Abstract
Neuro-inflammation and neuronal communication are considered as mis-regulated processes in the aetiology and pathology of bipolar disorder (BD). Which and when specific signal pathways become abnormal during the ontogeny of bipolar disorder patients is unknown. To address this question, we applied induced pluripotent stem cell (iPSC) technology followed by cortical neural differentiation on adipocyte-derived cells from BD type I patients (with psychotic episodes in psychiatric history) and healthy volunteers (controls). RNA sequencing in iPSC and cortical neural stem cell (NSC) lines were used to examine alterations between the transcriptomes from BD I and control samples during transition from the pluripotent stage towards the neural developmental stage. At the iPSC stage, the most highly significant differentially expressed gene (DEG) was the NLRP2 inflammasome (P=2.66 × 10−10). Also among 42 DEGs at the NSC stage, NLRP2 showed the strongest statistical significance (P=3.07 × 10−19). In addition, we have also identified several cytoskeleton-associated genes as DEGs from the NSC stage, such as TMP2, TAGLN and ACTA2; the former two genes are recognised for the first time to be associated with BD. Our results also suggest that iPSC-derived BD-cortical NSCs carry several abnormalities in dopamine and GABA receptor canonical pathways, underlining that our in vitro BD model reflects pathology in the central nervous system. This would indicate that mis-regulated gene expression of inflammatory, neurotransmitter and cytoskeletal signalling occurs during early fetal brain development of BD I patients.
Introduction
Bipolar disorder (BD) is a severe and chronic disorder characterised by the cyclic occurrence of episodes of mania and depression. BD is also associated with significant disability, morbidity, and cognitive impairment.1, 2 It is frequently comorbid with several medical conditions including cardiovascular and metabolic diseases.2, 3 However, the connections between extra- and intra-cerebral pathologies are largely unknown. Nevertheless, the co-occurrence of autoimmune diseases has been reported, that is, systemic lupus erythematosus,4 multiple sclerosis5, 6 and autoimmune thyroiditis,7 as well as altered levels of circulating inflammatory cytokines, including interleukin (IL)-6, TNF-α, IFN-γ and IL1-β.8 A review of immunological factors in the pathophysiology of BD describes a major imbalance in inflammatory cytokines.9 These findings suggest the presence of immunological activation in BD of adult individuals. However, when chronic inflammation starts during the lifespan of BD patients is unknown.
The molecular and cellular mechanisms contributing to BD initiation and progression are poorly understood. The heritability of BD is estimated to be as high as 90%, suggesting a strong genetic basis.10 In light of this high heritability, genome-wide association studies have been used to identify genetic pathways associated with BD.11 In fact, several abnormally regulated genes have been identified in adult BD patient samples.12 It is not known when a BD-associated mis-regulated gene expression would start during the lifespan of BD patients.
Combining iPSC technology13, 14, 15 and neural differentiation16 of cells from patients and healthy controls allows in vitro modelling of human neurodegenerative disorders,17, 18, 19, 20, 21, 22, 23 as well as complex genetic neuropsychiatric disorders.24, 25, 26, 27 However, the polygenic nature of BD and the pronounced genetic overlap between schizophrenia and BD type I (BD I, with manic episodes in psychiatric history)11, 28, 29 requires further investigations to find out when and which genes as well as which signalling pathways are abnormally regulated during neural development in BD. Thus, there is a need to investigate the global transcriptome of BD I patients and healthy controls during the early stages of neural development, that is, the neural stem cell (NSC) stage.
For this purpose, we used disease modelling using iPSC technology and cortical neural differentiation of adipocytes obtained from euthymic BD I patients under medical treatment as well as from healthy individuals. Subsequently, we performed RNA sequencing (Seq), providing high sensitivity with the capacity to detect low-copy transcripts. We established a human BD model system to understand when and how the BD I-associated genes manifest, by comparing RNAs at iPSC and NSC stages of six BD I patients and four healthy controls (including one from Cellartis DEF-hiPSC line). We found several differentially expressed genes (DEGs) involved in immune responses at the NSC stage. Remarkably, one of these genes, NLRP2, was identified as a DEG as early as in iPSC stage when compared with the controls. Furthermore, the DEGs were uploaded to a signalling pathway analysis, demonstrating dopamine and GABA receptor canonical pathways to be abnormally expressed already in the NSC stage. Our results not only provide further evidence that BD I is a developmental disorder with polygenetic disposition, but also reveal that mis-regulated expression of genes involved in inflammation and neurotransmitter systems occur already during the early stages of fetal neural development in BD I patient cells.
Materials and methods
Generation of hiPSC lines and culture conditions
Abdominal subcutaneous adipose tissue was isolated and primary adipocyte cell lines were established.30 Starting with adipocytes from patients (B1, B2, B3, B4, B5, B6) and controls (C1, C2 and C3), iPSCs were generated and characterised by Cellectis (formerly Cellartis, presently Takara Clontech, Shiga, Japan). As an additional and study-independent control, one Cellartis DEF-hiPSC ChiPSC4 line (C4) was used. All lines were cultured under feeder-free conditions in Cellartis DEF-CS (Takara Bio Europe, Gothenburg, Sweden) at 37 °C in a humidified atmosphere of 5% CO2 in air. Primary adipocytes were cultured as described earlier30 without supplement of insulin, dexamethasone or other reagents that maintain adipocyte characteristics.
To create an in vitro BD model system, adipocyte cell lines were thus originally derived from abdominal subcutaneous fat samples of altogether 35 BD (11 BD I, 7 BD II and 17 BD non ultra descriptus) patients and 38 healthy controls, as earlier described.30 From these cell lines, six BD I and three healthy controls were selected on technical grounds. The six BD I patients (three females, three males) donating abdominal adipocytes were all euthymic at the time of examination with GAF scores (±s.d.) 69.2±13.6, Young Mania Rating Scale 0.7±1.6 and Montgomery–Åsberg Depression Rating Scale 5.7±5.1. They all had suffered from at least one psychotic episode in their psychiatric history. All have Caucasian origins. Their age at first mood episode compared with their current age was 12/53, 18/39, 17/44, 15/47, 15/47 and 21/32 years. Four were currently medically mood-stabilised with lithium, four with antipsychotics and two with antiepileptics. Three controls were healthy females, age 27, 26 and 29 years. One ChiPSC4 line (C4) male control was added.
Directed differentiation of hiPSC
The hiPSCs lines were plated on COAT1 (Takara Bio Europe) coated 12-well plates in feeder-free conditioned Cellartis DEF-CS medium (Takara Bio Europe) until confluence. Neural induction was initiated by changing the culture medium to 1:1 mixture of N2 media, consisting of DMEM/F12 GlutaMAX (Life Technologies, Carlsbad, CA, USA), N2 supplement (Life Technologies), 5 μg ml−1 insulin (Sigma-Aldrich, St. Louis, MO, USA), 1 mm Ultra glutamine (Lonza, Basel, Switzerland), 100 μm non-essential amino acids (Gibco, Carlsbad, CA, USA), 100 μm 2-mercaptoethanol (Gibco), 50 U ml−1 penicillin and streptomycin (Lonza), and B27 media (Neurobasal; Life Technologies), consisting of B27 with vitamin A (Life Technologies), 2 mm Ultra glutamine (Lonza), 50 U ml−1 penicillin and streptomycin (Lonza) supplemented with 1 μm Dorsomorphin (Tocris Bioscience, Bristol, UK) and 10 μm SB431542 (Tocris Bioscience). Neural induction media was replaced every day for 8–10 days until a uniform neuro-epithelial sheet was observed, and then both iPSC and neuro-epithelial cells were frozen in RNA protect (Qiagen, Redwood City, CA, USA) for miRNA and transcriptome analysis. Neuro-epithelial cells were collected by dissociation with Dispase (Life Technologies) and aggregates were re-plated on laminin-coated plates and maintained in neural maintenance media. On appearance of rosette structures, NSCs were expanded by supplementing media with 20 ng ml−1 FGF2. After a further 4 days, FGF was withdrawn and the cultures were passaged using Accutase and maintained in a neural maintenance media until frozen at day 23–30 post initiation of neural induction. For neurogenesis, NSCs were cultured on poly-l-ornithine/laminin-coated dishes on a feeder layer of human astrocytes. Neural maintenance media was changed every second day.
