Abstract
Recently, 125 loci with genome-wide support for association with schizophrenia were identified. We investigated the impact of these variants and their accumulated genetic risk on brain activation in five neurocognitive domains of the Research Domain Criteria (working memory, reward processing, episodic memory, social cognition and emotion processing). In 578 healthy subjects we tested for association (i) of a polygenic risk profile score (RPS) including all single-nucleotide polymorphisms (SNPs) reaching genome-wide significance in the recent genome-wide association studies (GWAS) meta-analysis and (ii) of all independent genome-wide significant loci separately that showed sufficient distribution of all allelic groups in our sample (105 SNPs). The RPS was nominally associated with perigenual anterior cingulate and posterior cingulate/precuneus activation during episodic memory (PFWE(ROI)=0.047) and social cognition (PFWE(ROI)=0.025), respectively. Single SNP analyses revealed that rs9607782, located near EP300, was significantly associated with amygdala recruitment during emotion processing (PFWE(ROI)=1.63 × 10−4, surpassing Bonferroni correction for the number of SNPs). Importantly, this association was replicable in an independent sample (N=150; PFWE(ROI)<0.025). Other SNP effects previously associated with imaging phenotypes were nominally significant, but did not withstand correction for the number of SNPs tested. To assess whether there was true signal within our data, we repeated single SNP analyses with 105 randomly chosen non-schizophrenia-associated variants, observing fewer significant results and lower association probabilities. Applying stringent methodological procedures, we found preliminary evidence for the notion that genetic risk for schizophrenia conferred by rs9607782 may be mediated by amygdala function. We critically evaluate the potential caveats of the methodological approaches employed and offer suggestions for future studies.
Introduction
Schizophrenia, a severe and often chronic disease that affects ~1% of the population, has one of the highest heritability estimates in psychiatry (80%).1 Genome-wide association studies (GWAS) have been uncovering an increasing number of common variants underlying disease susceptibility, promising valuable insights into pathogenic biological pathways. The largest GWAS to date including 36 989 cases and 113 075 controls identified 125 genetic loci (of which 108 were independent) associated with schizophrenia.2
It was suggested that investigating important neurocognitive domains implemented within specific brain circuits could be a promising way for biological psychiatry. The Research Domain Criteria (RDoC) approach3, 4 postulates five major domains (negative valence, positive valence, cognition, social processes and arousal/regulation) containing several subdomains of which many are relevant to schizophrenia. Following the RDoC rationale, in order to uncover biological mechanisms underlying mental illness, these domains warrant investigation at different units of analyses, including genetics and brain circuits. In the last years, imaging genetics studies investigated the impact of genetic risk variants on domain-related brain circuits (for overviews see refs 5, 6, 7). In our own previous work we were able to identify potential intermediate phenotypes8, 9 in five RDoC subdomains (working memory (WM), episodic memory, reward processing (RP), social cognition and emotion processing) that were modulated by risk variants within schizophrenia-associated variants (for example, CACNA1C, ZNF804A).10, 11, 12, 13 Importantly, several of these findings were successfully replicated in independent samples.14, 15, 16
However, all of these studies as well as the overwhelming majority of all other published imaging genetics investigations (see Stein et al.17 for a notable exception, though using structural neuroimaging) have been performed with single genetic variants and usually with only one neurocognitive paradigm.
A crucial next step is to comprehensively investigate the impact of the 108 independent schizophrenia-associated loci uncovered recently.2 One approach complementing single single-nucleotide polymorphism (SNP) analyses is the use of polygenic risk profiles. As the accumulation of genetic variants is known to form a substantial proportion of genetic susceptibility to psychiatric disease,18 Purcell et al.19 proposed the use of a polygenic risk profile score (RPS), a sum across risk alleles of multiple SNPs weighted by their effect size in an independent study. Employing RPS is considered a feasible approach to investigate the combined genetic impact of multiple variants in small samples.20 However, investigating the linear combination of several hundreds or thousands of variants may come at the expense of losing specificity; that is, RPS may obscure information conveyed by single or subsets of genes. Therefore, we decided to employ two complementary exploratory analyses. We investigated a range of promising intermediate phenotypes evoking activation of dedicated brain circuits relevant for schizophrenia (WM, episodic memory, RP, social cognition and emotion processing). These five neurocognitive subdomains cover four of the five general RDoC domains, that is, negative valence, positive valence, cognition and social cognition. First, we tested for association with an RPS including all SNPs reaching genome-wide significance (P<5 × 10−8; combined risk of 125 SNPs) in the recent GWAS meta-analysis. Second, we tested the effects of all genome-wide significant single variants that showed sufficient distribution of all allelic groups within our sample in order to identify contributions of specific variants. Results were assessed within task-specific target areas located within widespread brain circuits (Supplementary Figure S2).
Materials and methods
Subjects
A total of 578 German volunteers who never suffered from psychiatric disorder (evidenced by SCID-I)21 were recruited at Mannheim, Berlin and Bonn as part of an ongoing study on neurogenetic mechanisms of unipolar depression, bipolar disorder and schizophrenia (http://www.ngfn.de/en/ schizophrenie.html; http://www.sys-med.de/en/consortia/integrament/). N=333 participants had no lifetime family history of schizophrenia or an affective disorder, and n=245 subjects had at least one first-degree relative affected by schizophrenia (n=72), bipolar disorder (n=71) or depression (n=102). Affected index patients did not suffer from any other psychotic or affective disorder, and the investigated relatives had no family history of multiple different psychiatric diagnoses (for example, cases of both, affective and psychotic disorders). All subjects had grandparents of European origin. Following application of exclusion criteria n=472–509 subjects were included in the analyses of the respective tasks (see Supplementary Material for details). N=150 controls recruited as part of a study on the neurogenetic mechanisms of alcohol dependence in Berlin and Bonn (http://www.ngfn.de/en/alkoholabh__ngigkeit.html)22 served as replication sample. All participants never suffered from psychiatric disorder according to the Structured Clinical Interview of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) Axis-I Disorders (SCID-I).21 Demographic characteristics of the respective subsamples are given in Supplementary Tables S1 and S2. The study was approved by local ethics committees of the universities of Heidelberg, Berlin and Bonn. All participants gave written informed consent to the study according to the Declaration of Helsinki.
DNA extraction and genotyping
Ethylenediaminetetraacetic acid anticoagulated venous blood samples were collected from all individuals. Lymphocyte DNA was isolated using the Chemagic Magnetic Separation Module I (Chemagen, Baesweiler, Germany) according to the manufacturer’s recommendations. A genome-wide data set was generated at the Department of Genomics, Life & Brain Center, University of Bonn using Illumina’s Human610Quad, Human660W-Quad and Infinium PsychArray-24 BeadChips (Illumina, San Diego, CA, USA). Quality control and imputation were performed with standard parameters used by the Psychiatric Genetics Consortium (PGC) Statistical Analyses Group and RPS were calculated using methods described by Purcell et al.19 (see Supplementary Material for details).
Functional imaging tasks
During functional magnetic resonance imaging (fMRI) subjects completed an associative episodic memory (EM) task requiring encoding, recall and recognition of face-profession pairs. The WM n-back task required continuous updating and retrieval of elements held in short-time memory. The RP monetary incentive delay task allowed the study of anticipation of monetary gains or losses. The Theory of Mind (ToM) task consisted of cartoon stories requiring subjects to take the protagonist’s perspective and judge changes in his/her affective states. The face-matching task (FMT) operationalized subjects’ implicit processing of negative emotions while viewing faces showing either fearful or angry expressions (for detailed descriptions see refs 10, 11, 12, 13 and Supplementary Material). All tasks were previously shown to robustly activate target structures and to possess excellent test–retest reliability in between-group designs.12, 16, 23, 24
Imaging parameters
Blood-oxygen-level dependent fMRI was performed on three Siemens Trio 3 T MR-Tomographs at the Life and Brain Center of the University of Bonn, the Central Institute of Mental Health Mannheim, and the Charité-Universitätsmedizin Berlin. At all sites, identical sequences and scanner protocols were employed (EM: 33 slices, axially tilted (−30°), slice thickness 2.4 mm+0.6 mm gap, field of view (FOV) 192 mm, repetition time (TR) 1.96 s, echo time (TE) 30 ms, flip angle 80°, all other fMRI tasks: 28 slices, slice thickness 4 mm+1 mm gap, FOV 192 mm, TR 2 s, TE 30 ms, flip angle 80°). Quality-control measurements were conducted at all sites on every day of data collection according to a multicenter quality-assurance protocol, revealing stable parameters over time.25 To account for any variance related to differences across sites, the site was used as a covariate for all statistical analyses.
