Summary
Long noncoding RNAs (lncRNAs) have been shown to play critical roles in the biology of various cancers. However, their expression patterns and biological functions in human colorectal cancer (CRC) remain largely unknown. The aim of this study was to explore lncRNA profiles in CRC and investigate key lncRNAs involved in CRC tumorigenesis and progression. The microarray data of six CRC and matched non-cancerous tissues revealed distinct lncRNA profiles, including 899 upregulated and 1,646 downregulated lncRNAs (p < 0.05, fold change > 2.0). Furthermore, we found that the lncRNA BC032913 was generally underexpressed in 115 CRC samples compared with normal tissues. Reduced BC032913 levels were significantly associated with an advanced tumor, lymph nodes, distant metastasis (TNM) stage and a higher risk of lymph node and distant metastases. BC032913 downregulation indicated poor overall survival in CRC patients. Moreover, BC032913 enhanced the mRNA and protein expression of TIMP3 and inhibited Wnt/β-catenin pathway activity, thus suppressing CRC metastasis in vitro and in vivo. Collectively, the obtained data show that BC032913 plays an inhibitory role in CRC aggression by upregulating TIMP3, followed by inactivation of the Wnt/β-catenin pathway. Our findings indicate that the novel lncRNA BC032913 could serve as a novel prognostic marker and effective therapeutic target for CRC.
Keywords: colorectal cancer metastasis, lncRNA BC032913, TIMP3
Introduction
Colorectal cancer (CRC) is one of the most common types of gastrointestinal tumors worldwide. It is estimated that 134,490 new cases of CRC and 49,190 CRC-related deaths occurred in 2016.1 In China, CRC ranks as the sixth most common malignancy and as the fifth leading cause of cancer-related deaths. This mortality is mainly caused by distant metastases.2 Therefore, understanding the mechanisms responsible for the development of metastasis and elucidating molecular biomarkers of CRC may improve CRC patient survival.
In mammals, up to 90% of the genomic DNA sequence can be transcribed into RNA. However, with the exception of the approximately 2% of genes that encode proteins (mRNA), the majority of the human genome consists of noncoding genes, suggesting that noncoding RNAs (ncRNAs) may play significant roles in diverse biological processes.3, 4 Among various types of ncRNAs, long noncoding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides and were previously considered to represent random transcriptional noise.5 In recent years, increasing evidence has demonstrated that lncRNAs participate in the pathogenesis of multiple diseases through various mechanisms of downstream gene regulation, such as epigenetic, post-transcriptional, and transcriptional regulation.6, 7, 8, 9 Interestingly, accumulating evidence has linked lncRNA dysregulation to multiple human malignancies.10, 11 For example, the well-known lncRNA HOTAIR (HOX antisense intergenic RNA) is overexpressed in breast, colon, liver, and pancreatic cancer and is involved in chromatin remodeling by recruiting polycomb repressive complex 2 (PRC2) to specific target genes.12, 13, 14, 15, 16 Additionally, the lncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) was initially found to be a negative prognostic factor for the survival of early-stage non-small-cell lung cancer (NSCLC) patients17 and has been implicated in alternative splicing regulation.18 Subsequently, it was reported that MALAT1 levels are elevated in various types of cancer, such as liver, breast, pancreas, colon, prostate, and bladder cancer.19, 20, 21, 22 Hence, lncRNA expression profiles could facilitate the diagnosis and prognosis of various human cancers, and functional lncRNAs may serve as potential therapeutic targets. However, the current understanding of lncRNA expression in CRC is limited, and only a few novel lncRNAs have been functionally characterized in CRC.23, 24, 25
In the present study, we examined the lncRNA expression profiles of six pairs of CRC samples compared with adjacent normal specimens: 899 and 1,646 lncRNAs were found to be up- and downregulated, respectively. We further discovered the novel lncRNA BC032913, which showed lower expression in CRC samples than in normal tissues. Reduced levels of BC032913 were significantly associated with an advanced tumor, lymph nodes, distant metastasis (TNM) stage and a higher risk of lymph node and distant metastases. BC032913 downregulation also indicated poor overall survival (OS), which suggested that BC032913 could serve as a prognostic factor for predicting CRC patient survival.
Previous studies have indicated that alternative factors are involved in CRC pathogenesis, including genetic alternations in oncogenes or tumor suppressor genes, as well as changes in the transforming growth factor β (TGF-β) and Wnt signaling pathways.26 As a putative tumor suppressor gene, the tissue inhibitor of metalloproteinases (TIMP) proteins are endogenous inhibitors of matrix metalloproteinases (MMPs), which contribute to tumor progression by degrading the extracellular matrix.27 TIMP-3, a member of the TIMP family, has been shown to inhibit tumor growth, angiogenesis, invasion, and metastasis.28 Many studies have found that TIMP3 expression levels are reduced in a number of cancer types.29, 30 In CRC patients, strong epithelial and stromal cytoplasmic staining of TIMP-3 indicates longer survival.31 It has been reported that TIMP3 suppresses colorectal tumor growth and metastasis in vitro and in vivo.32 Although TIMP3 plays a significant role in cancers, limited data are available regarding the mechanism of TIMP3 dysregulation. It has been documented that aberrant methylation of TIMP-3 occurs in primary cancers of the colon, lung, breast, kidney, and brain. Such tumor-specific methylation is associated with a lack of the TIMP-3 protein.33 Qin et al.34 have reported that miR-191 suppresses TIMP3 expression via direct targeting and, thereby, promotes the invasiveness of colorectal carcinoma cells. In the present study, we show, for the first time, that the novel lncRNA BC032913 directly enhances the transcriptional and translational levels of TIMP3.