Immunostaining
For immunofluorescence, iPSCs, neuro-epithelial cells and NSCs were collected by dissociation with Tryple Select (Life Technologies), Dispase (Life Technologies) and Accutase, respectively. After counting, the 5.0 × 106 iPSC were re-plated onto eight-well chamber slides coated with laminin 521 (BioLamina, Sundbyberg, Sweden), and cultured overnight in Cellartis DEF-CS (Takara Bio Europe). Neuro-epithelial aggregates were re-plated on laminin-coated coverslips and maintained in neural maintenance medium until appearance of rosette structures. NSCs were re-plated onto eight-well chamber slides coated with laminin and maintained in neural maintenance medium for 60 days. The cells were fixated with 4.0% paraformaldehyde/phosphate-buffered saline (PBS) for 20 min, washed in 0.1% Tween 20/PBS, and permeabilised with 0.25% Triton X-100/PBS for 10 min and blocked by 0.1% Tween 20/0.3 m glycine/10% normal donkey or goat serum/PBS for 1 h. The primary antibodies used were anti-Oct-3/4 (H-134) (1:200, sc-9081; Santa Cruz Biotechnology, Dallas, TX, USA), anti-SSEA4 (MC813) (1:50, ab16287, Abcam, Cambridge, UK), anti-sialyl-lactotetra (TR4) (1:500), anti-Pax6 (1:200, ab2237; Millipore, Solna, Sweden), anti-Otx2 (1:200, ab9566; Millipore), anti-Nestin (ab22035; Abcam), anti-Ki67 (1:200, sc-23900; Santa Cruz). For secondary antibodies, Alexa Fluor 488 conjugated donkey anti mouse IgG and Alexa Fluor 555 conjugated donkey anti rabbit IgG (1:1000, Life Technologies) were used, which were diluted in blocking solution and added for 2 and 1 h, respectively, each followed by washes in 0.1% Tween 20/PBS. The nuclei were counterstained with Hoechst. The slides were mounted and analysed on Nikon Eclipse Ni-E automated upright microscope (Tokyo, Japan) equipped with Orca Flash 4.0 camera (Hamamatsu Photonics, Shizuoka, Japan), using Plan Apo lambda × 20/0.75 or Plan Apo lambda 60 (Nikon) objectives.
Electrophysiology recordings
For electrophysiological experiments, NSC were plated on laminin-coated Ibidi μ-dishes (Ibidi, Munich, Germany) and maintained in a neural maintenance media for 30 to 127 days. The μ-dishes were mounted under a microscope (Nikon E600FN) where the cells were perfused (2–3 ml min−1) with artificial cerebrospinal fluid containing 1 mm NaH2PO4, 123 mm NaCl, 26 mm NaHCO3, 3 mm KCl, 2 mm MgCl2, 1 mm CaCl2 and 10 mm d-glucose. The artificial cerebrospinal fluid was continuously bubbled with gas containing 95% O2 and 5% CO2. Patch-clamp recordings were performed on cells visually identified using infrared differential interference contrast video microscopy. The data were acquired with a patch-clamp amplifier (EPC-10; Heka Elektronik, Lambrecht, Germany) at a sampling frequency of 20 kHz and filtered at 2.9 kHz. Patch pipettes (3–7 MΩ) were pulled using a horizontal Flaming/Brown micropipette puller (P-97; Sutter Instrument Company, Novato, CA, USA). The pipette solution contained 127 mm K-gluconate, 8 mm KCl, 10 mm Hepes, 15 mm phosphocreatine, 4 mm Mg-ATP and 0.3 mm Na-GTP (pH 7.2, 295 mOsm). Action potentials were recorded in current clamp at –70 mV and a series of 15 hyperpolarising and depolarising current injections (–20 to 50 pA and –20 to 120 pA, 300 ms duration) were applied. Analyses of action potentials were done using Igor Pro (WaveMetrics, Lake Oswego, OR, USA). To record spontaneous excitatory postsynaptic currents and inhibitory postsynaptic currents, the cells were voltage clamped at –70 and 0 mV, respectively. Analyses of spontaneous excitatory postsynaptic currents/inhibitory postsynaptic currents were performed using the Mini Analysis Program (Synaptosoft, Fort Lee, NJ, USA). Series resistance was monitored using a 10 mV hyperpolarising pulse and was not allowed to exceed 30 MΩ.
RNA extraction
RNA was isolated from iPSC and NSC using total RNA purification plus kit (Norgen, Thorold, ON, Canada). To ensure that the samples were not contaminated with genomic DNA, an additional DNase I digestion step was performed with RNase-free DNase (Norgen) according to the manufacturer’s instructions. RNA quantity and quality were determined using an ND-1000 spectrophotometer and Agilent RNA 6000 Nano Kit (Santa Clara, CA, USA), respectively. RNA integrity number values above 8.0 were accepted for further analysis. The RNA samples were stored at –80 °C.
Reverse transcription and real-time PCR
Complementary DNA synthesis was performed with iScript Reverse Transcription Supermix for reverse transcription and quantitative PCR (RT-qPCR; Bio-Rad). Endogenous messenger RNA (mRNA) levels were measured by RT-qPCR analysis using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. Each sample was analysed in triplicate using the following oligonucleotide pairs: RPLP F: 5′-CGACAATGGCAGCATCTACAAC-3′ and R: 5′-CGGACACCCTCCAGGAAG-3′ GUSB F: 5′-AATCACTATGCGCATCAACAACA-3′ and R: 5′-TTGGGATACTTGGAGGTGTCA-3′ and NLRP2 Hsa Prime PCR assay (Bio-Rad). All the primer pairs yielded a single product as confirmed by dissociation curve analyses, and gave no product in the no-template control. The data analysis was performed in GenEx 6.1 (TATAA Biocenter, Gothenburg, Sweden), using a preset calculation, subtraction of gDNA background with Validprime. The data were then exported to Excel and normalised with the ΔΔCt method, results presented as fold change.
Transcriptome sequencing and data analysis
Isolation of mRNA, library preparation and transcriptome sequencing using Illumina HiSeq2000 were performed by BGI Tech Solutions (Hong Kong). The data files made in BGI were analysed in-house as follows. Before mapping of the genome, adaptors were cut and the raw reads were subjected to quality control, trimming and filtering. The Trim Galore (0.3.3) tool, a wrapper tool around Cutadapt31 and FastQC, was used for this purpose with default settings (phred score cut-off=20). Trimmed reads were mapped to the human (hg19) reference genome using STAR (2.4.0f).32 HTSeq (0.5.3p3; ref. 33) was used to quantify number of reads/gene using a list of annotated genes downloaded from Ensembl (version 75).34 The DESeq2 package35 from Bioconductor (Cambridge, MA, USA) was used to determine DEGs between samples derived from BD patients and healthy controls. Furthermore, analysis of DEGs was performed using DESeq2 between iPSC and NSC stages for BD and control cell lines, respectively.
