Abstract
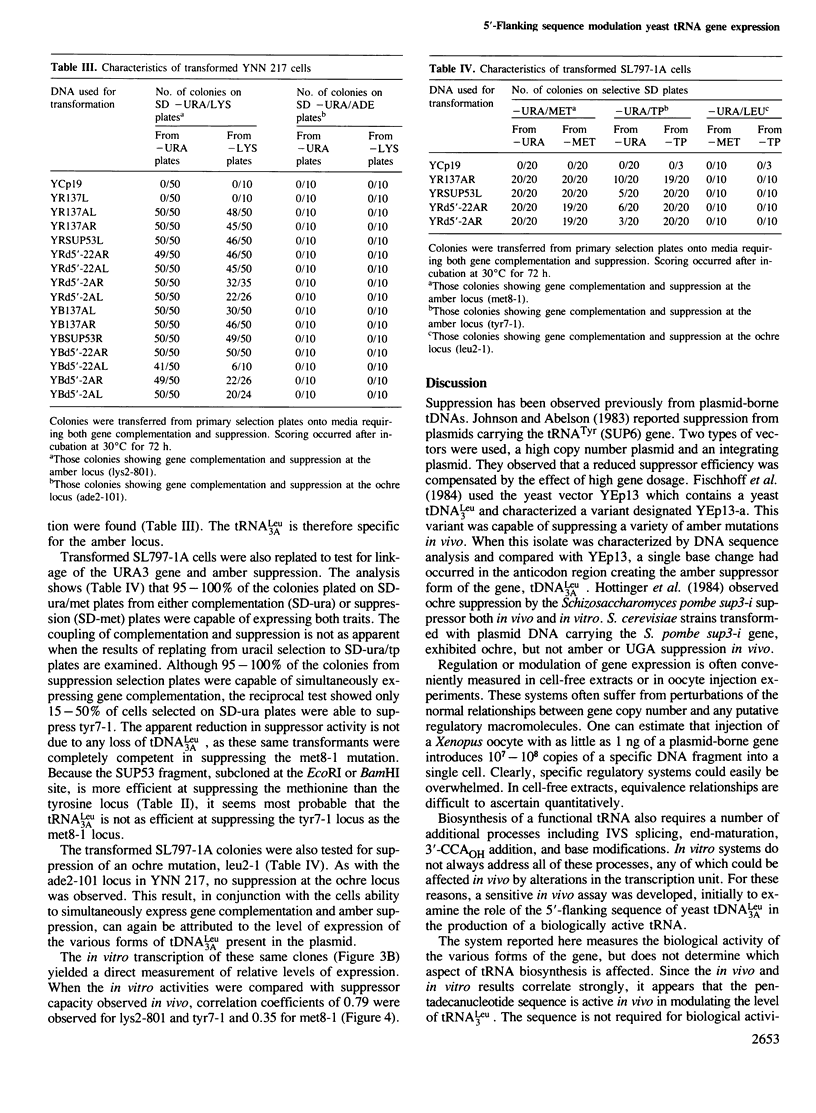
A pentadecanucleotide sequence, TTTCAACAAATAAGT, contiguous with the 5'-end of Saccharomyces cerevisiae tRNA-Leu3 coding sequence acts as a positive modulator of transcription in a homologous in vitro system. To determine whether modulation also takes place in vivo, the amber suppressor forms of tRNA-Leu3 genes with different 5'-flanking sequences were generated by site-specific mutagenesis and cloned into YCp19, a yeast vector maintained at 1-2 copies per cell. These plasmids were transformed into S. cerevisiae strains marked with amber mutations lys2-801, met8-1, and tyr7-1. The ability of the tRNA-Leu3 amber suppressor genes (tDNA-Leu3A) to suppress functionally lys2-801 and tyr7-1 mutations in the yeast host strain correlated well with template activities measured in vitro. We conclude that the plasmid-borne tRNA gene acts as an effective suppressor from the plasmid and the conserved pentadecanucleotide sequence modulated the expression of yeast tRNA-Leu3 in vivo as well as in vitro. This regulatory sequence if found associated with genes coding for a number of tRNAs which are abundant in yeast. We postulate that this sequence represents a mechanism by which production of specific tRNAs can be enhanced to match demand created by codon use preferences.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Fitzgerald-Hayes M., Carbon J. Structural analysis and sequence organization of yeast centromeres. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1175–1185. doi: 10.1101/sqb.1983.047.01.133. [DOI] [PubMed] [Google Scholar]
- Clancy S., Mann C., Davis R. W., Calos M. P. Deletion of plasmid sequences during Saccharomyces cerevisiae transformation. J Bacteriol. 1984 Sep;159(3):1065–1067. doi: 10.1128/jb.159.3.1065-1067.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Cooley L., Appel B., Söll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5' terminus of histidine tRNA. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Eigel A., Feldmann H. Ty1 and delta elements occur adjacent to several tRNA genes in yeast. EMBO J. 1982;1(10):1245–1250. doi: 10.1002/j.1460-2075.1982.tb00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischhoff D. A., Waterston R. H., Olson M. V. The yeast cloning vector YEp13 contains a tRNALeu3 gene that can mutate to an amber suppressor. Gene. 1984 Mar;27(3):239–251. doi: 10.1016/0378-1119(84)90069-6. [DOI] [PubMed] [Google Scholar]
- Goulian M., Kornberg A., Sinsheimer R. L. Enzymatic synthesis of DNA, XXIV. Synthesis of infectious phage phi-X174 DNA. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2321–2328. doi: 10.1073/pnas.58.6.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. D., Clarkson S. G., Tocchini-Valentini G. Transcription initiation of eucaryotic transfer RNA genes. Cell. 1982 May;29(1):3–5. doi: 10.1016/0092-8674(82)90083-6. [DOI] [PubMed] [Google Scholar]