The Wnt/β-catenin canonical signaling pathway, which was previously regarded as a key pathway involved in determining cell fate,35 has been found to be deregulated in various cancers.36 The Wnt signaling pathway has been reported be altered in 93% of colorectal tumors.37 In the absence of Wnt signaling, cytosolic β-catenin is rapidly phosphorylated and degraded by a multiprotein complex consisting of the core proteins glycogen synthase kinase 3β (GSK-3β), adenomatous polyposis coli (APC), and axin. In the presence of Wnt, GSK-3β is inactivated following binding to its cell surface receptor. Hence, non-phosphorylated β-catenin is released from the complex and translocates to the nucleus, which allows β-catenin to interact with the transcription factors TCF/LEF to transactivate target genes such as c-myc, cyclin D1, and CD44.38, 39, 40
Here we demonstrate for the first time that lncRNA BC032913 suppresses CRC metastasis through upregulating TIMP3 expression, followed by inhibition of the nuclear translocation of β-catenin, which inactivates the Wnt/β-catenin pathway.
Results
Overview of lncRNA Expression Profiles in CRC
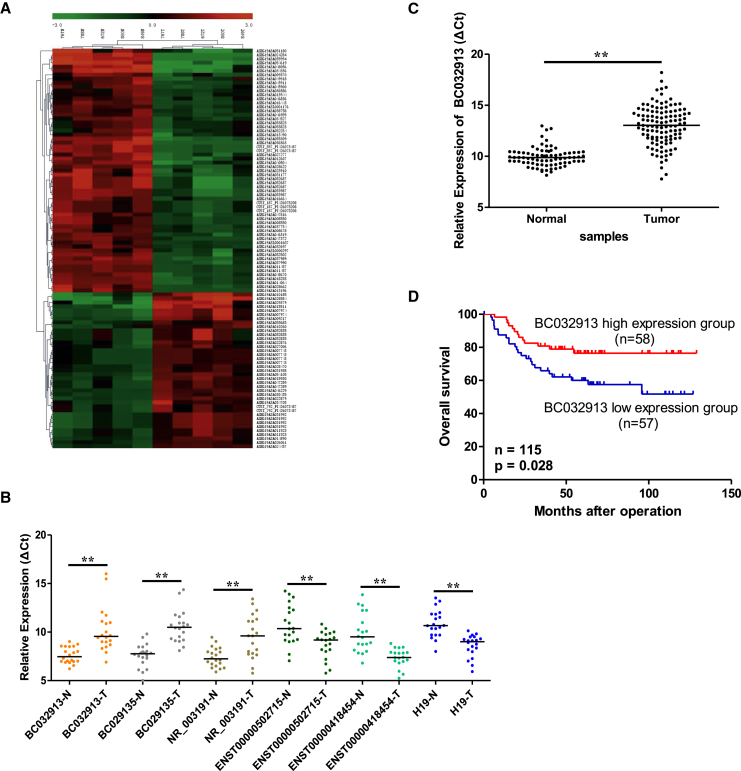
To identify lncRNAs that are specifically dysregulated in CRC, the lncRNA expression profiles of six CRC tissues and matched normal tissues were detected via microarray analysis. The basic information of the six patients is shown in Table S1. In each patient, the CRC tissues were confirmed to consist of more than 70% tumor cells, whereas the adjacent normal tissues were verified to contain no tumor cells through H&E staining (data not shown). In total, 2,545 lncRNAs were significantly differentially expressed (fold change > 2, p < 0.05) between the CRC and normal tissues. The 100 most differentially expressed lncRNAs were used for clustering analysis. The result of hierarchical clustering (Figure 1A) showed that CRC tissues were distinctly separated from normal tissues according to these distinguishable lncRNAs expression patterns. Among these lncRNAs, we prioritized lncRNAs that showed strong expression in normal tissues (normalized intensity > 5) and markedly dysregulated in CRC samples (cutoff fold change = 4). The five most upregulated and downregulated lncRNAs, respectively, were chosen for further validation by qPCR in an additional 20 pairs of CRC tissues and adjacent normal tissues. The results indicated that ENST00000502715 and ENST00000418454 were increased in CRC, whereas the levels of BC029135, NR_003191, and BC032913 were decreased (Figure 1B). Five of the ten lncRNAs were confirmed, except for the remaining lncRNAs that cannot be detected via qPCR in the tissues or had an inconsistent expression pattern compared with that of the microarray data. As a classic lncRNA marker, H19 is already known to be overexpressed in multiple cancers, including CRC. It has been shown in our microarray data that H19 was upregulated in CRC tissues (p = 0.0345, fold change = 3.722; Table S2). Therefore, the H19 level was also detected to underscore the validity of the chosen lncRNA expression (p < 0.01; Figure 1B). Detailed information for the above dysregulated lncRNAs is shown in Table S2.