Pathways and network analysis
Ingenuity pathway analysis software (Qiagen) was used to assess the disease-associated DEGs for the enrichment of gene networks, canonical pathways and biological processes relevant to the pathogenesis of BD. A list of the DEGs between BD and controls were uploaded to ingenuity pathway analysis, and analysis was performed at significance level of P<0.05 and log2-fold enrichment >1.8.
Results
iPSC generated from adipocyte cell lines from six BD I patients and four healthy controls
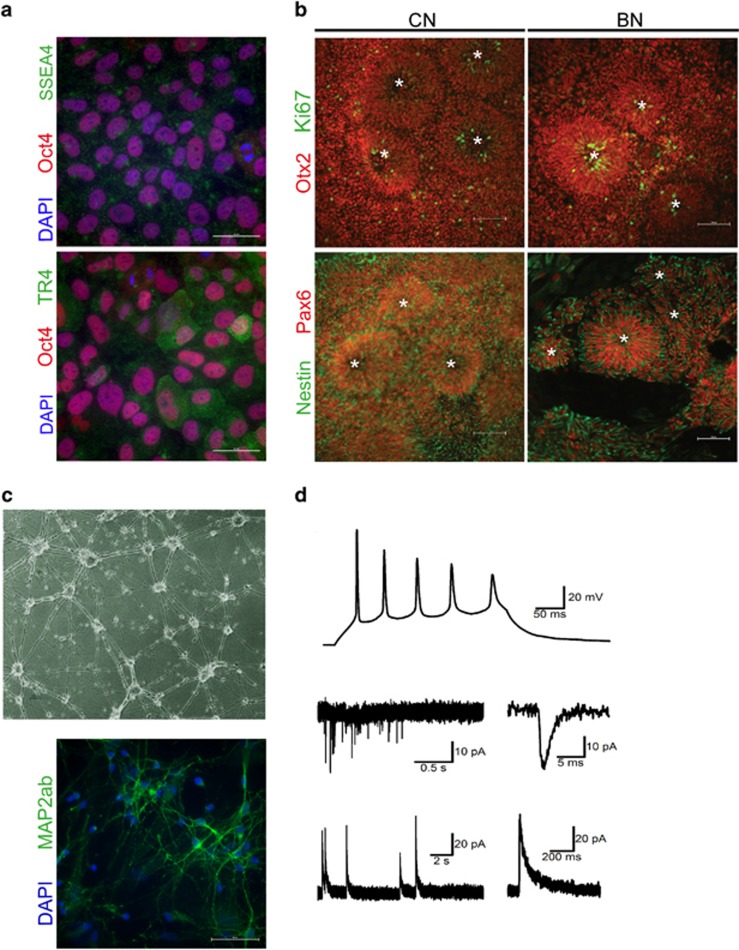
Nine of the established adipocyte cell lines mentioned above (the six BD I patients and four healthy controls) were selected for reprogramming into iPSC lines36 using a non-integral episomal reprogramming technology to avoid gene alteration.37, 38 Briefly, iPSC colonies were initially selected by morphology, passaged several times and expanded before characterisation. Pluripotency of all lines was confirmed by the presence of the pluripotency markers, that is, TRA-1-60, TRA-1-81, SSEA4, OCT4 and SOX2 (data not shown). As a further test of pluripotency, undirected differentiation of all nine iPSC lines into embryoid bodies and subsequent immunofluorescence analysis using β-tubulin, HNF4α and ASMA confirmed the presence of the cells from all three germ layers, that is, ectoderm, endoderm and mesoderm (data not shown). After expansion in our laboratory, presence of pluripotency markers Oct4, SSEA4 and sialy-lactotetra (TR4)39 was confirmed by immunofluorescence (Figure 1a). These results demonstrate the successful generation of pluripotent iPSC lines from BD patients and healthy controls.
Figure 1.
Directed differentiation of human iPSCs, into cortical stem and progenitor cells followed by cortical neurogenesis. (a) Representative images of iPSCs confirming presence of pluripotency markers Oct4 (red), SSEA4 (upper, green) and syalys-lactotetra (TR4) (lower, green). Nuclei were counterstained with Hoechst. Scale bars represent 50 μm. (b) Representative images of NSC forming rosettes derived from healthy control (CN) and bipolar patient (BN) specific iPSCs. NSCs exhibit a rosette-like growth pattern (indicated with a star in rosetta lumen) of Ki67 positive (upper, green) proliferating cells and stain positive for Otx2 (upper, red) and cortical stem cell marker Pax6 (lower, red) as well as Nestin (lower, green). Scale bars represent 100 μm. (c) NSCs can be differentiated into neurons expressing MAP2-AB (green). Nuclei were counterstained with Hoechst. Scale bars represent 50 μm. (d) Whole-cell current-clamp and voltage-clamp recordings were performed at different time points on NSCs differentiated into cortical-like neurons. (Upper) Example sweep showing evoked action potentials in response to a depolarising current pulse (300 ms). (Middle) Example sweep of voltage-clamp recordings showing spontaneous EPSCs recorded at –70 mV. (Lower) Example sweep of voltage-clamp recordings showing spontaneous IPSCs recorded at 0 mV. EPSC, excitatory postsynaptic current; iPSC, induced pluripotent stem cell; IPSC, inhibitory postsynaptic current; NSC, neural stem cell.
Characterisation of control and BD NSCs
To specifically direct differentiation of iPSC lines into cortical stem and progenitor cells (or neural lineage), all nine iPSC lines and the study-independent control line, ChiPSC4, were plated as single cells and subsequently neuralised by dual inhibition of SMAD signalling in combination with retinoid signalling.16 Eight to 10 days post neural induction, confirmation of directed iPSC differentiation was performed by expression of TERT as well as the transcription factors OCT4, NANOG, FOXG1 and PAX6.
Using this approach, decreased expression of pluripotency markers JARID1, NANOG and TERT was confirmed, while expression of PAX6 and FOXG1 was increased, indicating successful neural induction in all 10 iPSC lines (Table 1). In addition to the expression of unique cortical stem and progenitor cells transcription factors, all 10 lines were capable of forming neural rosette structures with a morphology characteristic of early neuro-epithelium and a feature of ES/iPSC-derived NSC.40, 41 Immunofluorescence analysis of these rosette structures confirmed their cortical identity by high levels of PAX6 and OTX2 as well as NESTIN (Figure 1b). In addition, there was no detectable difference in the labelling of either control or BD NSC by the cell cycle marker Ki67 (Figure 1b). These results demonstrate the successful differentiation of iPSCs into NSCs.
