Abstract
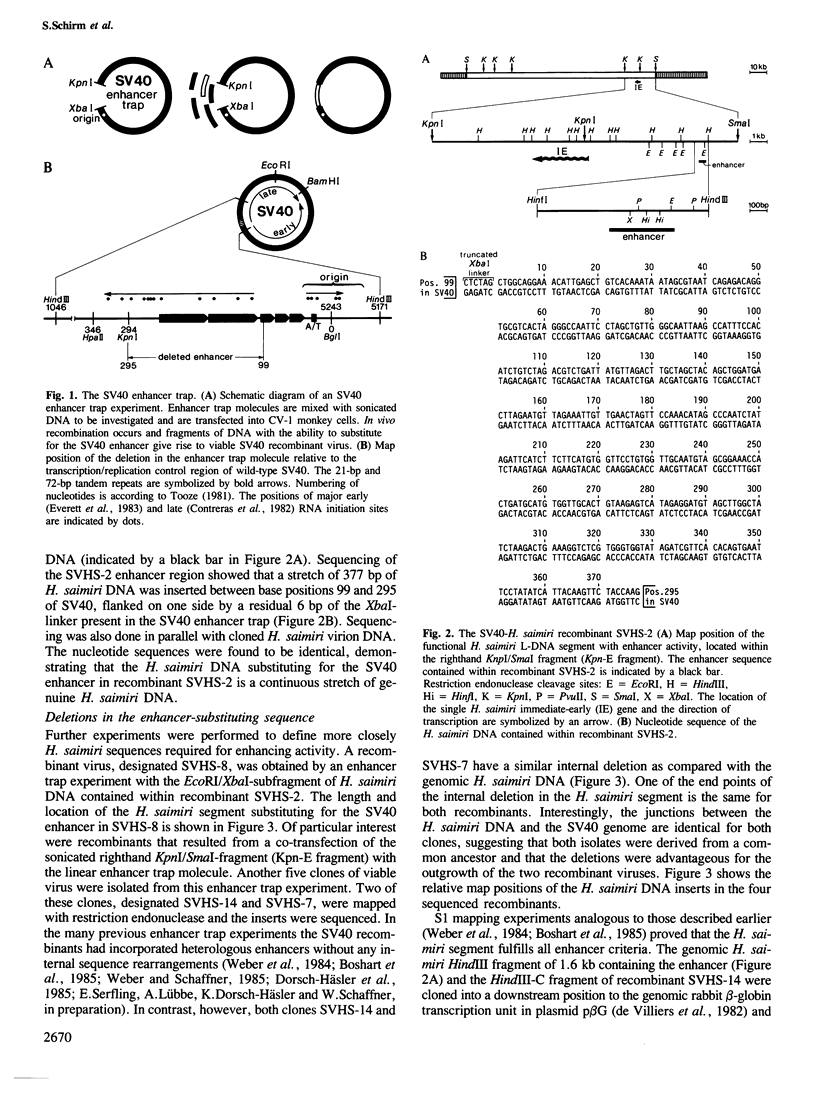
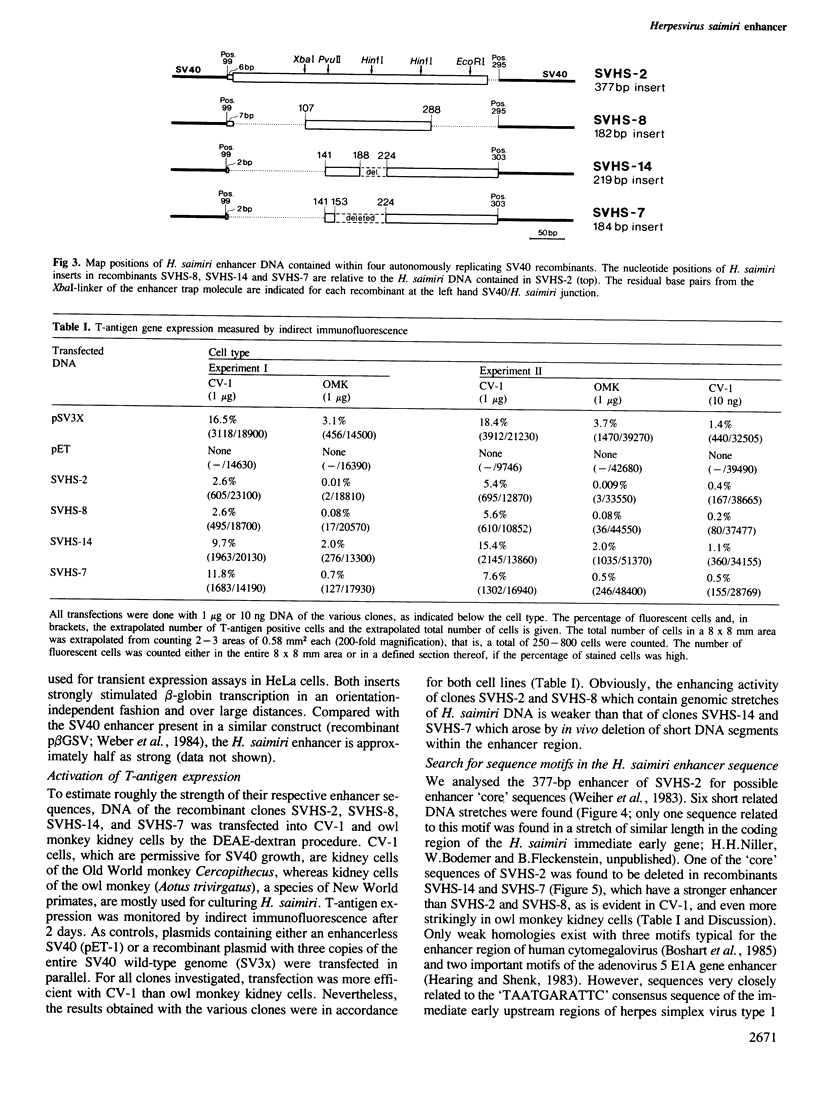
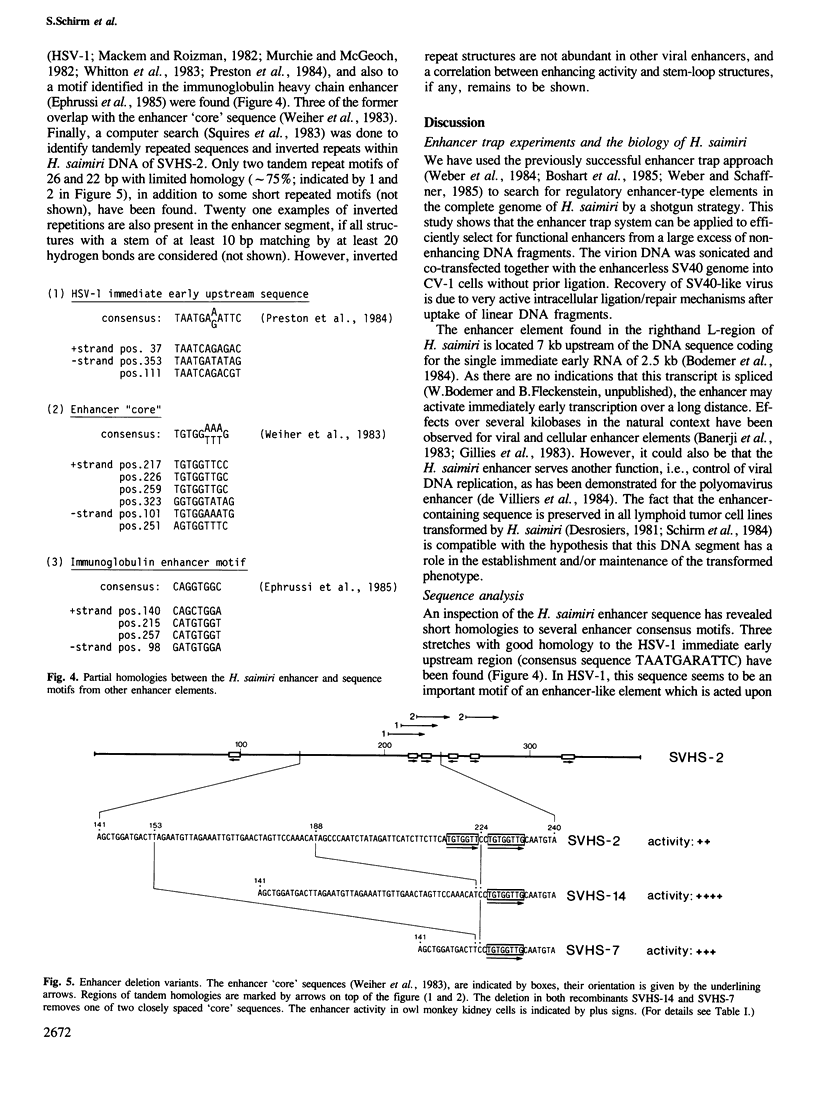
Herpesvirus saimiri, an oncogenic agent of New World primates, has a linear double-stranded DNA genome of approximately 155 kb. To test its genome for the presence of a transcription enhancer, we have mixed randomly fragmented H. saimiri DNA with non-infectious, linear SV40 DNA lacking the 72-bp repeat enhancer region (the so-called SV40 enhancer trap) and co-transfected this DNA mixture into monkey CV-1 cells. Viable SV40-like viruses were generated by intracellular ligation/repair processes with short H. saimiri DNA fragments. One recombinant, SVHS-2, had integrated a 377-bp enhancer segment from the righthand region of the H. saimiri genome, 7 kb upstream of DNA sequences encoding an immediate-early mRNA. This enhancer sequence is contained within the non-repetitive portions of the viral genome known to be preserved episomally in all lymphoid tumor cell lines. Further recombinant viruses (SVHS-14, SVHS-7, and SVHS-8) essentially contain subsets of the 377-bp insert. Unlike in the previous enhancer trap experiments, where heterologous enhancers were incorporated without any sequence alterations, SVHS-14 and SVHS-7 have suffered short internal deletions of a very similar segment of the H. saimiri insert. This renders the enhancer more active, implying that the deleted segment, while it may have a role in the herpesvirus infection cycle, exerts a negative effect within the isolated enhancer.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bodemer W., Knust E., Angermüller S., Fleckenstein B. Immediate-early transcription of Herpesvirus saimiri. J Virol. 1984 Aug;51(2):452–457. doi: 10.1128/jvi.51.2.452-457.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Fleckenstein B., Werner F. J., Mulder C. Structure of Herpesvirus saimiri genomes: arrangement of heavy and light sequences in the M genome. J Virol. 1976 Jul;19(1):154–161. doi: 10.1128/jvi.19.1.154-161.