Figure 1.
lncRNA Expression Profiles and lncRNA BC032913 Levels in CRC Tissues
(A) Hierarchical clustering of the lncRNA microarray expression data. Columns represent human tumor samples (five CRC tissues and matched normal tissues); rows represent the 100 lncRNAs (37 overexpressed and 63 underexpressed in CRC) that best distinguished primary tumors from normal tissues. (B) Six dysregulated lncRNAs were validated through real-time qPCR in 20 pairs of CRC and non-tumor samples. (C) BC032913 expression levels were significantly lower in 115 CRC tumor tissues compared with 79 matched normal tissues via real-time qPCR. GAPDH was used as an internal control (p < 0.01). The horizontal line inside the dots indicates the median. (D) Kaplan-Meier survival curves of patients with CRC based on BC032913 expression status. Patients in the low-level group (n = 57) had a significantly poorer prognosis than those in the high-level group (n = 58, p < 0.05). The p values were calculated using log-rank tests.
lncRNA BC032913 Expression in CRC Tissues and Its Correlation with CRC Patient Clinical Characteristics and Survival
One of the five verified lncRNAs, BC032913, showing the highest fold change differential expression (p = 0.0229, fold change = 25.038; Table S2), was picked out for further analysis. We found that BC032913 was markedly underexpressed in 115 CRC tissues compared with 79 paired normal tissues (p < 0.01; Figure 1C). To further investigate the clinical significance of BC032913 levels, patients were separated into “high-level” (n = 58) and “low-level” (n = 57) BC032913 groups according to the median value of BC032913 expression. As shown in Table 1, low BC032913 levels were positively correlated with a more advanced TNM stage (p = 0.02) and a higher risk of lymph node (p = 0.032) and distant metastases (p = 0.032). Kaplan-Meier survival analysis showed that the low-level group was associated with poor OS in CRC patients (p = 0.028’ Figure 1D). These clinical data suggested that BC032913 could affect CRC metastasis and serve as a novel prognostic marker for CRC. Hence, we selected BC032913 for further study.
Table 1.
Correlation between lncRNA BC032913 Expression and Clinicopathological Characteristics in 115 CRC Patients
| Variable | lncRNA BC032913 Expression |
p Value | |
|---|---|---|---|
| Low (n = 57) | High (n = 58) | ||
| Age (years) | |||
| ≤59 | 27 | 27 | 0.930 |
| >59 | 30 | 31 | |
| Gender | |||
| Male | 34 | 36 | 0.790 |
| Female | 23 | 22 | |
| Tumor Differentiation | |||
| Well to moderate | 44 | 42 | 0.555 |
| Poor | 13 | 16 | |
| TNM Stage | |||
| I–II | 22 | 35 | 0.020 |
| III–IV | 35 | 23 | |
| Tumor Size (cm) | |||
| ≤5 | 37 | 41 | 0.507 |
| >5 | 20 | 17 | |
| Tumor Location | |||
| Rectum | 28 | 29 | 0.925 |
| Colon | 29 | 29 | |
| CEA (ng/mL) | |||
| ≤5 | 33 | 35 | 0.789 |
| >5 | 24 | 23 | |
| CA199 (U/mL) | |||
| ≤35 | 39 | 43 | 0.498 |
| >35 | 18 | 15 | |
| Tumor Invasion | |||
| pT1pT2 | 11 | 11 | 0.964 |
| pT3pT4 | 46 | 47 | |
| Lymph Node Metastasis | |||
| Absent | 25 | 37 | 0.032 |
| Present | 32 | 21 | |
| Distant Metastasis | |||
| Absent | 41 | 51 | 0.032 |
| Present | 16 | 7 | |
The median expression level of lncRNA BC032913 was used as the cutoff. Patients with CRC were divided into BC032913 “low” (expression lower than the median) and “high” groups (expression higher than the median). Pearson’s χ2 tests were used to analyze correlations between BC032913 levels and clinical features. The results were considered statistically significant at p < 0.05. CEA, carcinoembryonic antigen; CA199, carbohydrate antigen 19-9.