Table 1. Fold changes in gene expression for some marker genes during neural induction shown for cell lines derived from BD patients and healthy controls.
| Gene |
DEG CNSC/CiPSC |
DEG BDNSC/BDiPSC |
||
|---|---|---|---|---|
| Fold change (log2) | P-value | Fold change (log2) | P-value | |
| Jarid1 | –2.15 | 3.60 × 10–32 | –2.49 | 7.01 × 10–65 |
| Nanog | –6.91 | 8.86 × 10–9 | –6.18 | 1.99 × 10–23 |
| Tert | –2.07 | 1.72 × 10–18 | –1.80 | 1.77 × 10–21 |
| Foxg1 | 7.92 | 7.68 × 10–12 | 8.19 | 6.82 × 10–19 |
| Lhx2 | 9.64 | 4.00 × 10–22 | 8.64 | 1.83 × 10–34 |
| Otx1 | 3.73 | 1.30 × 10–6 | 2.97 | 7.76 × 10–7 |
| Otx2 | 1.84 | 5.54 × 10–6 | 2.00 | 4.60 × 10–10 |
| Pax3 | 5.35 | 0.001 | 7.16 | 1.57 × 10–8 |
| Pax6 | 6.40 | 6.80 × 10–33 | 5.95 | 2.57 × 10–43 |
| Pax7 | 6.39 | 4.38 × 10–5 | 3.80 | 0.002 |
| Pou3f2 | 5.26 | 3.10 × 10–13 | 4.81 | 9.08 × 10–19 |
| Sox1 | 6.40 | 3.88 × 10–9 | 8.92 | 3.50 × 10–16 |
| Sox2 | 1.02 | 0.0007 | 0.83 | 0.0006 |
Abbreviations: BD, bipolar disorder; DEG, differentially expressed gene; iPSC, induced pluripotent stem cell; NSC, neural stem cell.
NSCs have the capacity to make functional neurons
To further investigate the in vitro BD model system, we asked whether generated NSCs are capable of differentiating into functional neurons. Thus, we differentiated the NSCs into cortical-like neurons expressing MAP-AB (Figure 1c), using an established protocol.16 Whole-cell current-clamp and voltage-clamp recordings were then performed at different time points between 51 and 147 days after start of neural induction. In 49 of 58 investigated cells (84%), action potential firing could be induced using a depolarising current injection (300 ms) (Figure 1d). In 16 of the 58 cells (28%), spontaneous synaptic events could also be detected. All the spontaneous synaptic events were seen in cells showing action potentials (16 of 49 cells, 33%).
Both excitatory postsynaptic currents and inhibitory postsynaptic currents could be detected in five cells, only excitatory postsynaptic currents in seven cells and only iPSCs in four cells (Figure 1d). Taken together, these data indicate that the generated NSCs can be further differentiated into functional neurons of both excitatory and inhibitory type and further strengthen the capacity of the generated in vitro BD model system.
Comparison of the transcriptome of BD and control cells at iPSC and NSC stages
Having established the capacity of the system, we addressed developmental differences between BD and control cell lines at the transcriptome level. To this end, RNA was isolated from both iPSC (BI, CI) and NSC (BN, CN) stages for the six BD I and four control lines and subjected to RNA-Seq analysis. All the samples from individual stages, that is, iPSC and NSC, clustered together, revealing clear similarity within iPSC and NSC stages regardless of origin (Supplementary Figure). These results indicate that samples from each stage are very closely related and that each stage is likely to exhibit functionally distinct and unique transcriptomes.
A total of 3302 and 3187 transcripts from healthy controls and BD I patients, respectively, were differentially expressed at log2-fold enrichment 2 or higher, with P-values in the control and BD cell lines between different stages (Supplementary Information 1 and 2). With neural induction, expression of genes associated with pluripotency, such as JARID1, NANOG and TERT, was downregulated, while expression of genes associated with neuronal patterning was increased to a similar extent in BD as well as in control cell lines (Table 1).
These data suggest that with neural induction, cells express different sets of signalling molecules compared with iPSCs. In addition, no significant difference of expression of genes involved in neuronal patterning or NSC self-renewal was observed between BD I patients and healthy controls.
NLRP2 is the most significant DEG, at both the iPSC and NSC stages, in discriminating BD from healthy controls
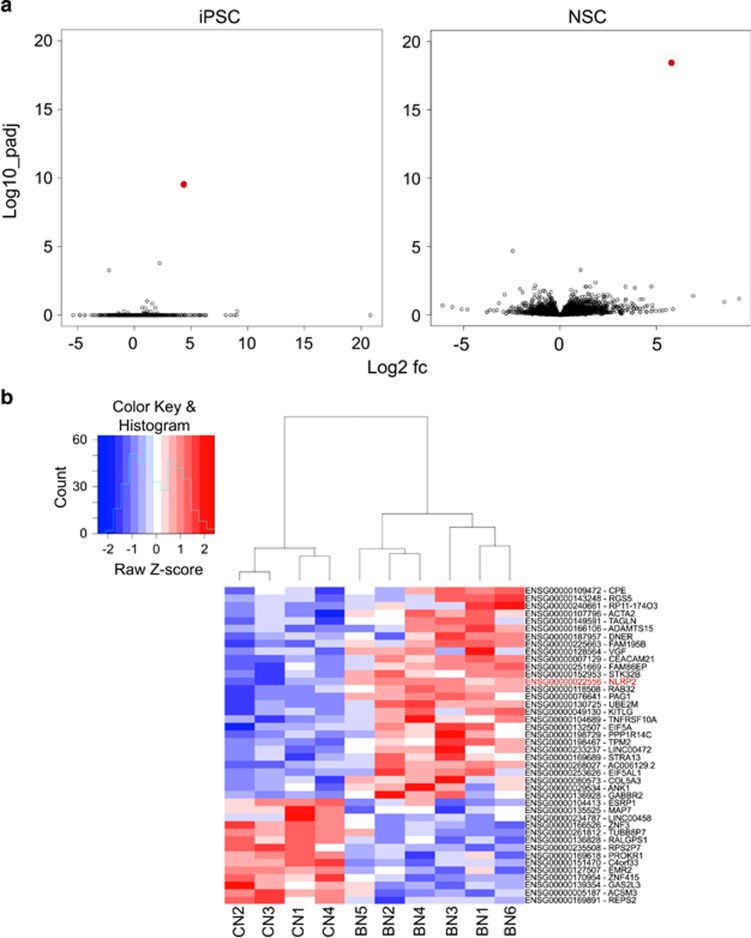
To identify BD-associated genes, we examined the average differences in gene expression in samples from BD I patients relative to the healthy control iPSC and NSC cell lines, respectively. Examination of volcano plots revealed that only three genes exhibited a significant difference in expression between the six BD I and four control iPSC lines (adjusted P<0.05; Figure 2a, iPSC). In contrast to the lack of multiple DEGs in the iPSC state, there were 42 genes with significant expression differences between BD and controls in the NSC stage (adjusted P<0.05; Figure 2a, NSC). The heatmap and cluster analysis of NSC stage DEGs show the diversity in the gene expression profiles (Figure 2b). To extract the genes with the strongest difference in gene expression between BD and controls, the cut-off value for log2-fold change was set to 1.8. In total, 15 unique genes showed >1.8 log2-fold change between BD and control NSC (adjusted P<0.05), respectively (Tables 2A and B). Interestingly, one of these genes was differentially expressed at both states, that is, NLRP2 (adjusted P=2.66 × 10−10).
Figure 2.