Functional image processing
Image processing and statistical analyses were conducted using statistical parametric mapping methods as implemented in SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/). Scans were subjected to a strict quality assessment before inclusion into further analyses. Following data-quality assessment and application of general exclusion criteria (see above) n=472–509 subjects were included in the respective analyses (see Supplementary Figure 1 and Supplementary Methods for details). During pre-processing, images were realigned to a mean image (movement parameters confined to <3 mm translation and <1.7° rotation between volumes), slice-time-corrected, spatially normalized to a standard stereotactic space (a brain template created by the Montreal Neurological Institute) with a voxel size of 3 × 3 × 3 mm and smoothed with a 9 mm full width at half maximum Gaussian filter. A first-level fixed-effects model was computed for each participant and task. Regressors were created from the time course of the experimental conditions of interest per task and convolved with a canonical hemodynamic response function. Movement parameters, and for the ToM task also instructions and button presses, were included in the first-level models as covariates of no interest. For each subject, individual contrast images of the task effect (WM: 2-back>0-back; EM: memory>control; RP: anticipation of monetary win/loss>anticipation of neutral outcomes; ToM: mentalizing>control; FMT: faces>shapes) were subsequently entered into group statistics.
Statistical group analyses
RPS: To test for genetic association with the intermediate phenotypes, the respective individual contrast images were analyzed using second-level multiple regression models for each task, including the RPS as regressor of interest, and age, sex, site, subgroup (no familial liability for psychiatric disorders, affected first-degree relative of patients with schizophrenia, bipolar disorder or depression), chip used for genotyping and the first three principal components (derived from a statistically independent set of common SNPs representing potential population stratification) as nuisance covariates. Effects of RPS on predefined regions of interest (ROIs) were assessed determining the familywise error (FWE)-corrected significance values of the maximally activated voxel within each ROI using undirected tests.
For the WM, EM and FMT tasks, ROIs were defined a priori using anatomical labels provided by the Wake Forest University Pick Atlas (www.fmri.wfubmc.edu/downloads). These were the bilateral dorsolateral prefrontal cortex (BA46 and BA9 with subtraction of medial voxels of BA9, see Esslinger et al.11 for details) for the WM task, the left and right hippocampus and perigenual anterior cingulate cortex (pgACC) for EM tasks and the bilateral amygdala, as well as the pgACC for the FMT (Supplementary Figure S2). As the AAL atlas does not provide anatomical labels for the ventral striatum, a spherical ROI was created for the RP task using a voxel located in the center of the ventral striatum (x=±9, y=11, z=−8) surrounded by a sphere of 10 mm. For the ToM task, four functional ROIs were created for those regions previously shown to be aberrantly activated in patients with schizophrenia (for example, Sugranyes et al.26; Walter et al.27) and to be genetically modulated.13, 16 As these regions were not covered by specific AAL ROIs, masks for the medial prefrontal cortex, bilateral temporal parietal junction (TPJ) and posterior cingulate cortex/precuneus (PCC/Pcu) were created based on coordinates reported by a meta-analysis on brain areas involved in ToM28 using the toolbox TWURoi (see Supplementary Material for details).
Single SNP fMRI analyses
Comparable to RPS analyses, second-level multiple regression models were computed for each SNP and each task with number of minor alleles as regressor of interest, and age, sex, site, subgroup, genotyping chip and the first three principal components of potential population stratification as nuisance covariates. We excluded SNPs with less than 10 subjects in one allelic group, resulting in 105 independent analyses. Similar to RPS analyses, we extracted FWE-corrected P-values of the maximally activated voxel within each ROI for each SNP from an undirected test. These significance values were then Bonferroni-corrected for the number of autosomal SNPs tested, that is, 105 (P<4.76 × 10−4).
We used an undirected F-test in all analyses, as, for the investigated RPS and the vast majority of SNPs, we had no hypotheses about the directionality of effects. Associations with the intermediate phenotype could be represented by reduced or increased activation, being either indicative of dysfunction, inefficiency or compensatory resilience mechanisms.
Results
Polygenic RPSs
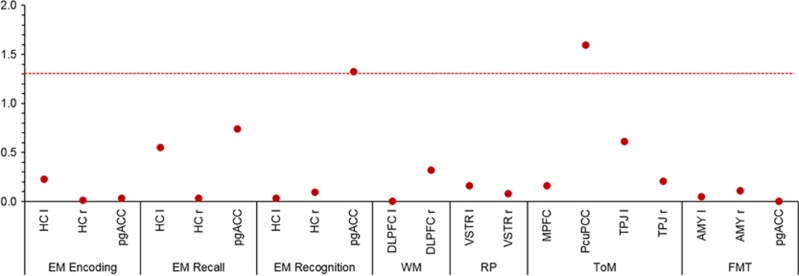
There were two significant associations between RPS and functional brain activation: accumulated genetic risk-predicted pgACC recruitment during EM recognition (x=−3, y=26, z=−11, F=13.72, Z=3.49, PFWE(ROI)=0.047) and PCC/Pcu activity during ToM (x=−9, y=−55, z=22, F=13.90, Z=3.52, PFWE(ROI)=0.025; Figure 1, Supplementary Figure S4).
Figure 1.
Results for the polygenic risk score (including all SNPs associated with schizophrenia at a genome-wide level, n=125) for regions of interest. Dashed line indicates –log10 P-value of familywise error correction for regions of interest in all tasks (PFWE(ROI)=0.05). x axis: region of interest, y axis: −log10 P-value for F-tests. AMY, amygdala; DLPFC, dorsolateral prefrontal cortex; EM, episodic memory; FMT, face-matching task; HC, hippocampus; l, left; pgACC, perigenual anterior cingulate cortex; MPFC, medial prefrontal cortex; Pcu/PCC, precuneus/posterior cingulate cortex; r, right; RP, reward processing; SNP, single-nucleotide polymorphism; ToM, Theory of Mind; TPJ, temporoparietal junction; VSTR ventral striatum; WM, working memory.
