Abstract
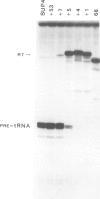
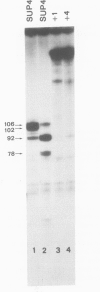
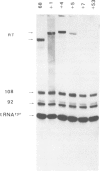
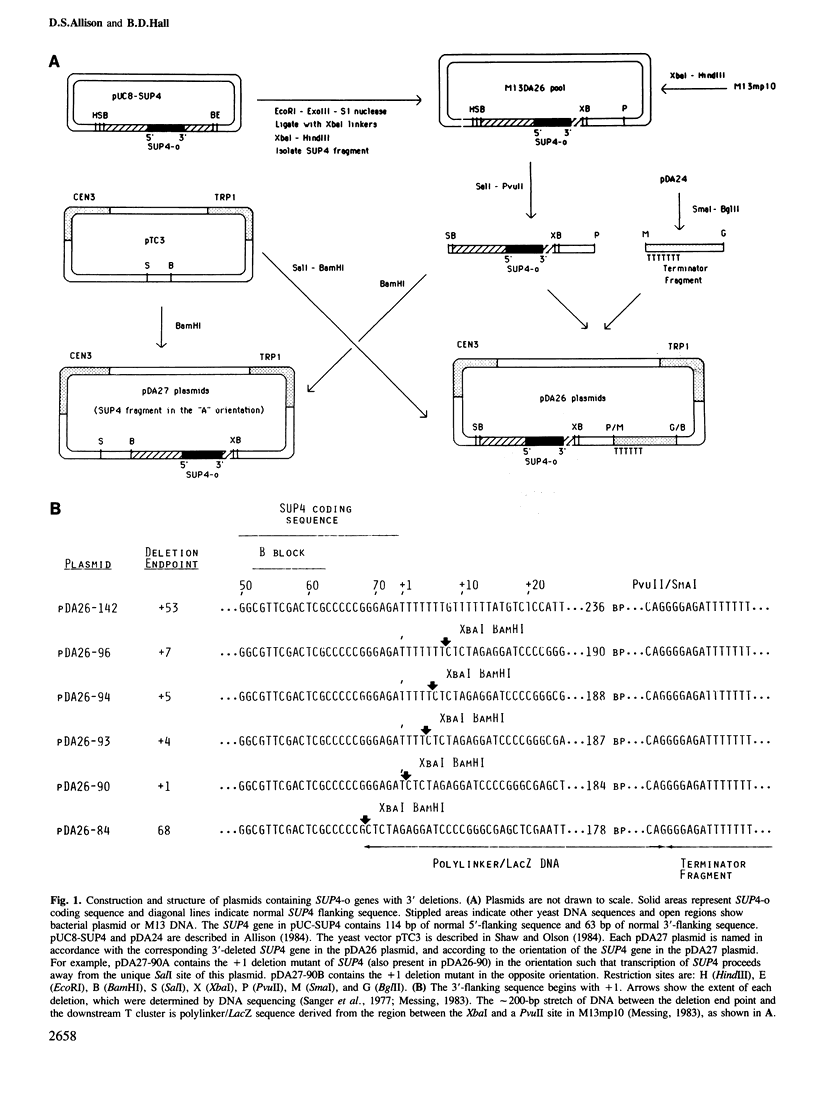
The SUP4-o gene of Saccharomyces cerevisiae codes for an altered tRNATyr capable of suppressing ochre mutations. We constructed mutant SUP4-o genes with deletions in the 3'-flanking sequence and tested each for its ability to suppress ochre mutations in transformed yeast cells. The effects of the different 3' deletions on various aspects of in vitro transcription and RNA processing were also determined, using a yeast cell-free extract. Deletions that leave five or fewer consecutive T residues in the 3'-flanking sequence of SUP4-o were found to result in decreased efficiency of transcription termination, both in vitro and in vivo. Unexpectedly, the suppression strength of each mutant SUP4-o gene is highly correlated with the relative extent of transcription termination at the 3' end of the gene. This result indicates that SUP4-o readthrough transcripts are not efficiently processed to functional suppressor tRNA in yeast cells. Deletions that extend into the T cluster in the 3'-flanking sequence also significantly decrease the ability of SUP4-o to compete for a transcription factor that is limiting in our extracts. This latter finding implies that the 3'-flanking sequence of SUP4 plays a role in transcription factor binding.