Differential gene expression signatures of BD-derived iPSC and NSC. (a) RNA-seq and DESeq analyses identified BD differentially expressed genes at both iPSC and NSC states. Volcano plots of log10 (adjusted P) versus the log2 (fold change between BD and controls) of all genes at respective stage. (b) Expression values of differentially expressed genes at NSC stage in 10 generated cell lines are shown in heat map format. The red, white and blue colours represent higher than average, close to average and lower than average expression of a particular gene, respectively, as measured by row standardised Z-scores. The rows are organised by hierarchical clustering using agglomerative clustering with complete linkage and Euclidian distance metric. BD, bipolar disorder; iPSC, induced pluripotent stem cell; NSC, neural stem cell.
Table 2. Differentially expressed genes between BD patients and healthy controls at (A) NSC and (B) iPSC stages, identified by RNA-seq.
| Symbol | Name | Fold change (log2) | Adjusted P-value |
|---|---|---|---|
| A | |||
| Upregulated DEGs at NSC stage | |||
| NLRP2 | NLR family, pyrin domain containing 2 | 5.816 | 3.07 × 10–19 |
| TPM2 | Tropomyosin-2 | 1.067 | 0.51 × 10–4 |
| TAGLN | Transgelin smooth muscle-alpha | 1.829 | 0.00439 |
| CEACAM21 | Carcinoembryonic antigen-related cell adhesion molecule 21 | 4.740 | 0.00826 |
| ADAMS15S | ADAM metallopeptidase with trombospondin type 1 motif, 15 | 3.397 | 0.00852 |
| ACTA2 | Actin alpha-2 sm | 1.568 | 0.00852 |
| VGF | VGF nerve growth factor inducible | 2.480 | 0.0312 |
| DNER | Delta/Noch-like EGF repeat containing | 2.255 | 0.0316 |
| ANK1 | Ankyrin 1 | 1.658 | 0.0330 |
| COL5A3 | Collagen, type V, alpha-3 | 2.008 | 0.0387 |
| RGS5 | Regulator of G-protein signalling 5 | 2.594 | 0.0427 |
| PAG1 | Phosphoprotein membrane anchor with glycosphingolipid microd. 1 | 2.282 | 0.0427 |
| GABBR2 | Gamma-aminobutyric acid (GABA) B receptor 2 | 1.865 | 0.0474 |
| PPP1R14C | Protein phosphatase 1, regulatory (inhibitor) subunit 14C | 2.268 | 0.0485 |
| Downregulated DEGs at NSC stage | |||
| RPS2P7 | Ribosomal protein S2 pseudogene 7 | –2.455 | 2.06 × 10–5 |
| ADGRE2 | Adhesion G protein-coupled receptor E2 | –2.075 | 0.0127 |
| MAP7 | Microtubulin-7 | –1.038 | 0.0197 |
| LINC00458 | Long intergenic non-protein coding RNA 458 | –1.941 | 0.0225 |
| TUBB8P7 | Tubulin, beta 8 class VIII pseudogene 7 | –2.872 | 0.0330 |
| B | |||
| Upregulated DEGs at iPSC stage | |||
| NLRP2 | NLR family, pyrin domain containing 2 | 4.370 | 2.66 × 10–10 |
| EEF1A1P16 | Eukaryotic translation elongation factor alpha 1 pseudogene 16 | 2.250 | 1.61 × 10–4 |
| Downregulated DEG at iPSC stage | |||
| RPS2P7 | Ribosomal protein S2 pseudogene 7 | –5.380 | 5.30 × 10–4 |
Abbreviations: BD, bipolar disorder; DEG, differentially expressed gene; iPSC, induced pluripotent stem cell; NSC, neural stem cell.
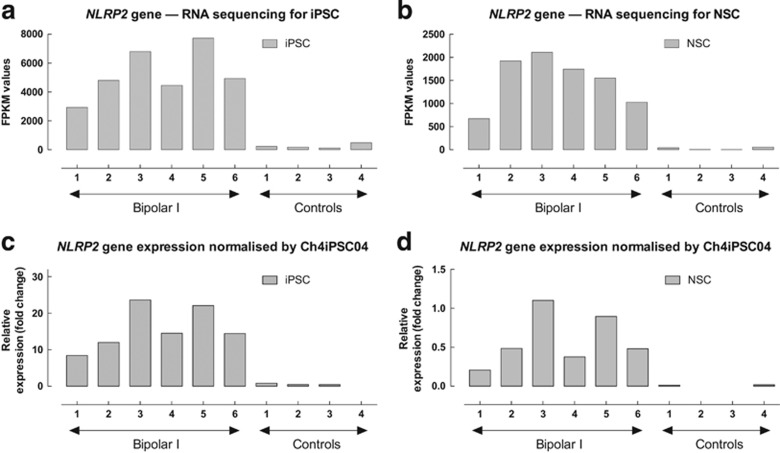
A closer look at the RNA sequencing results for NLRP2 gene expression revealed clear differences between BD I and control lines in both iPSC and NSC states (as illustrated in Figures 3a and b). Moreover, quantitative PCR analyses confirmed that expression of NLRP2 is upregulated in both BD iPSC and NSC lines (Figures 3c and d). These results demonstrate that it is possible to identify several DEGs in the NSC stage by using an in vitro BD model system, and among these genes NLRP2 was already affected at the iPSC state.
Figure 3.
Illustration of altered NLRP2 gene expression from six BD type I patients and four healthy controls. Relative changes in gene expression were calculated using the 2−ΔΔCt method with GUSB and RPLP as reference genes. (a) RNA sequencing for iPSC. (b) RNA sequencing for NSC. (c) qPCR validation of the NLRP2 gene in iPSC normalised by Ch4iPSC4. (d) qPCR validation of the NLRP2 gene in NSC normalised by Ch4iPSC4. BD, bipolar disorder; iPSC, induced pluripotent stem cell; NSC, neural stem cell; qPCR, quantitative PCR.
Several cytoskeleton-associated genes are over-represented as DEGs of BD-NSC lines
In the 42 DEGs in NSC stage, cytoskeleton-associated genes are over-represented at the high levels of significance. For we believe the first time, we here report TPM2 and TAGLN to be associated with BD. TPM2 encodes for tropomyosin beta chain type 2, and is known to stabilise actin filaments and regulate calcium-dependent muscle contraction (Table 2A). TAGLN encodes for transgelin smooth muscle-alpha, a gene that regulates actin cross-linking and gelling protein, but the functional role of this protein is unclear. ACTA2 (actin alpha-2 sm) has a role in cell motility, structure and integrity, being a major constituent of the contractile muscle. In addition, MAP7 (microtubulin-7), TUBB8P7 (tubulin beta 8 class VIII pseudogene 7), ANK1 (ankyrin 1) and COL5A3 (collagen type V alpha-3) were identified.
Canonical pathway analyses of DEGs between control and BD NSCs identify abnormalities in TREM1, dopamine and GABA receptor signalling
We next addressed whether common networks or pathways were over-represented in the list of genes associated with the top DEGs. Ingenuity pathway analysis (Qiagen)—an application enabling the discovery, visualisation and exploration of molecular interaction networks in gene expression data—was performed (cut-off values: adjusted P<0.05, log2-fold enrichment 1.8) revealing enrichment for relevant gene networks: the canonical pathway important at both iPSC and NSC stages was revealed to be TREM1 (triggering receptor expressed on myeloid cells 1; P=0.00376 and P=0.0442, respectively; Table 3). The top scoring canonical pathway correlated with inflammation markers, for example, intrinsic prothrombin activation pathway (P=0.0179) followed by GABA receptor (P=0.0418), and dopamine receptor (P=0.0486) signalling. Furthermore, neurological diseases (P=0.00160 to 0.0129) and psychological disorders (P=0.00323 to 0.00129) were the top scoring diseases.