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M., Weber F., Jahn G., Dorsch-Häsler K., Fleckenstein B., Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985 Jun;41(2):521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Contreras R., Gheysen D., Knowland J., van de Voorde A., Fiers W. Evidence for the direct involvement of DNA replication origin in synthesis of late SV40 RNA. Nature. 1982 Dec 9;300(5892):500–505. doi: 10.1038/300500a0. [DOI] [PubMed] [Google Scholar]
- Da Silva M. P. Cuidados de enfermagem nos pacientes com "shunt" e fistula artério-venosa. Rev Enferm Nov Dimens. 1976 Nov;2(5):290–294. [PubMed] [Google Scholar]
- Desrosiers R. C. Herpesvirus saimiri DNA in tumor cells--deleted sequences and sequence rearrangements. J Virol. 1981 Aug;39(2):497–509. doi: 10.1128/jvi.39.2.497-509.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Baty D., Chambon P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983 Apr 25;11(8):2447–2464. doi: 10.1093/nar/11.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk L. A., Wolfe L. G., Deinhardt F. Isolation of Herpesvirus saimiri from blood of squirrel monkeys (Saimiri sciureus). J Natl Cancer Inst. 1972 May;48(5):1499–1505. [PubMed] [Google Scholar]
- Fleckenstein B., Wolf H. Purification and properties of Herpesvirus saimiri DNA. Virology. 1974 Mar;58(1):55–64. doi: 10.1016/0042-6822(74)90140-8. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschka-Dierich C., Werner F. J., Bauer I., Fleckenstein B. Structure of nonintegrated, circular Herpesvirus saimiri and Herpesvirus ateles genomes in tumor cell lines and in vitro-transformed cells. J Virol. 1982 Oct;44(1):295–310. doi: 10.1128/jvi.44.1.295-310.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E., Schirm S., Dietrich W., Bodemer W., Kolb E., Fleckenstein B. Cloning of Herpesvirus saimiri DNA fragments representing the entire L-region of the genome. Gene. 1983 Nov;25(2-3):281–289. doi: 10.1016/0378-1119(83)90232-9. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson B., Khoury G., Vande Woude G., Gruss P. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature. 1982 Feb 18;295(5850):568–572. doi: 10.1038/295568a0. [DOI] [PubMed] [Google Scholar]