- Hottinger H., Stadelmann B., Pearson D., Frendewey D., Kohli J., Söll D. The Schizosaccharomyces pombe sup3-i suppressor recognizes ochre, but not amber codons in vitro and in vivo. EMBO J. 1984 Feb;3(2):423–428. doi: 10.1002/j.1460-2075.1984.tb01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. D., Ogden R., Johnson P., Abelson J., Dembeck P., Itakura K. Transcription and processing of a yeast tRNA gene containing a modified intervening sequence. Proc Natl Acad Sci U S A. 1980 May;77(5):2564–2568. doi: 10.1073/pnas.77.5.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. D., Raymond G. J. Three regions of a yeast tRNALeu3 gene promote RNA polymerase III transcription. J Biol Chem. 1984 May 10;259(9):5990–5994. [PubMed] [Google Scholar]
- Johnson P. F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983 Apr 21;302(5910):681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Kjellin-Straby K., Engelke D. R., Abelson J. Homologous in vitro transcription of linear DNA fragments containing the tRNAArg-tRNAAsp gene pair from Saccharomyces cerevisiae. DNA. 1984;3(2):167–171. doi: 10.1089/dna.1984.3.167. [DOI] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985 Feb;40(2):393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R., Messing J. Apple II computer software for DNA and protein sequence data. DNA. 1983;2(1):31–35. doi: 10.1089/dna.1.1983.2.31. [DOI] [PubMed] [Google Scholar]
- Liebman S. W., Srodulski Z., Reed C. R., Stewart J. W., Sherman F., Brennan G. Yeast amber suppressors corresponding to tRNA3Leu genes. J Mol Biol. 1984 Sep 15;178(2):209–226. doi: 10.1016/0022-2836(84)90140-2. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Raymond G. J., Johnson J. D. The role of non-coding DNA sequences in transcription and processing of a yeast tRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5969–5988. doi: 10.1093/nar/11.17.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Burke D. J., Cooley L., Söll D. The extent of a eukaryotic tRNA gene. 5'- and 3'-flanking sequence dependence for transcription and stable complex formation. J Biol Chem. 1984 Feb 10;259(3):1461–1467. [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., Dingermann T., Söll D. The minimum intragenic sequences required for promotion of eukaryotic tRNA gene transcription. Nucleic Acids Res. 1982 Sep 25;10(18):5393–5406. doi: 10.1093/nar/10.18.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring D. N., Venegas A., Rutter W. J. Yeast tRNA3Leu gene transcribed and spliced in a HeLa cell extract. Proc Natl Acad Sci U S A. 1981 Oct;78(10):5963–5967. doi: 10.1073/pnas.78.10.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Venegas A., Quiroga M., Zaldivar J., Rutter W. J., Valenzuela P. Isolation of yeast tRNALeu genes. DNA sequence of a cloned tRNALeu3 gene. J Biol Chem. 1979 Dec 25;254(24):12306–12309. [PubMed] [Google Scholar]
- Wallace R. B., Johnson P. F., Tanaka S., Schöld M., Itakura K., Abelson J. Directed deletion of a yeast transfer RNA intervening sequence. Science. 1980 Sep 19;209(4463):1396–1400. doi: 10.1126/science.6997991. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rey F. J., Donahue T. F., Fink G. R. sigma, a repetitive element found adjacent to tRNA genes of yeast. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4138–4142. doi: 10.1073/pnas.79.13.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]