Genome-wide significant SNPs
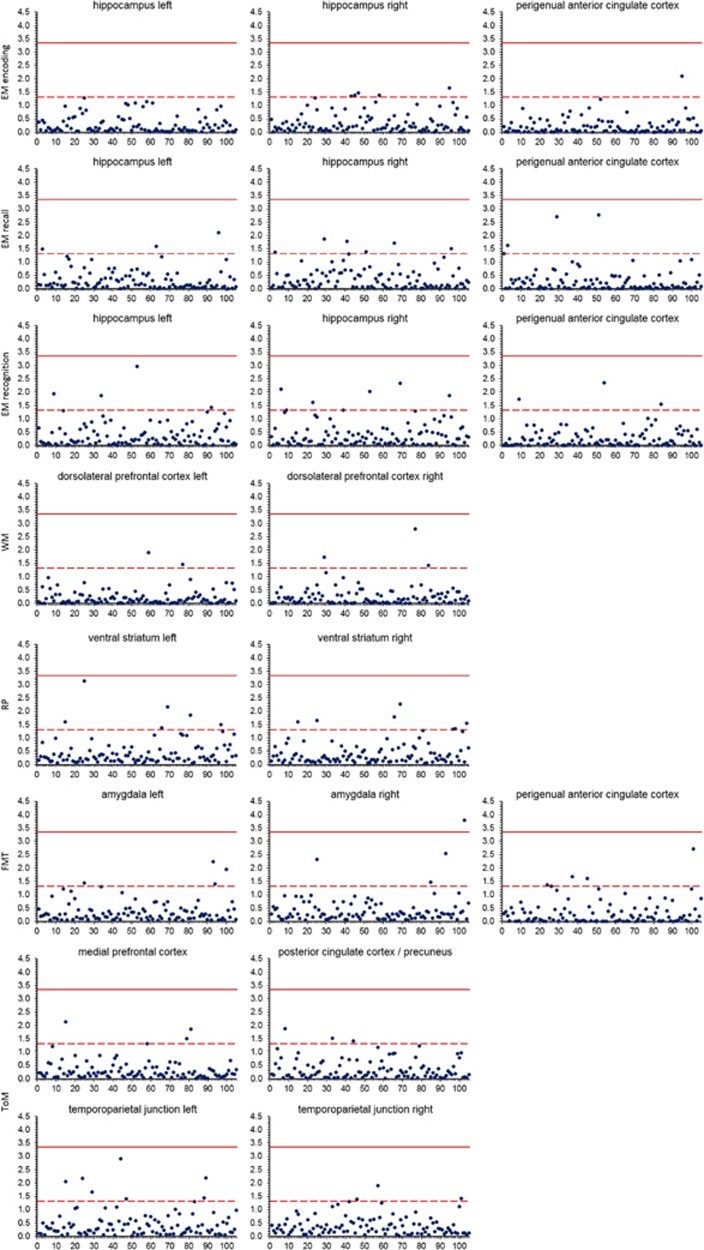
A median number of 4 (range 0–8) SNPs surpassed standard correction for multiple comparisons (PFWE(ROI)<0.05) across ROIs (Figure 2, Table 1, Supplementary Table S3). These included replicated risk variants for schizophrenia associated with imaging phenotypes before, for example, rs11696094 (ZNF804A; ToM, RP), rs2007044 (CACNA1C; EM, RP), rs1702294 (miR137; EM) and rs9636107 (TCF4; EM). However, only one single SNP association withstood correction for multiple comparisons (number of tests per ROI), that is, rs9607782 (chr22, 41.40–41.68 Mb, hg19.37; located in an linkage disequilibrium (LD) block with EP300, L3MBTL2, CHADL and RANGAP) predicted right amygdala activation during the FMT (x=27, y=5, z=−17, F=21.45, Z=4.43, PFWE(ROI)=1.63 × 10−4; Figure 3).
Figure 2.
Results of analyses of single SNPs reaching genome-wide significance for association with schizophrenia for regions of interest in all tasks. Dashed line indicates –log10 P-value of familywise error correction for respective region of interest. Red line indicates –log10 P-value of correction for multiple tests (P=4 × 10−4). x axis: number of SNP, ordered by position within chromosome, y axis: −log10 P-value of F-tests. EM, episodic memory; FMT, face-matching task; RP, reward processing; ToM, Theory of Mind; SNP, single-nucleotide polymorphism; WM, working memory.
Table 1. SNPs surpassing FWE ROI-corrected threshold of P<0.05 for fMRI tasks.
| ROI | Rank | SNP | Chr | Position | Gene | RA/NRA | Z | PFWE-ROI | Vx | MNI coords. (x, y, z) | PSx | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Episodic memory: encoding | |||||||||||||
| HC l | No significant findings | ||||||||||||
| HC r | 1 | rs9636107 | 18 | 53 195 247–53 200 117 | TCF4 | GA | 3.46 | 0.022 | 6 | 24 | −37 | 4 | 3.34e−12 |
| 2 | rs12704290 | 7 | 86 403 226–86 459 326 | GRM3 | GA | 3.31 | 0.035 | 1 | 15 | −37 | 10 | 3.33e−10 | |
| 3 | Chr6_84280274_D | 6 | 84 279 922–84 407 274 | SNAP91 | I2D | 3.24 | 0.042 | 2 | 36 | −16 | −17 | 8.15e−10 | |
| 4 | rs11191419 | 10 | 104 585 135–104 956 335 | ARL3, AS3MT, C10orf32, CNNM2, CYP17A1, INA, NT5C2, PCGF6, PDCD11, SFXN2, TAF5, TRIM8, USMG5, WBP1L | TA | 3.23 | 0.042 | 1 | 33 | −22 | −17 | 6.20e−19 | |
| 5 | rs11740474 | 5 | 153 671 057–153 688 217 | GALNT10 | TA | 3.21 | 0.046 | 1 | 18 | −13 | −17 | 3.15e−8 | |
| pgACC | 1 | rs9636107 | 18 | 53 195 247–53 200 117 | TCF4 | GA | 4.00 | 0.008 | 10 | 9 | 35 | 25 | 3.34e−12 |
| Episodic memory: recall | |||||||||||||
| HC l | 1 | rs715170 | 18 | 53 769 014–53 804 154 | LOC100505474 | CT | 3.73 | 0.008 | 8 | −33 | −10 | −17 | 1.27e−8 |
| 2 | rs12421382 | 11 | 109 285 471–109 610 071 | C11orf87 | CT | 3.38 | 0.026 | 3 | −30 | −37 | −5 | 3.70e−08 | |
| 3 | rs1498232 | 1 | 30 412 551–30 437 271 | — | TC | 3.29 | 0.032 | 3 | −18 | −13 | −20 | 2.86e−9 | |
| HC r | 1 | rs9841616 | 3 | 181 023 585–181 205 585 | FXR1, DNAJC19, SOX2−OT | TA | 3.56 | 0.014 | 17 | 36 | −13 | −23 | 2.35e−8 |
| 2 | rs2973155 | 5 | 152 505 619–152 711 619 | GRIA1* | CT | 3.51 | 0.017 | 8 | 33 | −7 | −20 | 1.11e−10 | |
| 3 | rs55661361 | 11 | 124 610 007–124 620 147 | ESAM, MSANTD2, NRGN, VSIG2 | GA | 3.50 | 0.019 | 6 | 39 | −22 | −17 | 2.80e−12 | |
| 4 | rs715170 | 18 | 53 769 014–53 804 154 | LOC100505474 | CT | 3.31 | 0.032 | 2 | 39 | −13 | −20 | 1.27e−8 | |
| 5 | rs3735025 | 7 | 137 039 644–137 085 244 | DGKI, PTN | TC | 3.22 | 0.042 | 2 | 36 | −10 | −26 | 3.28e−9 | |
| 6 | rs1498232 | 1 | 30 412 551–30 437 271 | — | TC | 3.20 | 0.043 | 1 | 18 | −28 | −8 | 2.86e−9 | |
| pgACC | 1 | rs3735025 | 7 | 137 039 644–137 085 244 | DGKI, PTN | TC | 4.36 | 0.002 | 4 | 6 | 2 | 31 | 3.28e−9 |
| 2 | rs9841616 | 3 | 181 023 585–181 205 585 | FXR1, DNAJC19, SOX2−OT | TA | 4.32 | 0.002 | 26 | −9 | 29 | −5 | 2.35e−8 | |
| 3 | rs1498232 | 1 | 30 412 551–30 437 271 | — | TC | 3.65 | 0.024 | 2 | 9 | 44 | −2 | 2.86e−9 | |
| 4 | rs4648845 | 1 | 2 372 401–2 402 501 | PLCH2 | TC | 3.43 | 0.048 | 1 | 3 | 32 | −2 | 8.70e−10 | |
| Episodic memory: recognition | |||||||||||||
| HC l | 1 | rs6984242 | 8 | 60 475 469–60 954 469 | CA8* | GA | 4.27 | 0.001 | 3 | −33 | −4 | −26 | 5.97e−9 |
| 2 | rs10803138 | 1 | 243 503 719–243 612 019 | AKT3, SDCCAG8 | GA | 3.64 | 0.012 | 2 | −24 | −10 | −26 | 2.03e−8 | |
| 3 | rs4391122 | 5 | 60 499 143–60 843 543 | ZSWIM6 | GA | 3.60 | 0.014 | 3 | −30 | −22 | −14 | 1.1e−14 | |
| 4 | rs4523957 | 17 | 2 095 899–2 220 799 | SGSM2, SMG6, SRR, TSR1 | TG | 3.30 | 0.037 | 1 | −24 | −4 | −26 | 2.86e−10 | |
| 5 | rs2909457 | 2 | 162 798 555–162 910 255 | DPP4, SLC4A10 | GA | 3.22 | 0.049 | 1 | −27 | −19 | −17 | 4.62e−8 | |
| HC r | 1 | rs2007044 | 12 | 2 321 860–2 413 760 | CACNA1C | GA | 3.93 | 0.005 | 16 | 36 | −37 | −8 | 3.22e−18 |