Table 3. Top canonical pathways enriched in BD patients as recognised by ingenuity pathway analysis at iPSC and NSC stages, respectively.
| Name | P-value |
|---|---|
| Top canonical pathways at iPSC stage | |
| TREM1 signalling | 0.00376 |
| Top canonical pathways at NSC stage | |
| Intrinsic prothrombin activation pathway | 0.0179 |
| GABA receptor signalling | 0.0418 |
| TREM1 signalling | 0.0442 |
| Dopamine receptor signalling | 0.0486 |
Abbreviations: BD, bipolar disorder; iPSC, induced pluripotent stem cell; NSC, neural stem cell; TREM1, triggering receptor expressed on myeloid cells 1.
NLRP2 is not abnormally regulated in BD patient-derived adipocytes
We showed that also in the in vitro BD model, NLRP2 transcripts are expressed higher in the BD-NSC-expressing cortical neuronal markers, when compared with those of healthy controls. We asked whether NLRP2 transcript levels also expressed higher in the primary adipocyte cell cultures from five BD patients and 11 healthy control lines by quantitative PCR. It was found to be undetectable or very low levels of the transcript without any differences between the patients and controls (data not shown), indicating that the NLRP2 transcript is not expressed in adipocytes. These results are in agreement with the GTEx portal (http://www.gtexportal.org), showing an absence of NLRP2 gene expression from subcutaneous adipose tissue, indicating that the NLRP2 gene is not expressed in adipocytes (see GTEx portal data set: www.gtexportal.org). These results demonstrate that in our in vitro BD model NLRP2 is expressed in early uncommitted stem cells—equivalent to the cells of the inner cell mass of a blastocyst—and in cortically committed neural stem cells.
Discussion
We established a human BD model system to understand when and which BD-associated genes manifest, by comparing RNA-expression levels at iPSC and NSC stages of BD I patients and healthy controls. The capacity of our in vitro iPSC-based BD model system regarding morphology, neuronal induction, NSC marker expression, and neuronal function are presented. Comparative RNA sequencing was applied and DEGs were identified at the iPSC and NSC stages.
Our study provides further evidence that BD represents a complex polygenetic developmental disorder. As the here-presented results in BD-specific altered RNA-expression levels and signal pathways indicate a high complexity, we will separately discuss each of our identified abnormalities with respect to clinical relevance for the aetiology, pathophysiology and diagnosis of BD. On the other hand, BD might be considered both a neurodegenerative and a neurodevelopmental disorder. Support for the former hypothesis is offered from reports of abnormal dendritic spine morphology and diminished hippocampal volume (as visualised by brain imaging in adult BD patients as well as in post-mortem tissue obtained from BD I patients), indicating neurodegenerative conditions.42, 43 However, owing to its well-documented heritability and alteration of dendritic spine morphology and immature axon growth, also seen in the autistic spectrum disorder,44, 45 as well as results from our current study, we suggest that bipolar patients have already a strong susceptibility toward the disorder in the early development of the nervous system in utero, or during the first few years of life. These considerations point at disease with very early onset, with its clinical phenotype becoming manifest much later.
Early elevated expression of NLRP2 and abnormal signalling pathway in immune responses: Does susceptibility for BD-associated inflammation start before birth?
Recently, the importance of inflammasomes in BD as well as in other central nervous system disorders (Alzheimer, stroke, brain and spinal cord injuries) have become recognised, indicated by high presence of inflammasomes in post-mortem brain samples.46, 47
NLRP2 belongs to the family of nucleotide-binding domain, leucine-rich repeat-containing proteins (NLRs, also known as NALP2, NBS1, PAN1 and PYPAF2), characterised by an N-terminal pyrin domain with innate immune functions, mediating activation of inflammasomes.48, 49, 50, 51 The NLRP2 inflammasome is a multiprotein complex, composed by NLRP2 and the adaptor protein apoptosis-associated speck-like protein, and it activates pro-inflammatory caspase-1, resulting in activation of IL-β.51, 52
In this pathological context, activation of inflammasomes has been described in microglia, macrophages and astrocytes,47 reviewed by Walsh et al.53 Interestingly, post-mortem frontal cortex obtained from BD patients shows a high amount of NLRP3 proteins (NLRP3 being a close relative to NLRP2), in comparison to bio-samples from healthy individuals.46 The presented highly significant difference in the NLRP2 transcriptome expression at the iPSC and NSC stages in our BD I patients cells indicates that inflammation-related process already starts at the very early embryonic development. In addition, the absence of high-level expression of NLRP2 transcripts in BD patient adipocytes indicates that the expression of NLRP2 differs among cell types. According to microarray data sets of NLRP2 expression, the human adult cortex (two donors at age 31) and the fetal telencephalon (two donors at ages 15 p.c.w. (post-conceptional weeks) and 16 p.c.w.) both express NLRP2 (Ref. http://human.brain-map.org/microarray/search/show?exact_match=true&seach_term=NLRP2&search_type=gene&donors=14380,15496,10021,9861,12876,15697, for fetal: 14751,12840). By the GTEx portal data set, NLRP2 was not detected in subcutaneous adipocyte but expressed in frontal cortex median RPKM: 0.731, sample #108: cortex RPKM: 0.635, sample #114: EBV transformed lymphocytes RPKM:2.970, sample #118. Thus, we need to search whether the described abnormal presence of proteins of the NLRP family in adult BD patients46 might have originated from an iPS cell stage, and whether abnormal regulation of NLRP genes during ontogeny induces a susceptibility that may become clinically relevant later during the patient’s lifespan.
As our finding of an early fetal expression of NLRP is completely new and unexpected, we can assume some prospective function of NLRP during human ontogeny. NLRP2 has been shown to be expressed in oocytes and granulosa cells and identified as a maternal effect gene, and the depletion of NLRP2 in zygotes led to early embryonic arrest.54 Our findings demonstrate markedly enhanced NLRP2 transcript levels as early as at the iPSC stage of cells generated from BD I patients, and clearly distinguished a difference from healthy controls in NSC (Figure 2). However, this alteration did not affect either pluripotency of iPSC lines or neural induction. These results agree with previous findings indicating that overexpression of NLRP2 in zygotes appears to lead to normal development before the blastocyst stage.54 However, further studies are required to describe the link of upregulated NLRP proteins during early human fetal neural development and adult brain neurones and glial cells.
Our study also revealed an enrichment of relevant gene networks in the TREM1 (Triggering Receptor Expressed on Myeloid cells 19) signalling pathway and showed that top scoring canonical pathway correlated with inflammation markers, for example, intrinsic prothrombin activation pathway, at both iPSC and NSC stages.
An increasing body of evidence points to the fact that inflammatory processes have critical roles in BD (see below). It is intriguing that a major modulator of inflammatory signalling as shown in this study is expressed at elevated levels in both iPSCs and NSCs generated from BD patients. Therefore, our results indicate that an abnormal regulation of inflammatory genes might start before birth, and thereby the susceptibility for BD-associated inflammation would more likely be directly genetically mediated, rather than environmentally induced.
Do abnormal dopamine and GABA neurotransmitter systems in BD-NSC result in impaired neurotransmitter-regulated neural development or neuronal function?