Full text
PDF

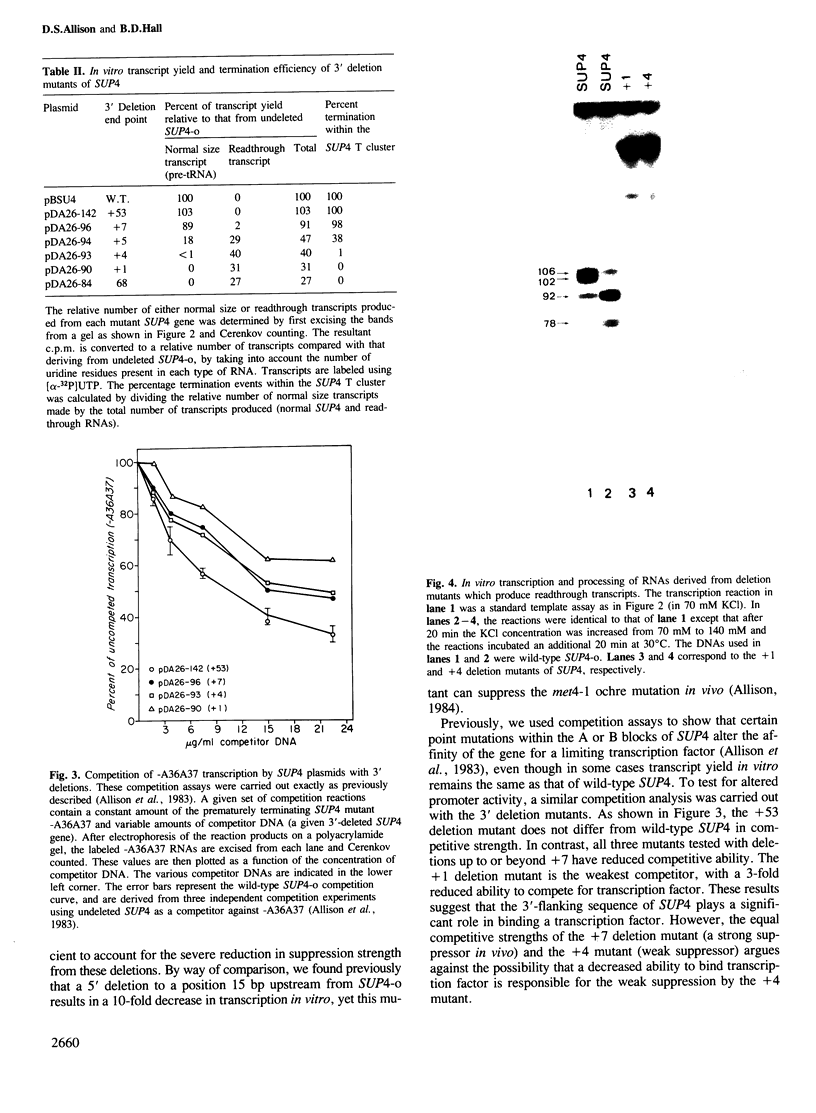




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adeniyi-Jones S., Romeo P. H., Zasloff M. Generation of long read-through transcripts in vivo and in vitro by deletion of 3' termination and processing sequences in the human tRNAimet gene. Nucleic Acids Res. 1984 Jan 25;12(2):1101–1115. doi: 10.1093/nar/12.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Hall B. D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984 Dec 1;3(12):2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon J. Yeast centromeres: structure and function. Cell. 1984 Jun;37(2):351–353. doi: 10.1016/0092-8674(84)90363-5. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Melton D. A., Cortese R. Promoter of a eukaryotic tRNAPro gene is composed of three noncontiguous regions. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1195–1199. doi: 10.1073/pnas.79.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Gage L. P. Transcription of a cloned Bombyx mori tRNA2Ala gene: nucleotide sequence of the tRNA precursor and its processing in vitro. Cell. 1979 Nov;18(3):817–828. doi: 10.1016/0092-8674(79)90134-x. [DOI] [PubMed] [Google Scholar]
- Gauss D. H., Sprinzl M. Compilation of tRNA sequences. Nucleic Acids Res. 1981 Jan 10;9(1):r1–23. [PMC free article] [PubMed] [Google Scholar]
- Goodman H. M., Olson M. V., Hall B. D. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbüchle O., Larson D., Hall G. I., Sprague K. U. The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell. 1979 Dec;18(4):1217–1229. doi: 10.1016/0092-8674(79)90234-4. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C., Leupold U. Suppressors in yeast. Curr Top Microbiol Immunol. 1974;64(0):1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Hofstetter H., Kressman A., Birnstiel M. L. A split promoter for a eucaryotic tRNA gene. Cell. 1981 May;24(2):573–585. doi: 10.1016/0092-8674(81)90348-2. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Schultz L. D., Shapiro R. A. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980 Mar;19(3):741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Specific transcription of homologous class III genes in yeast-soluble cell-free extracts. J Biol Chem. 1982 Jul 25;257(14):8432–8441. [PubMed] [Google Scholar]
- Klemenz R., Stillman D. J., Geiduschek E. P. Specific interactions of Saccharomyces cerevisiae proteins with a promoter region of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6191–6195. doi: 10.1073/pnas.79.20.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D. Mutations at the Saccharomyces cerevisiae SUP4 tRNA(Tyr) locus: isolation, genetic fine-structure mapping, and correlation with physical structure. Mol Cell Biol. 1982 Dec;2(12):1501–1513. doi: 10.1128/mcb.2.12.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., De Robertis E. M., Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980 Mar 13;284(5752):143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Ruet A., Camier S., Smagowicz W., Sentenac A., Fromageot P. Isolation of a class C transcription factor which forms a stable complex with tRNA genes. EMBO J. 1984 Feb;3(2):343–350. doi: 10.1002/j.1460-2075.1984.tb01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., Dingermann T., Schaack J., DeFranco D., Söll D. Transcription of eukaryotic tRNA genes in vitro. I. Analysis of control regions using a competition assay. J Biol Chem. 1983 Feb 25;258(4):2440–2446. [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Caspers P., Geiduschek E. P. Effects of temperature and single-stranded DNA on the interaction of an RNA polymerase III transcription factor with a tRNA gene. Cell. 1985 Feb;40(2):311–317. doi: 10.1016/0092-8674(85)90145-x. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Geiduschek E. P. Differential binding of a S. cerevisiae RNA polymerase III transcription factor to two promoter segments of a tRNA gene. EMBO J. 1984 Apr;3(4):847–853. doi: 10.1002/j.1460-2075.1984.tb01895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]