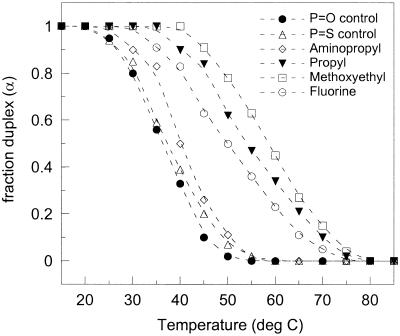
We have investigated the thermodynamic properties of six chemically different DNA–DNA and DNA–RNA duplexes, all with the following 10mer complementary base sequence: DNA 1, 5′-GTAGATCACT-3′; DNA 2, 3′-CATCTAGTGA-5′; or RNA 2, 3′-CAUCUAGUGA-5′. The DNA 1 strands were uniformly modified in the ribose moieties with either 2′-O-propyl, 2′-O-methoxyethyl, 2′-O-aminopropyl [these three with phosphodiester (P=O) backbones], or 2′-fluoro [with phosphorothioate (P=S) backbone] substituents and the control non-modified phosphodiester and phosphorothioate oligonucleotides. All oligonucleotides were hybridized to complementary non-modified DNA 2 or RNA 2 with a phosphodiester backbone. The far-UV CD spectra of all duplexes decreased, in a cooperative manner, upon heating due to thermally-induced strand–strand separation. The decrease in CD intensity at 280 nm (a peak that contains contributions mostly from nucleobase stacking interactions) was used to construct duplex melting curves that plot fraction of strands in duplex form versus temperature (Fig. 1).
Figure 1.
Thermal melting profiles (α versus temperature) for the studied 10mer backbone-modified DNA–DNA duplexes, as obtained from CD measurements (100 mM NaCl pH 7, 5 µM duplexes).
The curves were fitted to a two-state model of hybridization (using the Van’t Hoff equation) to yield the thermodynamic parameters Tm, ΔH° and ΔS° for each duplex and ΔG°(298 K) from ΔH° and TΔS°(298 K) (Table 1). This model assumes ΔCP = 0, which is most often true for normal DNA and RNA melting processes (21). The concentration dependence of Tm provides an alternative method for determining thermodynamics. In this method ΔH° and ΔS° are obtained from a 1/Tm against ln[CT/4] plot (22). To test the validity of the two-state assumption for one of the modified duplexes with hydrophobic substitutions, a 1/Tm versus ln[CT/4] plot was constructed for the methoxyethyl-modified DNA–DNA duplex (data not shown). CT could only be varied 10-fold (1–10 µM strands) due to instrument detection sensitivity as a lower limitation and Tm approaching 100°C as an upper constraint. Nevertheless, ΔH° estimated by this method was within 19% of that reported in Table 1 (–223 versus –187 kJ/mol), in accord with only a small, or no, ΔCP contribution. The helix–to–coil transitions of the duplexes were also monitored by the absorbance changes at 260 nm. All complexes showed broad monophasic melting transitions that agreed with the corresponding transitions monitored by CD. Absorbance hypochromicities of 10–20% were observed (Table 1).
Table 1. Thermodynamic parameters for formation of DNA–DNA and DNA–RNA duplexes from the single strands (DNA 1 + DNA 2/RNA 2) in 10 mM phosphate buffer pH 7.0, 100 mM NaCl (5 µM duplexes)a.
| Tm (°C)b | ΔH° (kJ/mol)b | TΔS° (kJ/mol)b | ΔG° (kJ/mol)b | ΔA260 (%)c | |||||
| DNA 1 sample |
DNA 2 |
RNA 2 |
DNA 2 |
RNA 2 |
DNA 2 |
RNA 2 |
DNA 2 |
RNA 2 |
DNA 2 |
| O-Propyl | 54.2 | 62.5 | –173.1 | –350.1 | –125.8 | –279.0 | –47.3 | –71.1 | 18.1 |
| O-Methoxyethyl | 57.9 | 72.4 | –187.0 | –312.9 | –136.4 | –238.0 | –50.6 | –74.9 | 19.4 |
| O-Aminopropyl | 39.7 | 41.1 | –223.5 | –208.4 | –180.9 | –165.8 | –42.6 | –42.6 | 12.6 |
| Control (P=O) | 36.3 | 31.6 | –260.0 | –207.9 | –218.6 | –171.4 | –41.4 | –36.5 | 15.3 |
| Fluoro (P=S) | 50.0 | n.d. | –152.5 | n.d. | –108.3 | n.d. | –44.2 | n.d. | 12.4 |
| Control (P=S) | 36.8 | 25.3 | –201.0 | –148.9 | –161.4 | –116.8 | –39.6 | –32.1 | 11.4 |
aDerived from Van’t Hoff plots for the formation of the duplexes using CD data.
bThe values given in parenthesis are the maximum uncertainties: Tm (± 1.5°C), ΔH° (± 10%), TΔS°(298 K) (± 10%) and ΔG°(298 K) (± 5%).
cThis column lists the observed absorbance hypochromicity [calculated as 100(A26080°C – A26020°C)/A26020°C] upon duplex formation.
n.d., not determined.