| 2 | rs1702294 | 1 | 98 374 984–98 559 084 | MIR137HG, MIR2682, MIR137, DPYD | CT | 3.78 | 0.008 | 6 | 36 | −34 | −8 | 3.36e−19 | |
| 3 | rs6984242 | 8 | 60 475 469–60 954 469 | CA8* | GA | 3.71 | 0.009 | 6 | 30 | −4 | −26 | 5.97e−9 | |
| 4 | rs9636107 | 18 | 53 195 247–53 200 117 | TCF4 | GA | 3.61 | 0.014 | 6 | 30 | −25 | −8 | 3.34e−12 | |
| 5 | rs4330281 | 3 | 17 221 366–17 888 266 | TBC1D5 | CT | 3.43 | 0.025 | 1 | 24 | −34 | 7 | 4.64e−9 | |
| 6 | chr5_140143664_I | 5 | 140 023 664–140 222 664 | AC005609.1, CD14, DND1, HARS, HARS2, IK, NDUFA2, PCDHA1, PCDHA10, PCDHA2, PCDHA3, PCDHA4, PCDHA5, PCDHA6, PCDHA7, PCDHA8, PCDHA9, TMCO6, WDR55, ZMAT2 | I12D | 3.24 | 0.047 | 2 | 30 | −13 | −23 | 4.85e−8 | |
| 7 | rs10803138 | 1 | 243 503 719–243 612 019 | AKT3, SDCCAG8, ANKRD63 | GA | 3.21 | 0.047 | 1 | 24 | −13 | −23 | 2.03e−8 | |
| pgACC | 1 | rs7819570 | 8 | 89 340 626–89 753 626 | MMP16 | TG | 4.15 | 0.005 | 29 | −6 | 41 | −5 | 1.22e−8 |
| 2 | rs10803138 | 1 | 243 503 719–243 612 019 | AKT3, SDCCAG8, | GA | 3.77 | 0.018 | 8 | 3 | 32 | −14 | 2.03e−8 | |
| 3 | rs190065944 | 15 | 78 859 610–78 859 610 | CHRNA5, CHRNA3, CHRNB4 | AG | 3.66 | 0.029 | 4 | 3 | 26 | −11 | 4.71e−8 | |
| Working memory | |||||||||||||
| DLPFC l | 1 | rs55833108 | 10 | 104 587 583–105 165 583 | ARL3, AS3MT, C10orf32, CNNM2, CYP17A1, INA, NT5C2, PCGF6, PDCD11, SFXN2, TAF5, TRIM8, USMG5, WBP1L | TG | 3.92 | 0.012 | 5 | −33 | 44 | 37 | 2.23e−8 |
| 2 | rs2068012 | 14 | 30 189 985–30 190 316 | PRKD1 | CT | 3.66 | 0.035 | 5 | −42 | 26 | 34 | 1.41e−8 | |
| DLPFC r | 1 | rs2068012 | 14 | 30 189 985–30 190 316 | PRKD1 | CT | 4.47 | 0.002 | 29 | 33 | 26 | 37 | 1.41e−8 |
| 2 | rs9841616 | 3 | 181 023 585–181 205 585 | FXR1, DNAJC19, SOX2−OT | TA | 3.81 | 0.019 | 5 | 30 | 38 | 37 | 2.35e−8 | |
| 3 | rs190065944 | 15 | 78 859 610–78 859 610 | CHRNA5, CHRNA3, CHRNB4 | AG | 3.63 | 0.038 | 3 | 27 | 56 | 34 | 4.71e−8 | |
| Reward processing | |||||||||||||
| VSTR l | 1 | rs2535627 | 3 | 52 541 105–52 903 405 | GLT8D1, GNL3, ITIH1, ITIH3, ITIH4, MUSTN1, NEK4, NISCH, NT5DC2, PBRM1, SMIM4, SPCS1, STAB1, TMEM110, TMEM110-MUSTN1 | TC | 4.13 | 0.001 | 20 | −6 | 2 | −8 | 4.26e−11 |
| 2 | rs2007044 | 12 | 2 321 860–2 413 760 | CACNA1C | GA | 3.46 | 0.007 | 35 | −3 | 14 | −5 | 3.22e−18 | |
| 3 | rs56205728 | 15 | 40 566 759–40 602 237 | ANKRD63, PAK6 PLCB2 | AG | 3.24 | 0.014 | 8 | −18 | 14 | −11 | 4.18e−9 | |
| 4 | rs11693094 | 2 | 185 601 420–185 785 420 | ZNF804A | CT | 3.05 | 0.026 | 3 | 0 | 8 | −8 | 1.53e−12 | |
| 5 | rs2905426 | 19 | 19 374 022–19 658 022 | NCAN, HAPLN4, TM6SF2, SUGP1, MAU2, GATAD2A, TSSK6, NDUFA13, YJEFN3, CILP2, PBX4 | GT | 2.97 | 0.032 | 3 | −18 | 8 | −5 | 3.63e−10 | |
| 6 | rs55661361 | 11 | 124 610 007–124 620 147 | ESAM, MSANTD2, NRGN, VSIG2 | GA | 2.82 | 0.042 | 1 | −9 | 17 | −5 | 2.80e−12 | |
| VSTR r | 1 | rs2007044 | 12 | 2 321 860–2 413 760 | CACNA1C | GA | 3.54 | 0.005 | 28 | 6 | 5 | −8 | 3.22e−18 |
| 2 | rs55661361 | 11 | 124 610 007–124 620 147 | ESAM, MSANTD2, NRGN, VSIG2 | GA | 3.16 | 0.016 | 5 | 12 | 20 | −5 | 2.80e−12 | |
| 3 | rs2535627 | 3 | 52 541 105–52 903 405 | GLT8D1, GNL3, ITIH1, ITIH3, ITIH4, MUSTN1, NEK4, NISCH, NT5DC2, PBRM1, SMIM4, SPCS1, STAB1, TMEM110, TMEM110-MUSTN1 | TC | 3.13 | 0.022 | 33 | 15 | 11 | −5 | 4.26e−11 | |
| 4 | rs11693094 | 2 | 185 601 420–185 785 420 | ZNF804A | CT | 3.05 | 0.026 | 2 | 0 | 8 | −8 | 1.53e−12 | |
| 5 | rs1023500 | 22 | 42 315 744–42 361 344 | CENPM, CYP2D6, FAM109B, NAGA, NDUFA6, SEPT3, SHISA8, SMDT1, SREBF2, TCF20, TNFRSF13C, WBP2NL | TC | 3.06 | 0.028 | 1 | 18 | 8 | −8 | 3.43e−8 | |
| 6 | rs2053079 | 19 | 30 981 643–31 039 023 | ZNF536 | GA | 2.85 | 0.046 | 2 | 12 | 8 | 1 | 4.49e−9 | |
| 7 | rs2905426 | 19 | 19 374 022–19 658 022 | CILP2, GATAD2A, HAPLN4, MAU2, NCAN, NDUFA13, PBX4, SUGP1, TM6SF2, TSSK6 | GT | 2.82 | 0.047 | 1 | 18 | 11 | −5 | 3.63e−10 | |
| Theory of Mind | |||||||||||||
| MPFC | 1 | 3rs11693094 | 2 | 185 601 420–185 785 420 | ZNF804A | CT | 3.81 | 0.007 | 5 | −6 | 41 | 37 | 1.53e−12 |
| 2 | Rs256205728 | 15 | 40 566 759–40 602 237 | ANKRD63, PAK6, PLCB2 | AG | 3.63 | 0.014 | 3 | 9 | 41 | 37 | 4.18e−9 | |
| 3 | rs26193698 | 14 | 99 707 919–99 719 219 | BCL11B | GA | 3.41 | 0.031 | 2 | −18 | 56 | 28 | 4.8e−9 | |
| 4 | rs1112691419 | 10 | 104 585 135–104 956 335 | WBP1L, CYP17A1, C10orf32, AS3MT, CNNM2 | TA | 3.25 | 0.049 | 1 | 18 | 44 | 34 | 6.20e−19 | |
| TPJl | 1 | rs133921127 | 6 | 73 132 701–73 171 901 | RIMS1* | CT | 4.29 | 0.001 | 26 | −39 | −58 | 34 | 2.69e−8 |
| 2 | rs7405404 | 16 | 13 728 459–13 761 359 | ERCC4* | TC | 3.90 | 0.006 | 11 | −48 | −67 | 22 | 1.01e−9 | |
| 3 | rs75968099 | 3 | 36 843 183–36 945 783 | TRANK1 | TC | 3.90 | 0.007 | 15 | −45 | −67 | 22 | 1.05e−13 | |