The balance of dopamine, glutamate and GABA systems is an essential key for normal brain development and maintenance.55 Our pathway analysis shows enrichment for disease-associated genes to relate relevant networks and pathways. Although the top scoring canonical pathway correlated with inflammation markers, it was followed by GABA receptor and dopamine receptor signalling pathways. Persistent abnormalities in dopaminergic and GABAergic neurotransmitter systems in neuronal progeny might lead to aberrant functional brain oscillatory patterns within brain networks in patients with BD and major depressive disorder (MDD).56 Moreover, ontogeny of cells expressing dopamine and/or GABA receptors have not been clearly demonstrated in human NSC, but genes like neurogenin2 and Nurr1 are expressed in mouse NSC, which could start to differentiate into both pathways.57, 58
Abnormal expression of cytoskeleton-associated genes: a prospective link between abnormal neural and cardiac development in BD and cardiovascular diseases?
We have also identified several cytoskeleton-associated genes as DEGs in the NSC stage. There is evidence that psychiatric disorders are connected with cytoskeletal genes by means of the transport of biological material, such as synaptic proteins along the axon.59, 60 The tubulin expression levels are often altered and dysfunctional in disease-specific regions such as the hippocampus and prefrontal cortex of schizophrenia and bipolar disorder patients.61 In our paper, TPM2 is the strongest DEG followed by TAGLN among the cytoskeletal genes, and both can regulate ACTA2. ANK1 (protein encoded by the gene ankyrin1) links the integral membrane proteins to the spectrin–actin cytoskeleton and ion channels, being important for cell motility and synaptic activation. Hereditary human diseases due to defects or gene variation of ankyrins (1/2/3) are known to be linked with diseases such as cardiac arrhythmia and bipolar disorder.30, 62 Bipolar patients with psychotic episode are comorbid with cardiovascular diseases.63 Thus, the consequences of overexpressed TPM2 and TAGLN, regulating actin filaments during the neural and cardiac development might give novel insights into the neural degeneration observed in bipolar disorder in post-mortem tissues,42 and might explain why BD is comorbid with cardiovascular diseases, and vice versa.
Inflammation in mood disorders and future directions
The role of immunological factors in the pathophysiology and diagnosis of BD I has been discussed in relation to schizophrenia, and emphasis is put on an imbalance of inflammatory cytokines.9 Anti-inflammatory drugs exert proven clinical effects as shown in several randomised controlled trials in MDD.64, 65, 66 Treatments targeted against NLRP inflammasomes have been suggested to become useful against neuro-inflammation and in treatment of MDD.47, 67 Moreover, some forms of depression can be considered to represent microglial aberrations that should be treated with microglial stimulators or inhibitors.68 Also, if the inflammasome activation actually precedes psychopathology and is not essentially deactivated by psychopharmacological treatment, then inflammasome measures such as NLRP2 may provide useful diagnostic biomarkers.
In conclusion, our discovery of an inflammasome activation in immature stem cells and neural cells at a very early stage of ontogeny indicates this feature of BD to have been set before environmental influences have had a reasonable chance to tune in. To this end, it might also be of importance to investigate the effects of different targeted treatments on the NLRP2 inflammasome. Studies are required to elucidate the role of NLRP2 in BD and its tentative use as a biomarker for this, and possibly other mood disorders.
Limitation of the study
Sample size is small, although difference of NLRP2 expression is highly significant between BD I and control groups. BD I is a complex and polygenic disorder, which may make these findings difficult to generalise to all the BD I population. This is an in vitro modelling human system, not an in vivo study.
Acknowledgments
The authors acknowledge grants from the Swedish Research Council, the Västra Götaland Region County Council (ALF), the Swedish Brain Foundation, the Åhlén Foundation, Uppsala university, the Assar Gabrielsson Foundation, the Fredrik and Ingrid Thuring Foundation, the Söderström-König Foundation, the Teodor Nerander Foundation (Fysiografen), the Wilhelm and Martina Lundgren Foundation, the Adlerbertska Forskningsfonden, the Psychiatry Research Foundation in Gothenburg, Alzheimerfonden and Kurt and Ingrid Dahrén’s Foundation.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
The authors declare no conflict of interest.
Supplementary Material
References
- Rolstad S, Jakobsson J, Sellgren C, Isgren A, Ekman CJ, Bjerke M et al. CSF neuroinflammatory biomarkers in bipolar disorder are associated with cognitive impairment. Eur Neuropsychopharmacol 2015; 25: 1091–1098. [DOI] [PubMed] [Google Scholar]
- Swartz HA, Fagiolini A. Cardiovascular disease and bipolar disorder: risk and clinical implications. J Clin Psychiatry 2012; 73: 1563–1565. [DOI] [PubMed] [Google Scholar]
- Fajutrao L, Locklear J, Priaulx J, Heyes A. A systematic review of the evidence of the burden of bipolar disorder in Europe. Clin Pract Epidemiol Ment Health 2009; 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachen EA, Chesney MA, Criswell LA. Prevalence of mood and anxiety disorders in women with systemic lupus erythematosus. Arthritis Rheum 2009; 61: 822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards LJ, Constantinescu CS. A prospective study of conditions associated with multiple sclerosis in a cohort of 658 consecutive outpatients attending a multiple sclerosis clinic. Mult Scler 2004; 10: 575–581. [DOI] [PubMed] [Google Scholar]
- Galeazzi GM, Ferrari S, Giaroli G, Mackinnon A, Merelli E, Motti L et al. Psychiatric disorders and depression in multiple sclerosis outpatients: impact of disability and interferon beta therapy. Neurol Sci 2005; 26: 255–262. [DOI] [PubMed] [Google Scholar]
- Kupka RW, Nolen WA, Post RM, McElroy SL, Altshuler LL, Denicoff KD et al. High rate of autoimmune thyroiditis in bipolar disorder: lack of association with lithium exposure. Biol Psychiatry 2002; 51: 305–311. [DOI] [PubMed] [Google Scholar]
- Bai YM, Su TP, Tsai SJ, Wen-Fei C, Li CT, Pei-Chi T et al. Comparison of inflammatory cytokine levels among type I/type II and manic/hypomanic/euthymic/depressive states of bipolar disorder. J Affect Disord 2014; 166: 187–192. [DOI] [PubMed] [Google Scholar]
- Altamura AC, Buoli M, Pozzoli S. Role of immunological factors in the pathophysiology and diagnosis of bipolar disorder: comparison with schizophrenia. Psychiatry Clin Neurosci 2014; 68: 21–36. [DOI] [PubMed] [Google Scholar]