Thermal melting points were increased by 18, 22 and 13°C for propyl-, methoxyethyl- and fluoro-modified 10mer DNA–DNA duplexes, respectively, as compared to non-modified phosphodiester and phosphorothioate control DNA–DNA duplexes (100 mM NaCl pH 7). However, the DNA–DNA duplex with ten aminopropyl modifications in one strand did not show much enhanced thermal stability (ΔTm = +3°C). In the case of DNA–RNA duplexes, thermal melting points were increased further: by 31, 41 and 10°C (100 mM NaCl pH 7) for the propyl-, methoxyethyl- and aminopropyl-modified DNA–RNA duplex, respectively, as compared to the non-modified DNA–RNA control (Table 1). For the mixed purine–pyrimidine sequence studied here, the non-modified DNA–RNA duplex is less stable than the non-modified DNA–DNA duplex (Tm of 32 and 36°C for DNA–RNA and DNA–DNA, respectively). That the DNA–RNA hybrid is less stable than the corresponding DNA–DNA duplex has also been reported for other mixed purine–pyrimidine sequences (23).
The stability of the modified DNA–DNA duplexes was also evaluated at a higher NaCl concentration (500 mM). In all cases, Tm values were higher at 500 mM NaCl (Table 2). The increases in stability per modification (relative to the controls) were very similar at high and low salt concentrations (Table 2).
Table 2. Thermal melting points for DNA 1–DNA 2 5 µM duplexes measured in 10 mM phosphate buffer pH 7.0 and either 100 or 500 mM NaCla.
| DNA 1 sample | Tm (°C) | ΔTm | Tm (°C) | ΔTmb |
| |
100 mM
NaCl |
100 mM NaCl |
500 mM
NaCl |
500 mM NaCla |
| O-Propyl | 54.2 | +1.8 | 59.7 | +1.6 |
| O-Methoxyethyl | 57.9 | +2.2 | 67.1 | +2.3 |
| O-Aminopropyl | 39.7 | +0.3 | 43.4 | ∼0 |
| Control (P=O) | 36.3 | – | 43.7 | – |
| Fluoro (P=S) | 50.0 | +1.3 | 58.4 | +2.0 |
| Control (P=S) | 36.8 | – | 38.7 | – |
aMaximum error in reported Tm values: ± 1.5°C.
bΔTm refers to [Tm(modified duplex) – Tm(control duplex)]/10, i.e. the difference in Tm per modification.
Secondary structure of single-stranded oligonucleotides
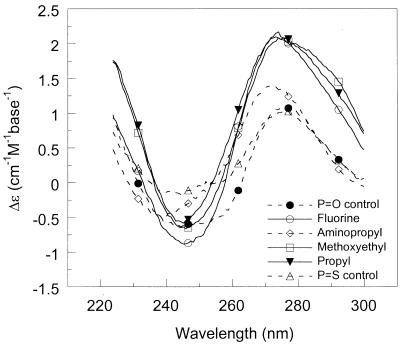
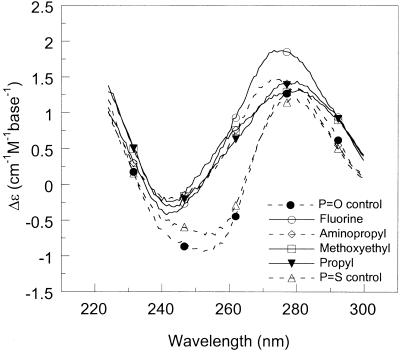
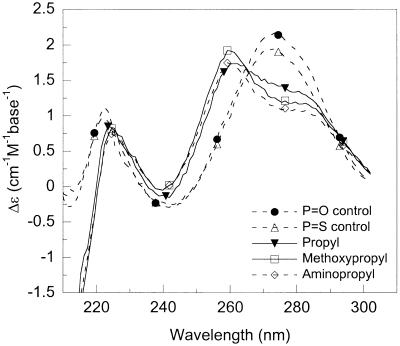
The shape and intensity of the CD bands in the far-UV region originates from mostly base–base stacking but also from interactions between the nucleobases with the DNA backbone ribose (24). It is possible, to a first approximation, to correlate the amplitude of CD signal in the 270–280 nm region with the amount of strand organization. The CD spectra of the DNA–DNA and DNA–RNA duplexes studied, as well as those of the single strands alone, are shown in Figure 2 (100 mM NaCl pH 7, 20°C). At the same concentration of nucleobases, fluoro-, propyl- and methoxyethyl-modified oligonucleotides have significantly larger CD intensities in the 270–280 nm region than aminopropyl- and non-modified strands. The increased CD is in accordance with preorganization of the sugar in a 3′-endo conformation, the favored structure with electronegative 2′-substitutions (10). In further support of a pre-organized structure, the single-stranded fluoro- and methoxyethyl-modified oligonucleotides alone exhibit cooperative thermal transitions (as monitored by CD and absorbance; Tm of 35–40°C). The estimated absorbance hypochromicity at 260 nm was 7 and 13% for fluoro- and methoxyethyl-modified single strands, respectively (data not shown).