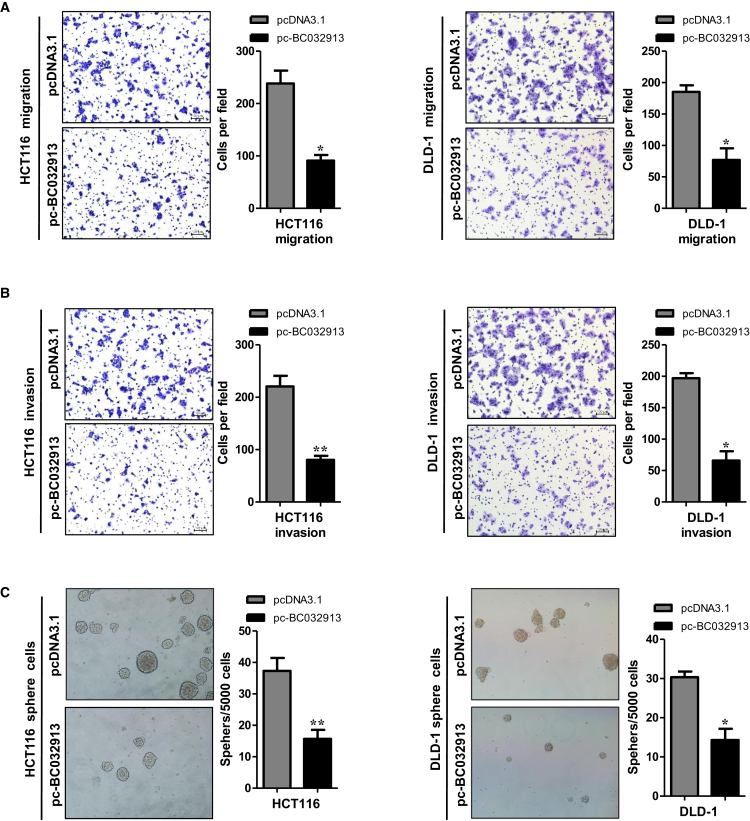
Overexpression of the lncRNA BC032913 Inhibits CRC Cell Migration, Invasion, and Sphere Formation In Vitro
Based on the above results, we hypothesized that BC032913 overexpression may affect the migration, invasion, and sphere-forming ability of CRC cells. Thus, we conducted migration, invasion, and sphere-forming assays, and the results demonstrated that ectopic BC032913 expression caused significant suppression of cell migration in both HCT116 and DLD-1 cells compared with cells transfected with an empty vector (p < 0.05; Figure 2A). Similarly, the invasion capacity of HCT116 and DLD-1 cells was markedly repressed after BC032913 overexpression (p < 0.05; Figure 2B). Overexpression of BC032913 resulted in the formation of fewer and smaller spheres (p < 0.05; Figure 2C). Collectively, these results demonstrate that BC032913 is a negative regulator of migration and invasion in CRC cells.
Figure 2.
Upregulation of lncRNA BC032913 Inhibits CRC Cell Migration, Invasion, and Sphere Formation In Vitro
(A) Compared with cells transfected with the empty vector, ectopic BC032913 expression caused significant suppression of cell migration in both HCT116 (left) and DLD-1 (right) cells. (B and C) BC032913 overexpression significantly inhibited the invasion ability (B) and sphere-forming capacity (C) of HCT116 (left) and DLD-1 (right) cells compared with the control cells. The numbers of cells that migrated, invaded, or formed spheres in the pc-BC032913 and pcDNA3.1 groups are shown at the right. The data obtained for the tumor spheres (>50 μm) were statistically analyzed. All data are presented as the mean ± SD obtained from three independent experiments.
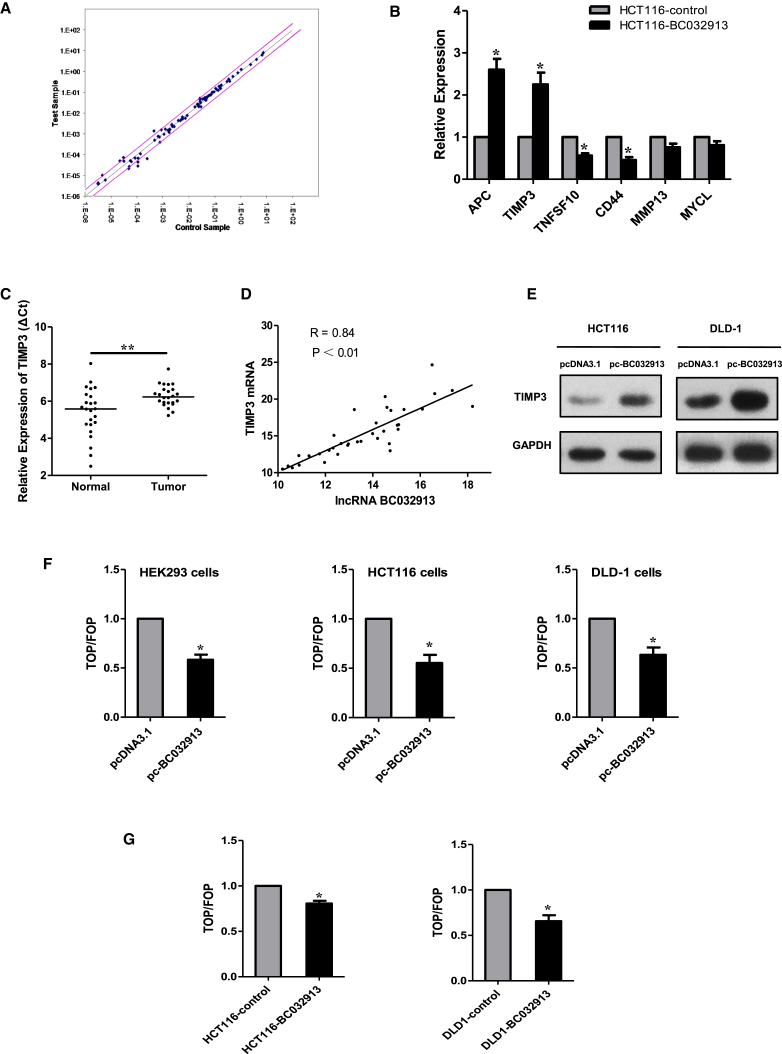
lncRNA BC032913 Upregulates TIMP3 and Inactivates Canonical Wnt/β-Catenin Signaling in CRC Cells
To explore the mechanism underlying the inhibition of an aggressive phenotype by BC032913, metastasis-related genes that were deregulated between HCT116 cells stably transfected with pLNCX2-BC032913 (HCT116-BC032913) or pLNCX2 (HCT116-control) were determined using a human tumor metastasis PCR array. Among the 84 metastasis-related genes, four were upregulated (MYCL, MMP13, TIMP3, and APC), and four (KISS1, TNFSF10, NR4A3, and CD44) were downregulated in HCT116-BC032913 cells (Figure 3A). The expression levels of APC, TIMP3, TNFSF10, and CD44 were further verified via real-time qPCR (p < 0.05; Figure 3B). Among the above genes, TIMP3 was significantly downregulated in 24 CRC specimens compared with matched normal tissues (p < 0.01; Figure 3C). Furthermore, we examined BC032913 expression and TIMP3 mRNA levels in 36 paired CRC specimens and observed a positive correlation (R = 0.84, p < 0.01; Figure 3D).