| 4 | rs11693094 | 2 | 185 601 420–185 785 420 | ZNF804A | CT | 3.78 | 0.009 | 24 | −57 | −61 | 19 | 1.53e−12 | |
| 5 | rs9841616 | 3 | 181 023 585–181 205 585 | FXR1, DNAJC19, SOX2−OT | TA | 3.54 | 0.022 | 2 | −60 | −37 | 4 | 2.35e−8 | |
| 6 | rs9922678 | 16 | 9 875 519–9 970 219 | GRIN2A | AG | 3.37 | 0.037 | 1 | −54 | −40 | 25 | 1.28e−8 | |
| 7 | rs12704290 | 7 | 86 403 226–86 459 326 | GRM3 | GA | 3.36 | 0.039 | 1 | −54 | −70 | 19 | 3.33e−10 | |
| 8 | rs12148337 | 15 | 70 573 672–70 628 872 | TLE3* | TC | 3.23 | 0.049 | 1 | −54 | −64 | 31 | 1.79e−8 | |
| TPJr | 1 | rs11139497 | 9 | 84 630 941–84 813 641 | TLE1 | AT | 3.70 | 0.013 | 13 | 51 | −37 | 25 | 3.61e−9 |
| 2 | rs7267348 | 20 | 48 114 136–48 131 649 | KCNB1, PTGIS | CT | 3.35 | 0.037 | 3 | 45 | −64 | 19 | 4.56e−8 | |
| 3 | chr7_2025096_I | 7 | 1 896 096–2 190 096 | MAD1L1 | D/I3 | 3.35 | 0.042 | 2 | 57 | −55 | 37 | 8.2e−15 | |
| PcuPCC | 1 | rs7523273 | 1 | 207 912 183–208 024 083 | C1orf132, CD46, CR1L | AG | 3.67 | 0.013 | 2 | −6 | −43 | 34 | 4.47e−8 |
| 2 | rs1501357 | 5 | 45 291 475–45 393 775 | HCN1 | CT | 3.42 | 0.031 | 1 | 3 | −46 | 22 | 5.05e−9 | |
| 3 | rs1339227 | 6 | 73 132 701–73 171 901 | RIMS1* | CT | 3.35 | 0.038 | 4 | −6 | −58 | 43 | 2.69e−08 | |
| Implicit emotion processing | |||||||||||||
| Amy l | 1 | rs8082590 | 17 | 17 722 402–18 030 202 | ATPAF2, DRG2, GID4, LRRC48, MYO15A, RAI1, SREBF1, TOM1L2 | GA | 3.48 | 0.006 | 27 | −24 | −4 | −20 | 1.77e−8 |
| 2 | rs6065094 | 20 | 37 361 494–37 485 994 | ACTR5, PPP1R16B, SLC32A1 | GA | 3.23 | 0.011 | 4 | −30 | −4 | −14 | 1.46e−11 | |
| 3 | rs2535627 | 3 | 52 541 105–52 903 405 | GLT8D1, GNL3, ITIH1, ITIH3, ITIH4, MUSTN1, NEK4, NISCH, NT5DC2, PBRM1, SMIM4, SPCS1, STAB1, TMEM110, TMEM110-MUSTN1 | TC | 2.85 | 0.037 | 3 | −24 | −1 | −20 | 4.26e−11 | |
| 4 | Chr18_52749216_D | 18 | 52 747 686–52 752 696 | TCF4 | I2D | 2.81 | 0.040 | 1 | −21 | −7 | −17 | 8.03e−11 | |
| 5 | rs4391122 | 5 | 60 499 143–60 843 543 | ZSWIM6 | GA | 2.74 | 0.050 | 1 | −21 | −4 | −17 | 1.1e−14 | |
| Amy r | 1 | rs9607782 | 22 | 41 408 556–41 675 156 | CHADL, EP300, L3MBTL2 | AT | 4.43 | 0.000 | 20 | 27 | 5 | −17 | 2.07e−11 |
| 2 | rs8082590 | 17 | 17 722,402–18,030,202 | ATPAF2 DRG2 GID4 LRRC48 MYO15A RAI1 SREBF1 TOM1L2 | GA | 3.70 | 0.003 | 44 | 30 | −4 | −20 | 1.77e−8 | |
| 3 | rs2535627 | 3 | 52 541 105–52 903 405 | GLT8D1, GNL3, ITIH1, ITIH3, ITIH4, MUSTN1, NEK4, NISCH, NT5DC2, PBRM1, SMIM4, SPCS1, STAB1, TMEM110, TMEM110-MUSTN1 | TC | 3.54 | 0.005 | 30 | 24 | 2 | −20 | 4.26e−11 | |
| 4 | rs8042374 | 15 | 78 803 032–78 926 732 | AC027228.1, AGPHD1, CHRNA3, CHRNA5, CHRNB4, IREB2, PSMA4 | AG | 2.88 | 0.034 | 1 | 27 | −1 | −29 | 2.44e−13 | |
| pgACC | 1 | rs7267348 | 20 | 48 114 136–48 131 649 | KCNB1, PTGIS | CT | 4.33 | 0.002 | 16 | −15 | 44 | 13 | 4.56e−8 |
| 2 | rs10043984 | 5 | 137 598 121–137 750 021 | CDC25C, CTNNA1, EGR1, ETF1, FAM53C, GFRA3, HSPA9, KDM3B, REEP2 | TC | 3.67 | 0.021 | 4 | 0 | 26 | −5 | 1.09e−8 | |
| 3 | Chr6_84280274_D | 6 | 84 279 922–84 407 274 | SNAP91 | I2D | 3.67 | 0.025 | 16 | 0 | 44 | −14 | 8.15e−10 | |
| 4 | rs75968099 | 3 | 36 843 183–36 945 783 | TRANK1 | TC | 3.51 | 0.042 | 1 | −15 | 44 | −5 | 1.05e−13 | |
| 5 | rs832187 | 3 | 63 792 650–64 004 050 | ATXN7, C3orf49, PSMD6, THOC7 | CT | 3.46 | 0.048 | 1 | 0 | 17 | 28 | 1.43e−8 | |
Abbreviations: A, adenine; BP, base pair; C, cytosine; Chr, chromosome; dir, direction of effect (−, decreased activation with increasing number of risk alleles; +, increased activation with increasing number of risk alleles); fMRI, functional magnetic resonance imaging; Freq RA, frequency of risk allele in the present sample; G, guanine; HC, hippocampus; L, left; MNI coords., MNI coordinates of peak activation in mm; MPFC, medial prefrontal cortex; NRA non-risk allele; PcuPCC, precuneus/posterior cingulate cortex; pgACC, perigenual anterior cingulate cortex; PFWE-ROI, familywise error corrected P-value within region of interest; R, right; RA, risk allele; ROI, region of interest; SNP, single-nucleotide polymorphism; T, thymine; TPJ, temporal parietal junction; Vx, number of voxels in significant cluster.
Single SNP fMRI analysis of variants that have been associated with schizophrenia at a genome-wide significant level. Positions from hg19 build 37; P-values for association with schizophrenia (pSx) from combined sample of ref. 2. Genes were identified using Ricopili (data set: PGC_SCZ52_may13). All genes located within regions containing variants in linkage disequilibrium of R2⩾0.6 with the SNP associated with schizophrenia2 are reported. Where chromosomal positions do not contain a gene, the nearest genes within a ±500 kb window is listed (indicated by *).