Figure 2.
CD spectra of (A) single-stranded oligonucleotides (DNA 1), (B) duplexes formed with the complementary non-modified DNA strand and (C) duplexes formed with the complementary non-modified RNA strand (100 mM NaCl pH 7, 20°C, 10 µM strands or 5 µM duplexes).
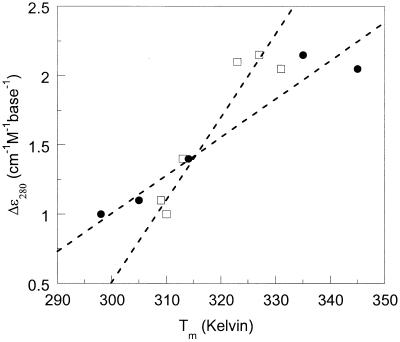
In Figure 3 we plotted the CD intensity at 280 nm (for single-stranded DNA 1 at 20°C) as a function of corresponding DNA–DNA and DNA–RNA duplex Tm values. There is a clear linear correlation between single-stranded CD intensity and duplex thermal stability (R2 is 0.89 and 0.93 for DNA–DNA and DNA–RNA Tm values, respectively). A larger CD intensity for the single strand correlates with an increased stability of the corresponding (DNA–DNA or DNA–RNA) duplex. There is no such relationship between duplex-CD intensity and duplex stability (data not shown).
Figure 3.
CD signal at 280 nm (20°C) for DNA 1 single strands plotted as a function of DNA–DNA (squares) and DNA–RNA (circles) duplex-melting temperatures (Tm values, Table 1). Linear fits (dashed lines) result in R2 = 0.89 (DNA–DNA) and R2 = 0.93 (DNA–RNA).
Duplex formation rates for sugar-modified DNA
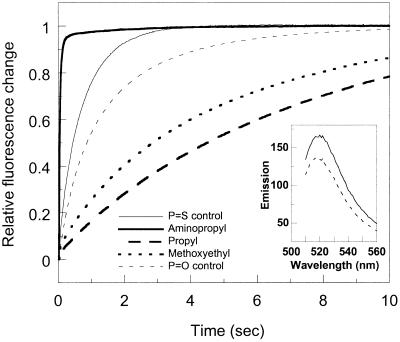
Hybridization kinetics for formation of duplexes containing modified oligomers (DNA 1) and fluorescein-labeled complementary strands (DNA 2 or RNA 2) were evaluated. Kinetics with unmodified control DNA 1 were also measured. Upon duplex formation, the fluorescein emission increases significantly, possibly due to a more hydrophobic environment in the duplex configuration as compared to in the single-stranded state (25). The increase in fluorescein emission was used to probe the DNA–DNA and DNA–RNA binding kinetics; hybridization was initiated by stopped-flow mixing. In all cases, the fluorescein emission increased by ∼30% upon hybridization. Examples of kinetic traces are shown in Figure 4. The decay data was analyzed in terms of second-order kinetics and the deduced rate constants are presented in Table 3 for two concentrations of NaCl: 100 and 500 mM (pH 7, 20°C).
Figure 4.
Formation kinetics (20°C) upon stopped-flow mixing of backbone-modified oligonucleotides (DNA 1) with complementary non-modified DNA (fluorescein–DNA2); for second-order rate constants see Table 2. Emission >510 nm was measured as a function of time; excitation at 496 nm. No fluorescence change occurred in the dead time of the instrument (2 ms). Inset, emission from single-stranded fluorescein–DNA2 (dashed line) and when in duplex with the aminopropyl-modified DNA 1 (solid line).