Figure 3.
lncRNA BC032913 Upregulates TIMP3 and Inactivates Canonical Wnt/β-Catenin Signaling in CRC Cells
(A) Metastasis-related genes that were deregulated between HCT116-BC032913 and HCT116-control were determined using a human tumor metastasis PCR array. The corresponding scatterplot showed that four genes were upregulated (MYCL, MMP13, TIMP3, and APC), and four other genes were downregulated (KISS1, TNFSF10, NR4A3, and CD44) in HCT116-BC032913 cells. The pink lines indicate the selected gene expression fold change (2.0) threshold. (B) APC, TIMP3, TNFSF10, and CD44 were further verified through real-time qPCR in HCT116-BC032913 and HCT116-control cells. (C) Compared with matched normal tissues, TIMP3 expression was significantly downregulated in 24 CRC specimens. The horizontal line inside the dots indicates the median. (D) There was a positive correlation (R = 0.84, p < 0.01) between BC032913 and TIMP3 mRNA levels in 36 paired CRC samples. Correlations were analyzed using Pearson’s correlation. (E) western blotting analysis showed that TIMP3 protein levels were elevated in both the HCT116-BC032913 and DLD-1-BC032913 cell lines compared with the corresponding control cell lines (HCT116-control/DLD1- control). GAPDH was used as a loading control. (F) In HEK293, HCT116, and DLD-1 cells, transiently expressed BC032913 markedly decreased TOPFlash activity. (G) In accordance with the above results, stably expressed BC032913 cells (HCT116-BC032913/DLD1-BC032913) significantly reduced TOPFlash activity compared with the control cells (HCT116-control/DLD1-control). Wnt pathway activities were measured by dividing the normalized TOPFlash value by the normalized FOPFlash value. The data are shown as the mean ± SD. The results are representative of three independent experiments (*p < 0.05, **p < 0.01).
Then we tested the ability of BC032913 to regulate the expression of endogenous TIMP3 proteins. Western blot analyses showed that the protein levels of TIMP3 were elevated in both the HCT116-BC032913 and DLD-1-BC032913 cell lines compared with the corresponding control cell lines (Figure 3E).
Simultaneously, we tested the effect of BC032913 on β-catenin/Wnt signaling using a standard TOPFlash (TOP)/FOPFlash (FOP) luciferase reporter assay in both HEK293/HCT116/DLD-1 cells transiently transfected with BC032913 (Figure 3F) and stably expressed BC032913 (HCT116-BC032913/DLD1-BC032913) cells (Figure 3G). As shown in Figures 3F and 3G, both transiently expressed and stably expressed BC032913 decreased TOP activity (p < 0.05).
The above data indicate that BC032913 could upregulate TIMP3 expression and inhibit the activity of the Wnt/β-catenin pathway.