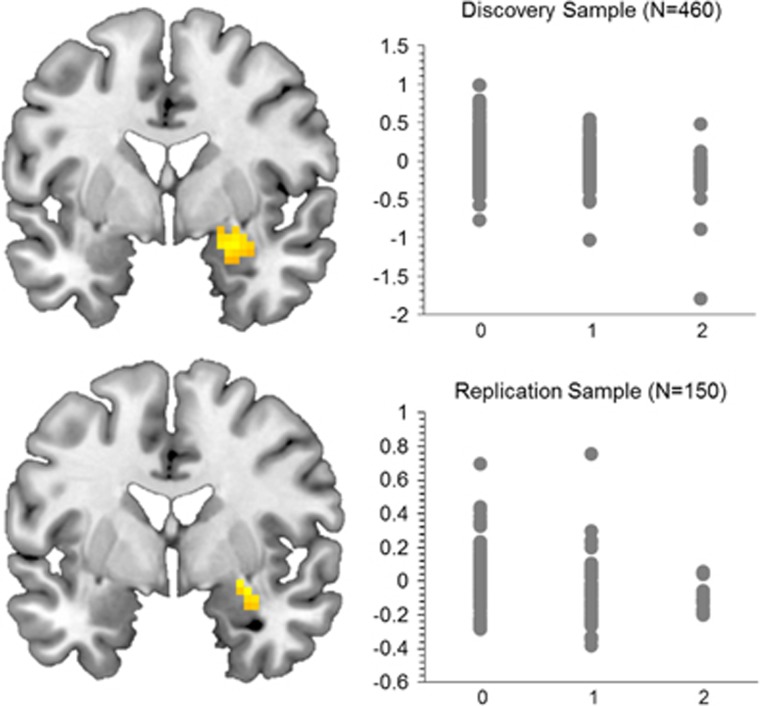
Figure 3.
Effects of rs9607782 on right amygdala activity during implicit emotion processing. Results are P<0.05 FWE-corrected across the ROI. The effect in the discovery sample additionally withstood Bonferroni correction for the number of SNPs tested (n=105), that is, PFWE<0.0004. N, sample size; FWE, familywise error; ROI, region of interest; SNP, single-nucleotide polymorphism.
Replication analysis
We could replicate the effect of rs9607782 on amygdala activation in an independent control sample using the identical FMT (x=30, y=−4, z=−11, F=10.94, Z=3.04, PFWE(ROI)=0.025; Figure 3). In both samples amygdala activity decreased with increasing number of risk alleles for schizophrenia.
Analyses of LD-independent non-schizophrenia-associated SNPs (n=105)
In order to assess the adequacy of correcting for the number of SNPs tested per ROI, we determined the number of significant effects of 105 common variants not significantly associated with schizophrenia on all neuroimaging phenotypes. Therefore, we generated a random set of 10 000 SNPs across the whole genome. From this set we randomly selected 105 variants with a P-value of >0.2 for association with schizophrenia and a minor allele frequency of >10%/<90% in the PGC_SCZ52 data set. We observed a median of three (range 0–7) significant hits applying a threshold of PFWE(ROI)<0.05 across the respective ROIs. None of these associations withstood correction for the number of variants tested per ROI (Supplementary Figure S3). The difference between the number of statistically meaningful associations per ROI using schizophrenia-related and -unrelated SNPs was marginally significant (P=0.06 after 10 000 permutations; one-tailed t-test). However, overall the P-values observed with schizophrenia-associated variants were significantly lower than among associations with unrelated variants (P<0.005 after 10 000 permutations; one-tailed t-test; see Supplementary Material for details).
Discussion
We investigated the impact of genetic risk for schizophrenia on a range of functional imaging phenotypes reliably activating distributed brain networks covering five RDoC subdomains with well-established relevance to the disease. We analyzed the effects of (i) a polygenic RPS and (ii) of 105 single SNPs genome-wide significantly associated with schizophrenia in face of benefits and drawbacks of both approaches. RPS offer assessment of accumulated genetic risk in a limited number of tests, but prohibit conclusions regarding specific contributions of SNPs and might conceal effects of some risk variants when combined with irrelevant signals. Single SNP analyses on the other hand have the disadvantage that the higher amount of tests (i) heightens the risk of false-positive results if not correctly accounted for (type I error) and (ii) increases the likelihood to find only those effects whose effect size is overestimated (winner’s curse).
The analysis of the polygenic RPS revealed two significant associations at a standard neuroimaging significance level (PFWE(ROI)<0.05). The RPS predicted pgACC activity during EM recognition. pgACC recruitment was previously found to be modulated by genetic risk for schizophrenia during active EM retrieval. Furthermore, we found an association between RPS and PCC/Pcu activity during ToM. Activation of the PCC/Pcu, one of the crucial mentalizing areas, has been associated twice with a risk variant within ZNF804A (rs1344706) in high LD with a SNP included in the RPS (rs11693094; R2=0.84). However, it must be emphasized that these results would not withstand multiple comparison correction for the total number of ROI analyses across all tasks (P<0.0025, that is, P<0.05 FWE across 20 ROI analyses). With such a stringent threshold none of the above-mentioned results would remain significant and, in fact, one false-positive finding would be expected in 20 independent tests (at P=0.05).
As we investigated 105 independent SNPs, we strictly corrected for this number within each ROI. Only one association withstood correction for multiple testing: rs9607782 was associated with right amygdala activity during implicit emotion processing. To the best of our knowledge, this SNP was not previously investigated in imaging genetics studies. Rs9607782 is located in an LD block with EP300, L3MBTL2, CHADL and RANGAP, and is in high LD (R2=0.77) with a missense mutation in EP300 (rs20551), suggesting functional relevance. Besides association with schizophrenia, mutations in the EP300 gene are responsible for the Rubinstein-Taybi syndrome, a developmental disorder that includes intellectual disability, impulsivity, distractibility and mood instability.29 EP300 encodes p300, a protein that functions as histone-acetyltransferase and is expressed in the brain in limbic and cortical regions. In mice, inhibition of p300 activity significantly impairs fear memory consolidation, and associated neural plasticity in the lateral amygdala.30 P300 inhibition further induced enhanced anxiety and mild cognitive impairment in a water maze task.31 Altered emotional significance detection and maladaptive appraisal have been associated with amygdala dysfunction in schizophrenia before.32, 33, 34 Thus, previous evidence supports the potential relevance of the observed association, particularly as we were able to replicate our result in an independent sample (Figure 3). This finding taps into the RDoC domains social cognition and negative valence. Importantly, the fMRI paradigm used to measure this potential intermediate phenotype is well qualified as an RDoC subdomain as it fulfills required criteria for good psychometric properties,24 association with psychopathology (association with schizophrenia was repeatedly shown on the behavioral and the brain level)35, 36 and heritability (with heritability estimates for emotion identification ranging between 0.21 and 0.43).37, 38
Other associations, although not surviving correction for multiple comparisons, include SNPs previously found to be related to imaging phenotypes from all RDoC domains assessed (see Table 1), for example, rs2007044 within CACNA1C was associated with hippocampal activity during EM,10, 14, 15, 39 but also with bilateral ventral striatal activation during RP. Rs11693094 within ZNF804A was associated with activity of ToM areas (medial prefrontal cortex, left temporal parietal junction) as shown before13, 16 and also with striatal activity. In addition, there were associations of variants within TCF4 (previously found to have an impact on hippocampal volume)40 with hippocampal and pgACC activity during EM as well as with amygdala recruitment during implicit emotion processing. rs1702294 within MIR137 (gene previously found to have an impact on frontalmediotemporal connectivity)41, 42 was associated with hippocampal activity, and rs2905426 within NCAN (previously associated with cortical thickness and folding in schizophrenia)43, 44 was associated with ventral striatal activation during RP.