Table 3. Second-order rate constants for formation of DNA–DNA and DNA–RNA duplexes from the single strands (using fluorescein-labeled DNA 2/RNA 2).
| DNA 1 sample | DNA 2 | DNA 2 | RNA 2 |
| |
100 mM NaCl (M–1s–1) |
500 mM NaCl (M–1s–1) |
100 mM NaCl (M–1s–1) |
| O-Propyl | 9.08 (± 0.2) × 104 | 4.46 (± 0.1) × 105 | 1.20 (± 0.02) × 105 |
| O-Methoxyethyl | 1.45 (± 0.03) × 105 | 5.91 (± 0.12) × 105 | 3.77 (± 0.07) × 105 |
| O-Aminopropyl | 5.60 (± 0.1) × 107 | 3.19 (± 0.06) × 107 | 4.90 (± 0.1) × 107 |
| Control (P=O) | 5.28 (± 0.1) × 105 | 7.22 (± 0.15) × 106 | 9.96 (± 0.02) × 105 |
| Fluoro (P=S) | 1.18 (± 0.02) × 106 | 2.47 (± 0.05) × 106 | n.d. |
| Control (P=S) | 1.01 (± 0.02) × 106 | 7.36 (± 0.15) × 106 | 1.50 (± 0.03) × 106 |
Measured in 10 mM phosphate buffer pH 7.0, 20°C, 100 or 500 mM NaCl using a stopped-flow analyzer in emission mode.
n.d., not done.
We found the rate constants for hybridization to the complementary DNA to be slower by factors of six and four for propyl- and methoxyethyl-modified oligonucleotides, respectively, when compared to the non-modified strand. The same qualitative result was observed for hybridization to complementary RNA (Table 3): 8-fold and 3-fold slower rates for propyl- and methoxyethyl-modified DNA as compared to the control. In contrast, at 100 mM NaCl pH 7, 20°C, the aminopropyl-modified DNA hybridized 100-fold faster than the control to complementary DNA and 50-fold faster than the control to complementary RNA (Table 3). Except for the aminopropyl-modified duplex, all duplexes had 5–10-fold higher rate constants at the higher salt concentration (Table 3).
DISCUSSION
The 2′-position of the carbohydrate moiety has proven to be a valuable position for oligonucleotide modifications for antisense technology (4,5,10,11). Many 2′-modifications exhibit high binding affinity to target RNA, enhanced chemical stability and nuclease resistance and increased lipophilicity. Here we have performed biophysical characterizations of binding affinity, hybridization rate and conformations of uniformly ribose-modified 2′-fluoro, 2′-O-propyl, 2′-O-methoxyethyl and 2′-O-aminopropyl decameric oligonucleotides using CD and fluorescence spectroscopy. The binding kinetics were probed using stopped-flow mixing of the modified oligonucleotides with fluorescein-labeled complements. The combined knowledge of basic biophysical behavior (as studied in this report) and biological properties (for example, resistance to degradation, membrane penetration and non-specific interactions) may point towards 2′ sugar-modified biopolymers that are well suited as gene-targeting tools.
Thermodynamics of sugar-modified duplexes
In order to determine the thermodynamic parameters associated with duplex formation for sugar-modified oligomers with unmodified DNA or RNA complement, thermal melting was monitored by far-UV CD. The transition profiles were analyzed assuming a two-state model with temperature-independent ΔH° and ΔS° values (21,22). We found the fluoro-, propyl- and methoxyethyl-modified DNA–DNA duplexes to have increased thermodynamic stability [as shown by increased Tm and ΔG°(298 K)] when compared to the non-modified control DNA–DNA duplex (Table 1). In the case of DNA–RNA duplexes, the thermodynamic stability was enhanced still further for propyl-, methoxyethyl- and aminopropyl-modified DNA–RNA duplexes. ΔTm per propyl modification was +1.8°C for DNA and +3.1°C for RNA binding. For each methoxyethyl modification, ΔTm was +2.2°C for DNA and +4.1°C for RNA binding. The aminopropyl modification showed only a minor increase in stability towards DNA binding (+0.3°C/modification), but a ΔTm/modification of 1.0°C for RNA binding. It has been reported that the exact value of ΔTm per substitution depends on the base sequence as well as the total number of modifications in the strand; only trends are consistent across all sequences studied (11). Our data agrees qualitatively with previous duplex stability results for duplexes incorporating these modifications (11). All modified oligonucleotides showed a stronger preference for RNA over DNA as the complement, as shown by higher ΔTm per modification. Moreover, also in accordance with earlier reports (11), the increased stability was weakest for the aminopropyl modification; only in duplex with RNA was it more stable than the control duplex.