- Craddock N, Sklar P. Genetics of bipolar disorder. Lancet 2013; 381: 1654–1662. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Badner JA, Cheng L, Zhang C, Zhang D, Cox NJ et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry 2013; 18: 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008; 451: 141–146. [DOI] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 2012; 7: 1836–1846. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 2010; 143: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Winner B, Gage FH. Pluripotent stem cells in neurodegenerative and neurodevelopmental diseases. Hum Mol Genet 2010; 19: R71–R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swistowski A, Peng J, Liu Q, Mali P, Rao MS, Cheng L et al. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells 2010; 28: 1893–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc Natl Acad Sci USA 2008; 105: 5856–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FFJr, Mattis VB, Lorson CL, Thomson JA et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 2009; 457: 277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 2009; 461: 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T et al. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum Mol Genet 2011; 20: 4530–4539. [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011; 473: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One 2011; 6: e23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa E, Sandler V, Shah A, Carroll R, Chang C, Rockowitz S et al. Development of patient-specific neurons in schizophrenia using induced pluripotent stem cells. J Neurogenet 2011; 25: 88–103. [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Tran N, Gage FH. Modeling psychiatric disorders at the cellular and network levels. Mol Psychiatry 2012; 17: 1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson E, Larsson H, Lichtenstein P. Common psychiatric disorders share the same genetic origin: a multivariate sibling study of the Swedish population. Mol Psychiatry 2016; 21: 717–721. [DOI] [PubMed] [Google Scholar]
- Hayashi A, Le Gal K, Södersten K, Vizlin-Hodzic D, Ågren H, Funa K. Calcium-dependent intracellular signal pathways in primary cultured adipocytes and ANK3 gene variation in patients with bipolar disorder and healthy controls. Mol Psychiatry 2015; 20: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011; 17: 10–12. [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S et al. Ensembl 2014. Nucleic Acids Res 2014; 42(Database issue): D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Kano K, Kondo D, Fukuda N, Iribe Y, Tanaka N et al. Mature adipocyte-derived dedifferentiated fat cells exhibit multilineage potential. J Cell Physiol 2008; 215: 210–222. [DOI] [PubMed] [Google Scholar]
- Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S et al. A more efficient method to generate integration-free human iPS cells. Nat Methods 2011; 8: 409–412. [DOI] [PubMed] [Google Scholar]
- Okita K, Yamakawa T, Matsumura Y, Sato Y, Amano N, Watanabe A et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 2013; 31: 458–466. [DOI] [PubMed] [Google Scholar]
- Saljo K, Barone A, Vizlin-Hodzic D, Johansson BR, Breimer M, Funa K et al. Comparison of the glycosphingolipids of human induced pluripotent stem cells and human embryonic stem cells. Glycobiology 2016; pii: cww125. [DOI] [PubMed]
- Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA 2004; 101: 12543–12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 2001; 19: 1129–1133. [DOI] [PubMed] [Google Scholar]
- Rajkowska G. Cell pathology in bipolar disorder. Bipolar Disord 2002; 4: 105–116. [DOI] [PubMed] [Google Scholar]
- Nugent AC, Milham MP, Bain EE, Mah L, Cannon DM, Marrett S et al. Cortical abnormalities in bipolar disorder investigated with MRI and voxel-based morphometry. Neuroimage 2006; 30: 485–497. [DOI] [PubMed] [Google Scholar]
- Kwan V, Meka DP, White SH, Hung CL, Holzapfel NT, Walker S et al. DIXDC1 phosphorylation and control of dendritic morphology are impaired by rare genetic variants. Cell Rep 2016; 17: 1892–1904. [DOI] [PubMed] [Google Scholar]
- Spence EF, Kanak DJ, Carlson BR, Soderling SH. The Arp2/3 complex is essential for distinct stages of spine synapse maturation, including synapse unsilencing. J Neurosci 2016; 36: 9696–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Andreazza AC, Elmi N, Chen W, Young LT. Nod-like receptor pyrin containing 3 (NLRP3) in the post-mortem frontal cortex from patients with bipolar disorder: a potential mediator between mitochondria and immune-activation. J Psychiatr Res 2016; 72: 43–50. [DOI] [PubMed] [Google Scholar]
- Minkiewicz J, de Rivero Vaccari JP, Keane RW. Human astrocytes express a novel NLRP2 inflammasome. Glia 2013; 61: 1113–1121. [DOI] [PubMed] [Google Scholar]
- Dorfleutner A, Chu L, Stehlik C. Inhibiting the inflammasome: one domain at a time. Immunol Rev 2015; 265: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol 2010; 10: 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140: 805–820. [DOI] [PubMed] [Google Scholar]
- Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med 2015; 21: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol 2012; 28: 137–161. [DOI] [PubMed] [Google Scholar]
- Walsh JG, Muruve DA, Power C. Inflammasomes in the CNS. Nat Rev Neurosci 2014; 15: 84–97. [DOI] [PubMed] [Google Scholar]
- Peng H, Chang B, Lu C, Su J, Wu Y, Lv P et al. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS One 2012; 7: e30344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin D, Aarum J, Schroeder JE, Jovanovic JN, Chuang TT. D1-like dopamine receptors regulate GABAA receptor function to modulate hippocampal neural progenitor cell proliferation. J Neurochem 2008; 107: 964–975. [DOI] [PubMed] [Google Scholar]
- Lener MS, Niciu MJ, Ballard ED, Park M, Park LT, Nugent AC et al. Glutamate and gamma-aminobutyric acid systems in the pathophysiology of major depression and antidepressant response to ketamine. Biol Psychiatry 2016; pii: S0006-3223(16)32377-0; doi: 10.1016/j.biopsych.2016.05.005 (e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Ruediger T, Bolz J. Neurotransmitters and the development of neuronal circuits. Adv Exp Med Biol 2007; 621: 104–115. [DOI] [PubMed] [Google Scholar]
- Vergano-Vera E, Diaz-Guerra E, Rodriguez-Traver E, Mendez-Gomez HR, Solis O, Pignatelli J et al. Nurr1 blocks the mitogenic effect of FGF-2 and EGF, inducing olfactory bulb neural stem cells to adopt dopaminergic and dopaminergic-GABAergic neuronal phenotypes. Dev Neurobiol 2015; 75: 823–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Golbaz K, Honer WG, Beasley CL. Deficits in axon-associated proteins in prefrontal white matter in bipolar disorder but not schizophrenia. Bipolar Disord 2016; 18: 342–351. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu SA et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry 2015; 20: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchisella F, Coffey ET, Hollos P. Microtubule and microtubule associated protein anomalies in psychiatric disease. Cytoskeleton (Hoboken) 2016; 73: 596–611. [DOI] [PubMed] [Google Scholar]
- Bennett V, Healy J. Organizing the fluid membrane bilayer: diseases linked to spectrin and ankyrin. Trends Mol Med 2008; 14: 28–36. [DOI] [PubMed] [Google Scholar]
- Poo M, Lam JW, Orida N, Chao AW. Electrophoresis and diffusion in the plane of the cell membrane. Biophys J 1979; 26: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N, Schwarz MJ, Dehning S, Douhe A, Cerovecki A, Goldstein-Muller B et al. The cyclooxygenase-2 inhibitor celecoxib has therapeutic effects in major depression: results of a double-blind, randomized, placebo controlled, add-on pilot study to reboxetine. Mol Psychiatry 2006; 11: 680–684. [DOI] [PubMed] [Google Scholar]
- Raison CL, Rutherford RE, Woolwine BJ, Shuo C, Schettler P, Drake DF et al. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA Psychiatry 2013; 70: 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler O, Benros ME, Nordentoft M, Farkouh ME, Iyengar RL, Mors O et al. Effect of anti-inflammatory treatment on depression, depressive symptoms, and adverse effects: a systematic review and meta-analysis of randomized clinical trials. JAMA Psychiatry 2014; 71: 1381–1391. [DOI] [PubMed] [Google Scholar]
- Singhal G, Jaehne EJ, Corrigan F, Toben C, Baune BT. Inflammasomes in neuroinflammation and changes in brain function: a focused review. Front Neurosci 2014; 8: 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yirmiya R, Rimmerman N, Reshef R. Depression as a microglial disease. Trends Neurosci 2015; 38: 637–658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.