Abstract
The nucleotide sequences of STE2 and STE3, cell type-specific sterile genes of Saccharomyces cerevisiae, were determined; major open reading frames encode 431 and 470 amino acids, respectively. STE2 and STE3 proteins seem to be folded in a similar fashion and are likely to be membrane-bound. Both consist of seven hydrophobic segments in each NH2-terminal region with a long hydrophilic domain in each COOH-terminal region. However, the two putative gene products do not exhibit extensive sequence homology. The STE2 protein has no obvious hydrophobic signal peptide; the NH2 terminus of the STE3 protein might serve as a signal peptide. The STE2 transcript, 1.7 kb, was detected in MATa strains but not in MATα strains, while the STE3 transcript, also 1.7 kb, was detected only in MATα cells. In STE2, two canonical TATA sequences are located 18 and 27 bp upstream of the mRNA start site, which has been mapped 32 bp before the initiator ATG codon, while STE3 contains a similar sequence (TATAGA), which is preceded by a long AT sequence, 140 bp upstream of the initiator ATG codon. Transcription of STE2 in a cells seems to be enhanced by exogenous α-factor.
Keywords: DNA sequence, molecular cloning, receptor, S. cerevisiae, mating pheromone
Full text
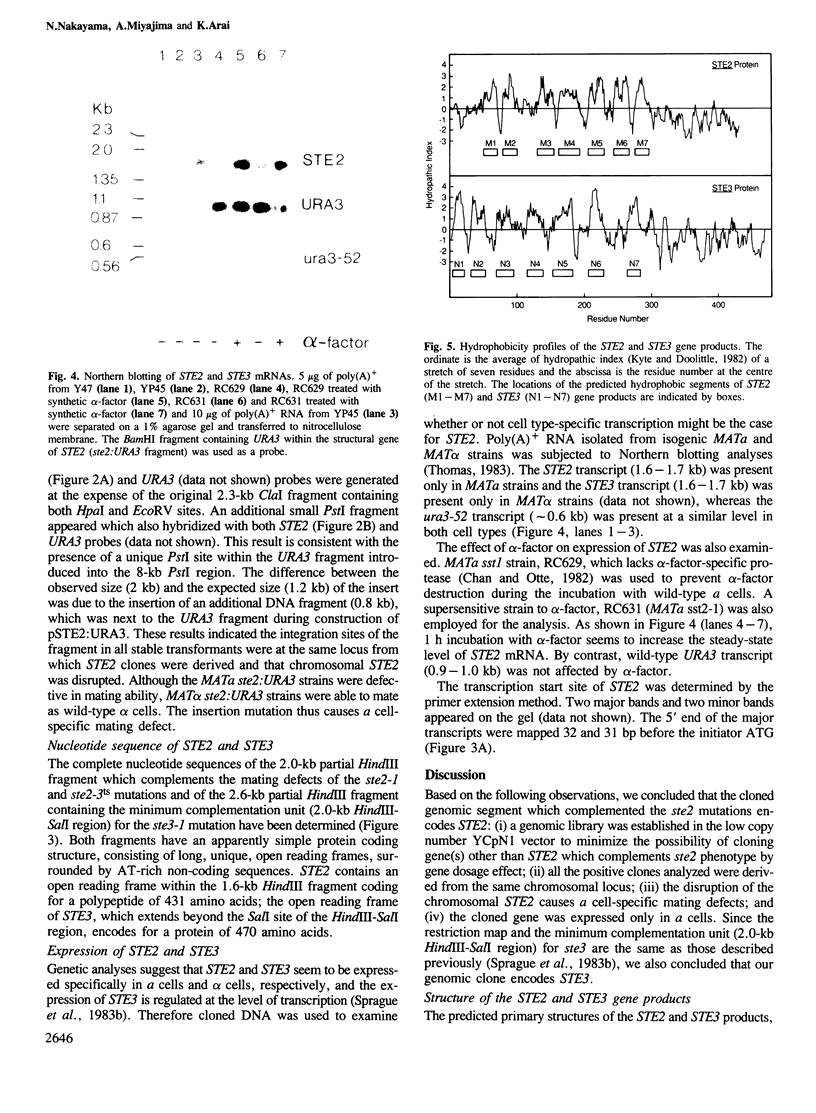
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Otte C. A. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol. 1982 Jan;2(1):21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Fairclough R. H., Finer-Moore J., Love R. A., Kristofferson D., Desmeules P. J., Stroud R. M. Subunit organization and structure of an acetylcholine receptor. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 1):9–20. doi: 10.1101/sqb.1983.048.01.004. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., Sprague G. F., Jr Induction of the yeast alpha-specific STE3 gene by the peptide pheromone a-factor. J Mol Biol. 1984 Oct 5;178(4):835–852. doi: 10.1016/0022-2836(84)90314-0. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980 Jun;85(3):811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975 Sep 4;257(5521):28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness D. D., Burkholder A. C., Hartwell L. H. Binding of alpha-factor pheromone to yeast a cells: chemical and genetic evidence for an alpha-factor receptor. Cell. 1983 Dec;35(2 Pt 1):521–529. doi: 10.1016/0092-8674(83)90186-1. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Herskowitz I. Structure of a yeast pheromone gene (MF alpha): a putative alpha-factor precursor contains four tandem copies of mature alpha-factor. Cell. 1982 Oct;30(3):933–943. doi: 10.1016/0092-8674(82)90298-7. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- MacKay V. L. Cloning of yeast STE genes in 2 microns vectors. Methods Enzymol. 1983;101:325–343. doi: 10.1016/0076-6879(83)01025-3. [DOI] [PubMed] [Google Scholar]
- Mackay V., Manney T. R. Mutations affecting sexual conjugation and related processes in Saccharomyces cerevisiae. I. Isolation and phenotypic characterization of nonmating mutants. Genetics. 1974 Feb;76(2):255–271. doi: 10.1093/genetics/76.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay V., Manney T. R. Mutations affecting sexual conjugation and related processes in Saccharomyces cerevisiae. II. Genetic analysis of nonmating mutants. Genetics. 1974 Feb;76(2):273–288. doi: 10.1093/genetics/76.