Complete, or partial, compensation between enthalpy (mainly hydrogen-bonding energies and van der Waals interactions) and entropy (mostly rearrangements of the molecules, solvent water and counter ions) has been observed for many different biological systems (26,27). Relatively small changes in overall binding free energy (ΔG°) are thought to be due to tighter binding (manifested in ΔH) in combination with a larger loss of degrees of freedom (seen in the ΔS term). A DNA–DNA association process per se is entropically unfavorable (reduction in the number of molecules), whereas formation of hydrogen bonds (base pairs) between the two strands and stacking of the bases are enthalpically favored. A 2′-ribose modification introduced in an oligonucleotide may contribute to either, or both, of the enthalpy and entropy parts of the change in free energy relative to the unmodified duplex. The substituent may form hydrogen bonds, electrostatic and/or hydrophobic interactions when binding to the non-modified complementary DNA or RNA. The 2′-ribose modifications investigated here (Scheme 1) are chemically different: the 2′-fluoro is a small, highly electronegative atom whereas the 2′-O-propyl and 2′-O-methoxyethyl modifications are long, rather hydrophobic alkyl/ethylene glycol chains. Both the propyl and aminopropyl modifications lack the gauche effect in the alkyl chain that is observed in the methoxyethyl side chain. The 2′-O-aminopropyl modification is, however, longer than the propyl side chain and may be extensively hydrated. Moreover, it has its carbon-chain ending with an amino group so that it has a positive charge at pH 7; this may substantially increase its electrostatic attraction for complementary DNA or RNA.
Table 1 shows that an increased thermodynamic stability [ΔG°(298K)] of a DNA–DNA duplex results from a larger decrease in unfavorable entropy than there is a decrease in favorable enthalpy. The higher stability of the fluoro-, propyl- and methoxyethyl-modified DNA–DNA duplexes is thus mostly an entropic effect. In agreement, the ribose modifications that stabilize the corresponding DNA–DNA or DNA–RNA duplexes the most (2′-fluoro, 2′-O-propyl and 2′-O-methoxyethyl) are the oligonucleotides with most organized structure (and therefore low initial entropy) in their single-stranded states according to CD (Figs 2A and 3).
Since the modified DNA strands bind more strongly to the RNA than the DNA complement (Table 1), the pre-organized structure appears to be more compatible with RNA-type duplexes. In accordance with this, the improved hybridization of 2′-fluoro and 2′-O-alkyl/ethylene glycol substituted oligonucleotides to complementary RNA has been attributed to the tendency of these substituents to shift the conformational equilibrium in the sugar towards a C3′-endo conformation, consistent with the A-form geometry of RNA duplexes (10,11). This is also evident from Figure 2C, which shows that the modified DNA 1–RNA 2 duplexes adopt structures characteristic of RNA (A-form) duplexes (positive CD peak centered at 260 nm), whereas the non-modified DNA–RNA duplex has a structure similar to B-form DNA (positive CD peak centered at 275 nm). In terms of thermodynamic parameters, the data in Table 1 suggest that the increased stability of the RNA–DNA duplexes with propyl and methoxyethyl modifications is the net result of large increases in favorable enthalpy of hybridization (tight binding) that is not compensated for by increases in unfavorable entropy (due to pre-organization).
Kinetics of duplex formation with modified oligomers
Association rates of phosphodiester and phosphorothioate oligonucleotides with unstructured targets (such as the single-stranded DNA 2 and RNA 2 used here) are typically of the order 106–107 M–1s–1 and are essentially independent of oligonucleotide length or base sequence (4). The association rate constant we measured between the non-modified phosphodiester and the fluorescein-labeled complementary DNA strands, 5.3 × 105 M–1s–1, is 14-fold faster than the rate constant determined for the same 10mer duplex by the Biacore technique (1.2 × 104 M–1s–1) (28). In another study, however, a 10mer DNA–DNA duplex formed with a rate constant of 1 × 107 M–1s–1 (29). These results, together with others, indicate that only comparisons of hybridization rates within the same experimental system are valid. The rate constants determined for the oligonucleotides studied here varied <5% from experiment to experiment. When comparing hybridization of DNA 1 to the DNA versus the RNA complement, the rates differed no more than 2-fold, RNA binding being slightly faster most often (Table 3).