- Luthman H., Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983 Mar 11;11(5):1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackem S., Roizman B. Structural features of the herpes simplex virus alpha gene 4, 0, and 27 promoter-regulatory sequences which confer alpha regulation on chimeric thymidine kinase genes. J Virol. 1982 Dec;44(3):939–949. doi: 10.1128/jvi.44.3.939-949.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie M. J., McGeoch D. J. DNA sequence analysis of an immediate-early gene region of the herpes simplex virus type 1 genome (map coordinates 0.950 to 0.978). J Gen Virol. 1982 Sep;62(Pt 1):1–15. doi: 10.1099/0022-1317-62-1-1. [DOI] [PubMed] [Google Scholar]
- Picard D., Schaffner W. Correct transcription of a cloned mouse immunoglobulin gene in vivo. Proc Natl Acad Sci U S A. 1983 Jan;80(2):417–421. doi: 10.1073/pnas.80.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. M., Cordingley M. G., Stow N. D. Analysis of DNA sequences which regulate the transcription of a herpes simplex virus immediate early gene. J Virol. 1984 Jun;50(3):708–716. doi: 10.1128/jvi.50.3.708-716.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi S., Schaffner W. Transformation of frog embryos with a rabbit beta-globin gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5051–5055. doi: 10.1073/pnas.78.8.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Müller I., Desrosiers R. C., Fleckenstein B. Herpesvirus saimiri DNA in a lymphoid cell line established by in vitro transformation. J Virol. 1984 Mar;49(3):938–946. doi: 10.1128/jvi.49.3.938-946.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C. H., De Felice M., Devereux J., Calvo J. M. Molecular structure of ilvIH and its evolutionary relationship to ilvG in Escherichia coli K12. Nucleic Acids Res. 1983 Aug 11;11(15):5299–5313. doi: 10.1093/nar/11.15.