All of these effects would stand out as significant hits in association studies on imaging phenotypes that typically apply a threshold of PFWE(ROI)<0.05. However, an increasing number of statistical tests always comes at the expense of an increasing risk of type I error if not corrected for properly. In total, we found a median of 4 and up to 8 significant hits per ROI, which again, given a significance threshold of P<0.05, is compatible to chance findings in 105 tests. In fact, these numbers correspond well with those found by Sullivan who observed similar numbers in simulated genetic data containing no valid associations.45 In his data, an uncorrected significance level of P<0.05 yielded a proportion of false-positive findings of 96.8% (at least one positive finding in 968 of 1000 simulations). When Sullivan corrected for the number of SNPs he tested (which is in accordance to what we did) he still found a false-positive proportion of 31.4%.45 Thus, accounting for the number of SNPs may still be insufficient and additional correction for the number of ROIs could be necessary. Indeed, Sullivan reported that correction for the total number of independent tests resulted in the appropriate false-positive proportion of 5%. Application of such strict correction for our single SNP analyses (in our case P<2.3 × 10−5) would result in no finding being interpretable as statistically significant. On the other hand, the combination of FWE correction across ROIs with Bonferroni correction for the number of tests could be too conservative for fMRI data, taking into account that task-related ROI activation may be highly correlated, that is, is not truly independent. By testing associations of 720 SNPs not associated with schizophrenia with functional brain correlates of WM and implicit emotion processing Meyer-Lindenberg et al.46 showed that the false-positive proportion was no higher than 1.0–4.1% applying FWE correction alone. If this applies to our data, those SNPs surpassing the standard threshold of PFWE(ROI)<0.05 may be true findings. Hence, to evaluate whether our findings contain true signal, we applied a similar strategy by testing the association of 105 SNPs not associated with schizophrenia or any mental function with our imaging phenotypes. Whereas the statistical difference for the total number of statistical meaningful associations among schizophrenia-related and -unrelated variants fell short of the significance threshold, we observed significantly higher probabilities for associations of disease-related than -unrelated variants. These results at least tentatively suggest further true signal in our data. Nevertheless, we believe that in face of the large amount of analyses we carried out in total, independent replication is essential and should be made mandatory in imaging genetics research, just as in psychiatric genetics research.
Our findings point to several issues relevant to the field of imaging genetics. It is foreseeable that larger genetic studies will discover more genome-wide significant genetic variants associated with psychiatric disorders. A brute force approach, that is, testing and correcting for all significantly associated variants known, will require a magnitude of statistical power difficult to achieve in functional imaging. There are three principal ways to respond to this conundrum. First, increase sample sizes (the mantra of genetics), for example, through consortial data accumulation worldwide. This is a reasonable path that has already been taken by the ENIGMA consortium47 to which we have contributed too.17, 48 However, costs and data availability limit this path and, in the foreseeable future, (relatively) large sample sizes will likely be restricted to structural and resting state data. The aggregation of task-related functional data will be even more difficult because of demands on harmonization of experimental procedures and psychometric task properties. Second, restrict analyses to theory-driven a priori hypotheses. This is formally correct but the possibility remains that ‘suitable’ hypotheses are established post hoc after extensive data mining. Hence, this is difficult to control without prior registration of hypotheses, in particular as more and more groups are performing genome-wide genotyping. That said, exploratory analyses should still be accepted if labeled appropriately and substantiated by replication studies. Third, and most forward-looking, the use of data reduction, feature selection and multivariate methods.49 Data reduction methods might include polygenic RPSs, gene set enrichment analyses or pathway analyses on the genetic side, and network analyses, independent component analyses, or graph theory on the imaging side. In addition, multivariate machine-learning methods might be useful to map high-dimensional genetic and neuroimaging data sets—this, however, also requires relatively large data sets.
Apart from these three principal approaches we would like to encourage the field to publish replication studies and in particular negative findings to enable future meta-analytical approaches. In addition, standards should be developed calling on imaging genetic studies to additionally publish whole-brain-effect sizes to facilitate replication and meta-analysis.
Our results suggest that risk scores aggregated across multiple independent loci may be less helpful than hoped for intermediate phenotype characterization. There is some evidence that brain effects are not as linear as associations with a (linear) polygenic RPS would imply.50 Further, it should be emphasized that comparable polygenic scores do not necessarily include the same combination of risk variants. Each individual will rather have a unique combination of risk variants, and it is thus not reasonable to assume that similar scores will have an impact on the same neural region or circuit. A possible solution would be genotyping a large sample of individuals and deeply phenotyping (using, for example, neuroimaging) a subset at the upper and lower tail of the distribution. This does not speak against the usefulness of RPS in general, but in intermediate phenotype research, more hypothesis-driven approaches to RPS definition may prove more fruitful.
Coming back to our own study presented here, we remind that our data are limited by the combined analyses of subjects with and without familial liability for psychiatric disorders. Still, all subjects had a negative lifetime history of psychiatric disorders as evidenced by a respective diagnostic interview and we accounted for subgroup status in all of our analyses.
Please note also that, in order to limit our variables on the imaging side, we restricted our analyses to regional effects using a ROI approach and did not test for other possible measures that focus on connectivity11 or network parameters.51, 52
In summary, although it unquestionably is a big challenge to acquire enough functional neuroimaging data for strict correction of both high-dimensional data sets (that is, imaging and genetics) using a brute force approach, we plead for the continuation of acquiring, analyzing and publishing imaging genetics results. These should include theory-driven restricted analyses, as well as stringent significance level adjustment and independent replication. Implementing these methodological requirements, our most robust finding was an association between a variant near EP300 with amygdala function during implicit emotion processing, a finding that is supported by preclinical and clinical findings and points to a disease-relevant mechanism potentially mediating aberrant anxiety processing and/or fear memory.
Acknowledgments
This work was supported by the German Ministry for Education and Research (BMBF) grants NGFNplus MooDS (Systematic Investigation of the Molecular Causes of Major Mood Disorders and Schizophrenia), NGFN-Plus Genetics of Alcohol Addiction/ e:Med Alcoholism (FKZ 01GS08159, 01ZX1311E), the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med program (grant number O1ZX1314B) and the German Research Foundation (DFG) grant SFB 636-B7. MMN is a member of the DFG funded Excellence-Cluster Immuno-Sensation. FD received financial support from the local Bonfor programme. We thank Swapnil Awasthi for help with genetic analyses.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
AML received consultancy fees from Astra Zeneca, Elsevier, F Hoffmann-La Roche, Gerson Lehrman Group, Lundbeck foundation, Outcome Europe Sárl, Outcome Sciences, Roche Pharma, Servier International and Thieme Verlag, and lecture fees—including the travel fees—from Abbott, Astra Zeneca, Aula Médica Congresos, BASF, Groupo Ferrer International, Janssen-Cilag, Lilly Deutschland, LVR Klinikum Düsseldorf, Servier Deutschland and Otsuka Pharmaceuticals. HW received a speaker honorarium from Servier. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 2003; 60: 1187–1192. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SE, Cuthbert BN. Research domain criteria: cognitive systems, neural circuits, and dimensions of behavior. Dialogues Clin Neurosci 2012; 14: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak MJ, Cuthbert BN. The NIMH research domain criteria initiative: background, issues, and pragmatics. Psychophysiology 2016; 53: 286–297. [DOI] [PubMed] [Google Scholar]
- Bhat S, Dao DT, Terrillion CE, Arad M, Smith RJ, Soldatov NM et al. CACNA1C (Ca(v)1.2) in the pathophysiology of psychiatric disease. Progr Neurobiol 2012; 99: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung R, Prata DP. What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol Med 2015; 45: 2461–2480. [DOI] [PubMed] [Google Scholar]