Our kinetic results have several important implications. The data show that there is no simple direct correlation between hybridization speed (kinetics) and duplex stability (thermodynamics) among this set of DNA–DNA/RNA duplexes. At our experimental conditions, the speed of hybridization to the complementary DNA or RNA is slower for propyl- and methoxyethyl-modified oligonucleotides than for the non-modified, fluoro- and aminopropyl-modified strands (Table 3). Most reported studies of modified oligonucleotides focus on thermodynamics, however, it was reported that also 2′-O-methyl modifications in one strand reduced the hybridization speed for a 15mer mixed sequence DNA–DNA duplex (8). 2′-Modifications are most likely to reside in the minor groove of the oligonucleotide–DNA/RNA duplexes formed (10). The crystal structure of a fully methoxyethyl-substituted duplex revealed that the side chains were situated in the minor groove, stabilizing extensive hydration of the groove and backbone (13). The trapping of water molecules between oxygen acceptors of substituent and backbone, as found in the crystal structure, suggests an important role of water for the increased duplex stability.
Molecular dynamics simulations have suggested that methoxyethyl-modified oligonucleotides have the same organized structure in the single-stranded state as in the duplex, due to multiple Gauche and extended hydration effects (10,13). This is in agreement with our CD data which indicates significant preorganized structure in the methoxyethyl-modified single strand. Re-arrangements of the extended hydration shell around the substituents upon forming the minor groove of the new hybrid may therefore be responsible for the observed slow binding kinetics of 2′-O-methoxyethyl- and 2′-O-propyl-modified oligonucleotides to the complementary strand. Since the duplex stability is high but hybridization speed slow, it follows that the dissociation rate must be even slower in these cases. The fluoro-modified single strand is also prestructured in a 3′-endo sugar conformation (10,13), but lacks a long substituent; we found its binding kinetics to the complement to be similar to that of the control non-modified DNA. This result supports the idea that alkyl chain interactions (steric effects and/or hydration) slow down binding in the case of the propyl- and methoxyethyl-modified strands.
Increasing the NaCl concentration from 100 to 500 mM caused an increase in DNA–DNA association rates for propyl-, methoxyethyl- and the non-modified oligonucleotides (Table 3). This behavior may be explained by the decrease in electrostatic repulsion between the anionic DNA strands in high concentrations of NaCl (30). Higher thermodynamic stability is generally observed for DNA–DNA duplexes at higher NaCl concentration (30), as is also the case here (Table 2). In a quartz crystal microbalance investigation of hybridization between 10mer (non-modified) DNA strands, a 6-fold increase in association rate constant was found when going from 100 to 500 mM NaCl (31). This result is in excellent agreement with the 4–7-fold increases in rate constants recorded here (Table 3). In contrast, the aminopropyl-modified strand shows somewhat slower DNA–DNA binding kinetics at the higher salt concentration. This observation may be related to a difference in co-existing ions on the aminopropyl-modified strand as compared to the other modified DNA oligomers. Since the aminopropyl modification is positively charged at pH 7, this substituent may have more condensed anions at the increased NaCl concentration that has a negative effect on DNA–DNA duplex-formation speed (ions must be released before hybridization) (30).
The association reaction between the aminopropyl-modified and the complementary DNA/RNA strands is much faster (∼5 × 107 M–1s–1) than the binding rate constants for all the other DNA–DNA pairs, regardless of ionic strength (Table 3). Although the thermodynamic stability of the aminopropyl-modified DNA–RNA duplex is not greatly increased, the positive charges in the oligonucleotide may speed up association by electrostatic attraction yielding high local concentration of strands. We observed little preorganized structure for the aminopropyl-modified strand by CD (Fig. 2A); this indicates no entropic advantage to forming a duplex but instead allows for rapid association without structural constraints. Since thermodynamic stability is not much increased for the aminopropyl-modified duplex (as compared to the control duplex), it follows that the dissociation rate must also be extremely rapid. NMR studies of an aminopropyl-modified duplex have shown that the aminopropyl chain and the phosphate have uncorrelated motions and the amine does not bind via inner-sphere coordination to the flanking phosphate (16). This finding, that the modifications themselves are not strongly interacting with the duplex backbone, supports the observation that dissociation can be rapid.