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro G. J., Sherr C. J., Sen A., King N., Daniel M. D., Fleckenstein B. Endogenous New World primate type C viruses isolated from owl monkey (Aotus trivirgatus) kidney cell line. Proc Natl Acad Sci U S A. 1978 Feb;75(2):1004–1008. doi: 10.1073/pnas.75.2.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Schaffner W. Enhancer activity correlates with the oncogenic potential of avian retroviruses. EMBO J. 1985 Apr;4(4):949–956. doi: 10.1002/j.1460-2075.1985.tb03723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., de Villiers J., Schaffner W. An SV40 "enhancer trap" incorporates exogenous enhancers or generates enhancers from its own sequences. Cell. 1984 Apr;36(4):983–992. doi: 10.1016/0092-8674(84)90048-5. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Werner F. J., Bornkamm G. W., Fleckenstein B. Episomal viral DNA in a Herpesvirus saimiri-transformed lymphoid cell line. J Virol. 1977 Jun;22(3):794–803. doi: 10.1128/jvi.22.3.794-803.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J. L., Rixon F. J., Easton A. J., Clements J. B. Immediate-early mRNA-2 of herpes simplex viruses types 1 and 2 is unspliced: conserved sequences around the 5' and 3' termini correspond to transcription regulatory signals. Nucleic Acids Res. 1983 Sep 24;11(18):6271–6287. doi: 10.1093/nar/11.18.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Olson L., Tyndall C., Schaffner W. Transcriptional 'enhancers' from SV40 and polyoma virus show a cell type preference. Nucleic Acids Res. 1982 Dec 20;10(24):7965–7976. doi: 10.1093/nar/10.24.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981 Dec 11;9(23):6251–6264. doi: 10.1093/nar/9.23.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W., Tyndall C., Lupton S., Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984 Nov 15;312(5991):242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]