- Hess JL, Quinn TP, Akbarian S, Glatt SJ. Bioinformatic analyses and conceptual synthesis of evidence linking ZNF804A to risk for schizophrenia and bipolar disorder. Am J Med Genet B 2015; 168B: 14–35. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 2003; 160: 636–645. [DOI] [PubMed] [Google Scholar]
- Lancaster T, Doherty J, Linden DE, Hall J. Imaging genetics of schizophrenia. In: Bigos KL, Hariri AR, Weinberger DR (eds). Neuroimaging Genetics: Principles and Practices. Oxford University Press: Oxford; New York, NY, USA, 2016. [Google Scholar]
- Erk S, Meyer-Lindenberg A, Schnell K, Opitz von Boberfeld C, Esslinger C, Kirsch P et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch Gen Psychiatry 2010; 67: 803–811. [DOI] [PubMed] [Google Scholar]
- Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C et al. Neural mechanisms of a genome-wide supported psychosis variant. Science 2009; 324: 605. [DOI] [PubMed] [Google Scholar]
- Grimm O, Heinz A, Walter H, Kirsch P, Erk S, Haddad L et al. Striatal response to reward anticipation: evidence for a systems-level intermediate phenotype for schizophrenia. JAMA Psychiatry 2014; 71: 531–539. [DOI] [PubMed] [Google Scholar]
- Walter H, Schnell K, Erk S, Arnold C, Kirsch P, Esslinger C et al. Effects of a genome-wide supported psychosis risk variant on neural activation during a theory-of-mind task. Mol Psychiatry 2011; 16: 462–470. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Linden DE, Lancaster T, Mohnke S, Grimm O et al. Replication of brain function effects of a genome-wide supported psychiatric risk variant in the CACNA1C gene and new multi-locus effects. NeuroImage 2014; 94: 147–154. [DOI] [PubMed] [Google Scholar]
- Krug A, Witt SH, Backes H, Dietsche B, Nieratschker V, Shah NJ et al. A genome-wide supported variant in CACNA1C influences hippocampal activation during episodic memory encoding and retrieval. Eur Arch Psychiatr Clin Neurosci 2014; 264: 103–110. [DOI] [PubMed] [Google Scholar]
- Mohnke S, Erk S, Schnell K, Schutz C, Romanczuk-Seiferth N, Grimm O et al. Further evidence for the impact of a genome-wide-supported psychosis risk variant in ZNF804A on the Theory of Mind Network. Neuropsychopharmacology 2014; 39: 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 2012; 44: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc Natl Acad Sci USA 1967; 58: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima D, Breen G. Polygenic risk scores in imaging genetics: usefulness and applications. J Psychopharmacol 2015; 29: 867–871. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, M G, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Non-patient Edition. (SCID-I/NP). Biometrics Research, New York State Psychiatric Institute: New York, NY, USA, 2002. [Google Scholar]
- Spanagel R, Bartsch D, Brors B, Dahmen N, Deussing J, Eils R et al. An integrated genome research network for studying the genetics of alcohol addiction. Addict Biol 2010; 15: 369–379. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Schmierer P, Grimm O, Tost H, Muhleisen T et al. Functional impact of a recently identified quantitative trait locus for hippocampal volume with genome-wide support. Transl Psychiatry 2013; 3: e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plichta MM, Schwarz AJ, Grimm O, Morgen K, Mier D, Haddad L et al. Test-retest reliability of evoked BOLD signals from a cognitive-emotive fMRI test battery. NeuroImage 2012; 60: 1746–1758. [DOI] [PubMed] [Google Scholar]
- Friedman L, Glover GH. Report on a multicenter fMRI quality assurance protocol. J Magn Reson Imaging 2006; 23: 827–839. [DOI] [PubMed] [Google Scholar]
- Sugranyes G, Kyriakopoulos M, Corrigall R, Taylor E, Frangou S. Autism spectrum disorders and schizophrenia: meta-analysis of the neural correlates of social cognition. PLoS One 2011; 6: e25322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter H, Ciaramidaro A, Adenzato M, Vasic N, Ardito RB, Erk S et al. Dysfunction of the social brain in schizophrenia is modulated by intention type: an fMRI study. Soc Cogn Affect Neurosci 2009; 4: 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Overwalle F. Social cognition and the brain: a meta-analysis. Hum Brain Mapp 2009; 30: 829–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven WM, Tuinier S, Kuijpers HJ, Egger JI, Brunner HG. Psychiatric profile in rubinstein-taybi syndrome. A review and case report. Psychopathology 2010; 43: 63–68. [DOI] [PubMed] [Google Scholar]
- Maddox SA, Watts CS, Schafe GE. p300/CBP histone acetyltransferase activity is required for newly acquired and reactivated fear memories in the lateral amygdala. Learn Mem 2013; 20: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viosca J, Lopez-Atalaya JP, Olivares R, Eckner R, Barco A. Syndromic features and mild cognitive impairment in mice with genetic reduction on p300 activity: differential contribution of p300 and CBP to Rubinstein-Taybi syndrome etiology. Neurobiol Dis 2010; 37: 186–194. [DOI] [PubMed] [Google Scholar]
- Gur RC, Gur RE. Social cognition as an RDoC domain. Am J Med Genet B 2016; 171B: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mier D, Lis S, Zygrodnik K, Sauer C, Ulferts J, Gallhofer B et al. Evidence for altered amygdala activation in schizophrenia in an adaptive emotion recognition task. Psychiatr Res 2014; 221: 195–203. [DOI] [PubMed] [Google Scholar]
- Underwood R, Kumari V, Peters E. Cognitive and neural models of threat appraisal in psychosis: a theoretical integration. Psychiatr Res 2016; 239: 131–138. [DOI] [PubMed] [Google Scholar]
- Kohler CG, Walker JB, Martin EA, Healey KM, Moberg PJ. Facial emotion perception in schizophrenia: a meta-analytic review. Schizophr Bull 2010; 36: 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chan RC, McAlonan GM, Gong QY. Facial emotion processing in schizophrenia: a meta-analysis of functional neuroimaging data. Schizophr Bull 2010; 36: 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins ME, Tepper P, Gur RC, Ragland JD, Klei L, Wiener HW et al. Project among African-Americans to explore risks for schizophrenia (PAARTNERS): evidence for impairment and heritability of neurocognitive functioning in families of schizophrenia patients. Am J Psychiatry 2010; 167: 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RE, Nimgaonkar VL, Almasy L, Calkins ME, Ragland JD, Pogue-Geile MF et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry 2007; 164: 813–819. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Schmierer P, Mohnke S, Grimm O, Garbusow M et al. Hippocampal and frontolimbic function as intermediate phenotype for psychosis: evidence from healthy relatives and a common risk variant in CACNA1C. Biol Psychiatry 2014; 76: 466–475. [DOI] [PubMed] [Google Scholar]
- Wirgenes KV, Sonderby IE, Haukvik UK, Mattingsdal M, Tesli M, Athanasiu L et al. TCF4 sequence variants and mRNA levels are associated with neurodevelopmental characteristics in psychotic disorders. Transl Psychiatry 2012; 2: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zhang X, Hou B, Li J, Qiu C, Qin W et al. The impact of MIR137 on dorsolateral prefrontal-hippocampal functional connectivity in healthy subjects. Neuropsychopharmacology 2014; 39: 2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothersill O, Morris DW, Kelly S, Rose EJ, Fahey C, O'Brien C et al. Effects of MIR137 on fronto-amygdala functional connectivity. NeuroImage 2014; 90: 189–195. [DOI] [PubMed] [Google Scholar]
- Schultz CC, Muhleisen TW, Nenadic I, Koch K, Wagner G, Schachtzabel C et al. Common variation in NCAN, a risk factor for bipolar disorder and schizophrenia, influences local cortical folding in schizophrenia. Psychol Med 2014; 44: 811–820. [DOI] [PubMed] [Google Scholar]
- Muhleisen TW, Mattheisen M, Strohmaier J, Degenhardt F, Priebe L, Schultz CC et al. Association between schizophrenia and common variation in neurocan (NCAN), a genetic risk factor for bipolar disorder. Schizoph Res 2012; 138: 69–73. [DOI] [PubMed] [Google Scholar]
- Sullivan PF. Spurious genetic associations. Biol Psychiatry 2007; 61: 1121–1126. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Nicodemus KK, Egan MF, Callicott JH, Mattay V, Weinberger DR. False positives in imaging genetics. NeuroImage 2008; 40: 655–661. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav 2014; 8: 153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N et al. Common genetic variants influence human subcortical brain structures. Nature 2015; 520: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Calhoun VD. A review of multivariate analyses in imaging genetics. Front Neuroinform 2014; 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Bandettini PA. The effect of stimulus duty cycle and "off" duration on BOLD response linearity. NeuroImage 2005; 27: 70–82. [DOI] [PubMed] [Google Scholar]
- Braun U, Schafer A, Walter H, Erk S, Romanczuk-Seiferth N, Haddad L et al. Dynamic reconfiguration of frontal brain networks during executive cognition in humans. Proc Natl Acad Sci USA 2015; 112: 11678–11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bertolino A, Walter H, Schneider M, Schafer A, Taurisano P et al. Altered functional subnetwork during emotional face processing: a potential intermediate phenotype for schizophrenia. JAMA Psychiatry 2016; 73: 598–605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.