CONCLUSIONS
To better understand and predict the behavior of 2′ sugar-modified oligonucleotides, biophysical characterization of their fundamental hybridization properties is required. We found that RNA duplexes with 2′-ribose modifications uniformly in one strand had significantly increased thermal stability as compared to both corresponding modified DNA duplexes and unmodified duplexes. The high stability of the duplexes formed with 2′-O-propyl- and 2′-O-methoxyethyl-modified oligonucleotides correlates with preorganized structure (3′-endo sugar conformation) in the single-stranded states. Despite the higher equilibrium stability, perhaps as a consequence of substituent hydration and structural rearrangements, the hybridization kinetics to complementary DNA or RNA was slower for 2′-O-propyl- and 2′-O-methoxyethyl-modified oligonucleotides than for 2′-fluoro and non-modified oligonucleotides. Although DNA-duplex stability was not increased by the presence of 2′-O-aminopropyl attachments and the RNA duplex showed only a slight increase in stability, the hybridization speed was significantly faster for the oligonucleotide with this charged modification as compared to all other modifications, presumably due to favorable electrostatic attraction.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the Cancer Association of Greater New Orleans (CAGNO) (Research Grant to P.W.S; Student Fellowship to A.S.), the Dreyfus Foundation (New Faculty Award to P.W.S.) and the Keck Foundation (instrumentation) for financial support.
References
- 1.Uhlmann E. and Peyman,A. (1993) Oligonucleotide analogs containing dephospho-internucleoside linkages. Methods Mol. Biol., 20, 355–389. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen P.E. (1991) Sequence-selective DNA recognition by synthetic ligands. Bioconjug. Chem., 2, 1–12. [DOI] [PubMed] [Google Scholar]
- 3.Zamecnik P.C. and Stephenson,M.L. (1978) Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl Acad. Sci. USA, 75, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crooke S.T. (2000) Progress in antisense technology: the end of the beginning. Methods Enzymol., 313, 3–45. [DOI] [PubMed] [Google Scholar]
- 5.Mesmaeker A., Haner,R., Martin,P. and Moser,H. (1995) Antisense oligonucleotides Acc. Chem. Res., 28, 366–374. [Google Scholar]
- 6.Lamond A.I. and Sproat,B.S. (1993) Antisense oligonucleotides made of 2′-O-alkyl RNA: their properties and applications in RNA biochemistry. FEBS Lett., 325, 123–127. [DOI] [PubMed] [Google Scholar]
- 7.Ratilainen T., Holmen,A., Tuite,E., Haaima,G., Christensen,L., Nielsen,P.E. and Norden,B. (1998) Hybridization of peptide nucleic acid. Biochemistry, 37, 12331–12342. [DOI] [PubMed] [Google Scholar]
- 8.Torigoe H., Shimizume,R., Sarai,A. and Shindo,H. (1999) Triplex formation of chemically modified homopyrimidine oligonucleotides: thermodynamic and kinetic studies. Biochemistry, 38, 14653–14659. [DOI] [PubMed] [Google Scholar]
- 9.Wang S. and Kool,E.T. (1995) Relative stabilities of triple helices composed of combinations of DNA, RNA and 2′-O-methyl-RNA backbones: chimeric circular oligonucleotides as probes. Nucleic Acids Res., 23, 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manoharan M. (1999) 2′-carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation. Biochim. Biophys. Acta, 1489, 117–130. [DOI] [PubMed] [Google Scholar]
- 11.Freier S.M. and Altmann,K.H. (1997) The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res., 25, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawasaki A.M., Casper,M.D., Freier,S.M., Lesnik,E.A., Zounes,M.C., Cummins,L.L., Gonzalez,C. and Cook,P.D. (1993) Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem., 36, 831–841. [DOI] [PubMed] [Google Scholar]
- 13.Teplova M., Minasov,G., Tereshko,V., Inamati,G.B., Cook,P.D., Manoharan,M. and Egli,M. (1999) Crystal structure and improved antisense properties of 2′-O-(2- methoxyethyl)-RNA. Nat. Struct. Biol., 6, 535–539. [DOI] [PubMed] [Google Scholar]