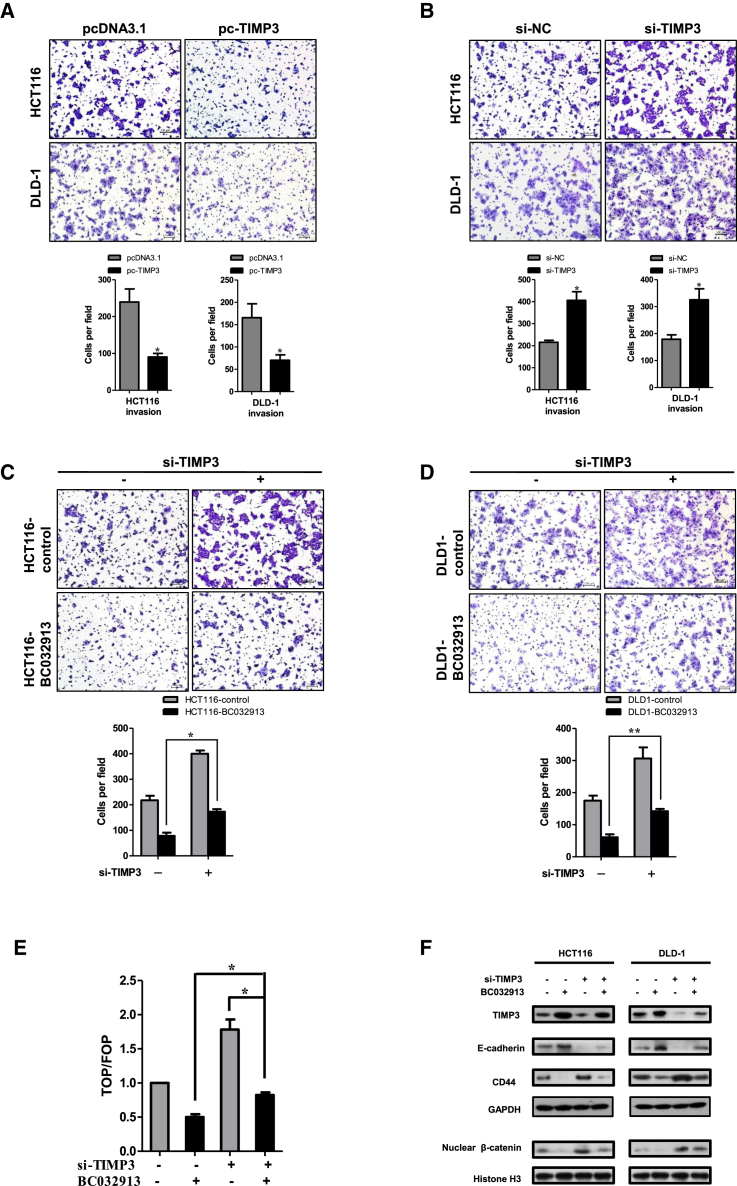
lncRNA BC032913 Upregulates TIMP3 to Inhibit Wnt/β-Catenin Pathway-Mediated CRC Cell Metastasis In Vitro
First, it was confirmed that overexpression of TIMP3 suppressed the invasion capability (p < 0.05; Figure 4A), whereas knockdown of TIMP3 promoted invasion (p < 0.05; Figure 4B) in HCT-116 and DLD-1 cells. Subsequently, we tested whether TIMP3 knockdown could reverse the effect of BC032913. Compared with the negative control (HCT116-control/DLD1-control), stably expressed BC032913 (HCT116-BC032913/DLD1-BC032913) markedly reduced the number of invasive cells (p < 0.05; Figures 4C and 4D). However, the BC032913-induced suppression of invasion was partially abrogated by transfection with TIMP3 small interfering RNAs (siRNAs) (si-TIMP3) (Figures 4C and 4D).
Figure 4.
lncRNA BC032913 Inactivated the Wnt/β-Catenin Pathway through Upregulating TIMP3 Expression in CRC Cell Metastasis
(A) Compared with the control cells, TIMP3 knockdown significantly enhanced the invasion ability of the cells. (B) Overexpression of TIMP3 resulted in fewer invasive cells than cells transfected with the empty vector. (C and D) TIMP3 siRNA (si-TIMP3) partially abrogated the suppression effect induced by BC032913 in both HCT116-BC032913 (C) and DLD-1-BC032913 (D) cells. The data obtained from the tumor spheres (>50 μm) were statistically analyzed. (E) As detected by luciferase reporter assays, ectopic expression of BC031913 caused significant suppression of TOP/FOP reporter activity. The addition of si-TIMP3 to HCT-116 cells transiently transfected with BC032913 augmented Wnt/β-catenin pathway activity to some extent. (F) western blot results showing the augment of TIMP3 and E-cadherin levels and reduction of nuclear β-catenin and CD44 expression by BC032913. Histone H3 was used as an endogenous control for the cell nuclear fraction. These changes in expression were reversed in HCT116 /DLD-1 cells that were cotransfected with pc-BC032913 and si-TIMP3. All experiments were performed in triplicate, and some results are shown as the mean ± SD (*p < 0.05, **p < 0.01).
To further verify the effect of the BC032913-TIMP3 axis on the Wnt/β-catenin signaling pathway, a TOP/FOP luciferase reporter assay was used to investigate the activation of Wnt/β-catenin signaling (p < 0.05; Figure 4E). The results showed a clear decrease in luciferase activity in the presence of BC032913 and partial rescue by TIMP3 knockdown.
Moreover, the western blot analysis shown in Figure 4F revealed that BC032913 increased TIMP3 and E-cadherin expression levels and decreased nuclear β-catenin and CD44 expression levels compared with the corresponding negative control. The above changes were reversed after the introduction of si-TIMP3.
These results provide evidence that TIMP3 participates in the regulation of the Wnt signaling pathway by BC032913 in CRC cell metastasis.