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manney T. R. Expression of the BAR1 gene in Saccharomyces cerevisiae: induction by the alpha mating pheromone of an activity associated with a secreted protein. J Bacteriol. 1983 Jul;155(1):291–301. doi: 10.1128/jb.155.1.291-301.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manney T. R., Woods V. Mutants of Saccharomyces cerevisiae resistant to the alpha mating-type factor. Genetics. 1976 Apr;82(4):639–644. doi: 10.1093/genetics/82.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller A. M., MacKay V. L., Nasmyth K. A. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985 Apr 18;314(6012):598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- Miyajima A., Miyajima I., Arai K., Arai N. Expression of plasmid R388-encoded type II dihydrofolate reductase as a dominant selective marker in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Mar;4(3):407–414. doi: 10.1128/mcb.4.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima A., Nakayama N., Miyajima I., Arai N., Okayama H., Arai K. Analysis of full-length cDNA clones carrying GAL1 of Saccharomyces cerevisiae: a model system for cDNA expression. Nucleic Acids Res. 1984 Aug 24;12(16):6397–6414. doi: 10.1093/nar/12.16.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J., Hogness D. S. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983 Oct;34(3):807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Numa S., Noda M., Takahashi H., Tanabe T., Toyosato M., Furutani Y., Kikyotani S. Molecular structure of the nicotinic acetylcholine receptor. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 1):57–69. doi: 10.1101/sqb.1983.048.01.008. [DOI] [PubMed] [Google Scholar]
- Rose M., Casadaban M. J., Botstein D. Yeast genes fused to beta-galactosidase in Escherichia coli can be expressed normally in yeast. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2460–2464. doi: 10.1073/pnas.78.4.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Chen E. Y., Lugovoy J. M., Chang C. N., Hitzeman R. A., Seeburg P. H. Saccharomyces cerevisiae contains two discrete genes coding for the alpha-factor pheromone. Nucleic Acids Res. 1983 Jun 25;11(12):4049–4063. doi: 10.1093/nar/11.12.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. J. DNA sequence analysis by primed synthesis. Methods Enzymol. 1980;65(1):560–580. doi: 10.1016/s0076-6879(80)65060-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Blair L. C., Thorner J. Cell interactions and regulation of cell type in the yeast Saccharomyces cerevisiae. Annu Rev Microbiol. 1983;37:623–660. doi: 10.1146/annurev.mi.37.100183.003203. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Jensen R., Herskowitz I. Control of yeast cell type by the mating type locus: positive regulation of the alpha-specific STE3 gene by the MAT alpha 1 product. Cell. 1983 Feb;32(2):409–415. doi: 10.1016/0092-8674(83)90460-9. [DOI] [PubMed] [Google Scholar]
- Strathern J., Hicks J., Herskowitz I. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J Mol Biol. 1981 Apr 15;147(3):357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- Strazdis J. R., MacKay V. L. Induction of yeast mating pheromone a-factor by alpha cells. Nature. 1983 Oct 6;305(5934):543–545. doi: 10.1038/305543a0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Wilson K. L., Herskowitz I. Negative regulation of STE6 gene expression by the alpha 2 product of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2420–2427. doi: 10.1128/mcb.4.11.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]