- 14.Guinosso C., Hoke,G., Frier,S., Martin,J., Ecker,D., Mirabelli,C., Crooke,S. and Cook,P. (1991) Synthesis and biophysical and biological evaluation of 2′-modified antisense oligonucleotides. Nucl. Nucl., 10, 259–262. [Google Scholar]
- 15.Martin P. (1996) Stereoselective synthesis of 2′-O-(2-methoxyethyl)ribonucleosides. Helv. Chim. Acta, 79, 1930–1938. [Google Scholar]
- 16.Griffey R.H., Monia,B.P., Cummins,L.L., Freier,S., Greig,M.J., Guinosso,C.J., Lesnik,E., Manalili,S.M., Mohan,V., Owens,S. et al. (1996) 2′-O-aminopropyl ribonucleotides: a zwitterionic modification that enhances the exonuclease resistance and biological activity of antisense oligonucleotides. J. Med. Chem., 39, 5100–5109. [DOI] [PubMed] [Google Scholar]
- 17.Crooke S., Graham,M., Zukerman,J., Brooks,D., Conklin,B., Cummins,L., Greig,M., Guinosso,C., Kornburst,D., Manoharan,M. et al. (1996) Pharmacokinetic properties of several novel oligonucleotide analogs in mice. J. Pharmacol. Exp. Ther., 277, 923–937. [PubMed] [Google Scholar]
- 18.Lesnik E.A., Guinosso,C.J., Kawasaki,A.M., Sasmor,H., Zounes,M., Cummins,L.L., Ecker,D.J., Cook,P.D. and Freier,S.M. (1993) Oligodeoxynucleotides containing 2′-O-modified adenosine: synthesis and effects on stability of DNA:RNA duplexes. Biochemistry, 32, 7832–7838. [DOI] [PubMed] [Google Scholar]
- 19.Griffey R.H., Monia,B.P., Cummins,L.L., Freier,S., Greig,M.J., Guinosso,C.J., Lesnik,E., Manalili,S.M., Mohan,V., Owens,S. et al. (1996) 2′-O-aminopropyl ribonucleotides: a zwitterionic modification that enhances the exonuclease resistance and biological activity of antisense oligonucleotides. J. Med. Chem., 39, 5100–5109. [DOI] [PubMed] [Google Scholar]
- 20.Monia B.P., Lesnik,E.A., Gonzalez,C., Lima,W.F., McGee,D., Guinosso,C.J., Kawasaki,A.M., Cook,P.D. and Freier,S.M. (1993) Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem., 268, 14514–14522. [PubMed] [Google Scholar]
- 21.Applequist J. and Damle,V. (1963) Theory of the effects of concentration and chain length on helix-coil equilibria in two-stranded nucleic acids. J. Chem. Phys., 39, 2719–2721. [DOI] [PubMed] [Google Scholar]
- 22.Puglisi J.D. and Tinoco,I.,Jr (1989) Absorbance melting curves of RNA. Methods Enzymol., 180, 304–325. [DOI] [PubMed] [Google Scholar]
- 23.Lesnik E.A. and Freier,S.M. (1995) Relative thermodynamic stability of DNA, RNA and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry, 34, 10807–10815. [DOI] [PubMed] [Google Scholar]
- 24.Rodger A. and Norden,B. (1997) Circular Dichroism and Linear Dichroism. Oxford University Press, Oxford, UK.
- 25.Morrison L.E. and Stols,L.M. (1993) Sensitive fluorescence-based thermodynamic and kinetic measurements of DNA hybridization in solution. Biochemistry, 32, 3095–3104. [DOI] [PubMed] [Google Scholar]
- 26.Ratilainen T., Holmen,A., Tuite,E., Nielsen,P.E. and Norden,B. (2000) Thermodynamics of sequence-specific binding of PNA to DNA. Biochemistry, 39, 7781–7791. [DOI] [PubMed] [Google Scholar]
- 27.Borea P.A., Gilli,G., Bertolasi,V. and Ferretti,V. (1987) Stereochemical features controlling binding and intrinsic activity properties of benzodiazepine-receptor ligands. Mol. Pharmacol., 31, 334–344. [PubMed] [Google Scholar]
- 28.Jensen K.K., Orum,H., Nielsen,P.E. and Norden,B. (1997) Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry, 36, 5072–5077. [DOI] [PubMed] [Google Scholar]
- 29.Lima W.F., Monia,B.P., Ecker,D.J. and Freier,S.M. (1992) Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry, 31, 12055–12061. [DOI] [PubMed] [Google Scholar]
- 30.Tomac S., Sarkar,M., Ratilainen,T., Wittung,P., Nielsen,P., Norden,B. and Graslund,A. (1996) Ionic effects on the stability conformation of peptide nucleic acid complexes. J. Am. Chem. Soc., 118, 5544–5552. [Google Scholar]
- 31.Okahata Y., Kawase,M., Niikura,K., Ohtake,F., Furusawa,H. and Ebara,Y. (1998) Kinetic measurements of DNA hybridization on an oligonucleotide-immobilized 27-MHz quartz crystal microbalance. Anal. Chem., 70, 1288–1296. [DOI] [PubMed] [Google Scholar]