Ectopic Expression of lncRNA BC032913 Inhibits CRC Metastasis In Vivo
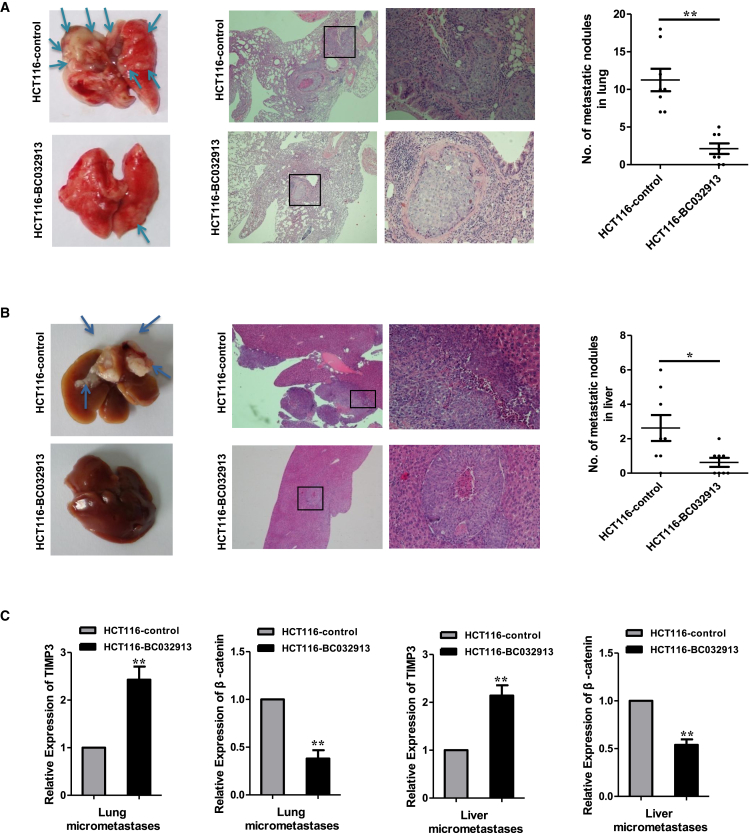
To investigate whether BC032913 was responsible for CRC metastasis in vivo, we subcutaneously injected HCT116 cells stably overexpressing BC032913 or empty vector into the tail vein of nude mice. Compared with the control, fewer and much smaller lung micrometastases were observed in the HCT116-BC032913 group (p < 0.01; Figure 5A). In the liver metastasis model, the above cells were injected into the spleens of nude mice. In contrast with the HCT116-control group, most mice in the HCT116-BC032913 group formed liver metastases (p < 0.05; Figure 5B). Furthermore, we assessed the expression of TIPM3 and β-catenin by real-time qPCR in lung and liver metastases originating from mice in the HCT116-control and HCT116-BC032913 groups. The mRNA levels of TIMP3 and β-catenin were increased and decreased, respectively, in mice in the HCT116-BC032913 group compared with mice in the HCT116-control group (p < 0.01; Figure 5C). Collectively, these findings are consistent with the in vitro results.
Figure 5.
Ectopic lncRNA BC032913 Expression Inhibits CRC Metastasis In Vivo
(A and B) The indicated treatments were tested in hepatic (A) and lung (B) metastasis models. Left: representative images of the metastatic nodules are indicated by arrows. Representative H&E staining results of the lung or liver metastatic nodules are shown in the center. The statistical results are shown on the right (n = 8, *p < 0.05, **p < 0.01). (C) As assessed by real-time qPCR, the mRNA levels of TIMP3 and β-catenin were increased and decreased, respectively, in lung and liver metastases originating from mice in the HCT116-BC032913 group compared with that in the HCT116-control group (p < 0.01). The data are shown as the mean ± SD.
In conclusion, lncRNA BC032913 inhibited CRC metastasis through inactivation of the Wnt/β-catenin pathway, which was followed by modulation of TIMP3, nuclear β-catenin, E-cadherin, and CD44 expression.
Discussion
Emerging research has shown that lncRNAs play important roles in cancer carcinogenesis and progression, and aberrant lncRNA expression has been identified in CRC.41, 42, 43 However, only a few studies have characterized novel lncRNAs in CRC. For example, RP11-462C24, an independent predictor of survival, is downregulated with the development of CRC.23 CCAT144 and CRNDE45 are increased in CRC samples, whereas lncRNA-42225 is decreased. CCAT2 is elevated in microsatellite-stable CRC and promotes tumor growth, metastasis, and chromosomal instability. It has further been shown that MYC, miR-17-5p, and miR-20a are upregulated by CCAT2 through TCF7L2-mediated transcriptional regulation.46 Nevertheless, the field of lncRNA research remains in its infancy, and few CRC-related lncRNAs have been well studied.
Here we identified significantly dysregulated lncRNAs in six pairs of CRC and adjacent normal samples using a microarray (fold change > 2, p < 0.05). Among these lncRNAs, 899 were upregulated, whereas 1646 were downregulated. Five selected lncRNAs were further validated in an additional 20 pairs of CRC tissues via real-time qPCR (Figure 1), and we found that the novel lncRNA BC032913 was markedly downregulated in 115 CRC tissues. Reduced BC032913 expression indicated poor OS in CRC patients, suggesting that BC032913 could be employed as a novel prognostic marker of CRC. Low BC032913 levels were positively correlated with an advanced TNM stage and a higher risk of lymph node and distant metastases, suggesting that the downregulation of BC032913 in CRC may facilitate the invasive/metastatic phenotype (Figure 1; Table 1). Therefore, transwell and sphere formation assays were performed to elucidate the function of this lncRNA in CRC cell lines in vitro. BC032913 overexpression caused significant suppression of cell metastasis in both HCT116 and DLD-1 cells compared with cells transfected with an empty vector (Figure 2). These findings may provide a novel path toward a better understanding of the molecular basis of CRC.
BC0329133 is transcribed from Homo sapiens DPP10 antisense RNA 1 (DPP10-AS1) located on chromosome 2, with a length of 744 bp, which is also known as LOC389023. Shulha et al.47 reported that LOC389023, a novel cis-bound antisense RNA, was highly enriched in nuclear RNA fractions extracted from samples of prenatal and normal adult human prefrontal cortex (PFC) but not cerebellar cortex. In humans, LOC389023 recruits polycomb 2 (PRC2) and other transcriptional repressors in cis, thereby inhibiting the expression of the sense transcript DPP10. This research suggests that BC032913 may also play a functional role in CRC through regulating DPP10 transcription. Hence, we performed real-time qPCR and found that DPP10 mRNA levels were decreased in CRC tissues (data not shown), which was in accordance with previously reported results.48 Nevertheless, DPP10 expression could not be detected through real-time qPCR in the normal colon epithelial cell line FHC or in human CRC cell lines (HCT116, DLD-1, SW480, SW620, HT29, and COLO205) (data not shown). Based on the obtained data, we inferred that the function of BC032913 in CRC may not depend on DPP10.
No research on the function and mechanism of BC032913 in cancer has been conducted to date. In the present study, we found that underexpression of BC032913 in CRC cells was important for the acquisition of an aggressive phenotype. To explore the underlying mechanism, metastasis-related genes that were deregulated in HCT116-BC032913 versus HCT116-control cells were identified using a human tumor metastasis PCR array. BC032913 was verified to elevate TIMP3 at both the mRNA and protein levels. Subsequently, we demonstrated that BC032913 inactivated the Wnt/β-catenin pathway by upregulating TIMP3 (Figures 3 and 4).
Relevant studies have shown that TIMP-mediated inhibition of Wnt-β-catenin signaling is independent of matrix metalloproteinase-mediated mechanisms. For instance, TIMP-1 increases the stability and nuclear localization of β-catenin in human mesenchymal stem cells (hMSCs), demonstrating that TIMP-1 acts an inhibitor of Wnt-β-catenin signaling.49 It has been reported that TIMP-2 inhibits the growth of lung cancer cells through enhancing the expression of the E-cadherin/β-catenin complex.50 TIMP-2 elevates E-cadherin expression and inhibits the Wnt/β-catenin pathway, thus suppressing the proliferation of melanoma cells with high activity of β-catenin.51 In mammary epithelial cells, extracellular TIMP3 functions to inactivate the Wnt-β-catenin pathway, followed by increasing phosphorylated β-catenin levels and decreasing nuclear β-catenin levels.52 In accord with the above studies, TIMP3, upregulated by BC032913, inhibits nuclear translocation of β-catenin, followed by inactivating the Wnt/β-catenin pathway in our research.
A downstream target gene of the Wnt pathway, CD44, was also identified to be downregulated by BC032913 using a human tumor metastasis PCR array (Figure 3B). Hence, we hypothesized that the BC032913/TIMP3 axis may inhibit CRC metastasis by inactivating Wnt/β-catenin signaling and subsequently blocking CD44 transcription. The data revealed that the BC032913/TIMP3 axis decreased the levels of nuclear β-catenin and CD44, which are important molecules in the Wnt/β-catenin pathway (Figure 4F).
CD44, a member of cell adhesion molecules, is involved in the progression and metastasis of multiple types of cancer.53 Recent accumulating evidence has demonstrated a direct link between epithelial-mesenchymal transition (EMT) and CD44, both of which contribute to cancer metastasis.54, 55, 56 To elucidate the relationship between CD44 and EMT markers in CRC, western blot assays were performed using HCT116 and DLD-1 cells. As shown in Figure 4F, BC032913 promoted E-cadherin expression levels and suppressed CD44 expression levels compared with the corresponding negative control. Moreover, TIMP3 knockdown reversed the effect of BC032913 on the expression of E-cadherin and CD44. These results indicated that the BC032913/TIMP3 axis inhibited CRC cell metastasis ability by upregulating an EMT marker (E-cadherin) and downregulating CD44 and nuclear β-catenin.
Additionally, CD44 was recently identified as a positive feedback regulator of Wnt/β-catenin signaling. However, distinguishing the role of CD44 as a target gene or as a Wnt regulator might be difficult.57 Therefore, further studies are necessary to determine whether the BC032913/TIMP3 axis decreases CD44 to inhibit Wnt/β-catenin activity or suppresses Wnt/β-catenin signaling and. thus. downregulates CD44.
In conclusion, our data suggest that the BC032913/TIMP3 axis inhibits CRC metastasis by inactivating Wnt/β-catenin signaling and suppressing EMT. However, further studies are needed to confirm the relationship between E-cadherin/CD44 and Wnt signaling in colorectal cells.
The introduction of si-TIMP3 partially abrogated the BC032913-induced suppression of CRC cell metastasis (Figure 4). These data suggest that, in addition to TIMP3, other unknown mechanisms might be involved in BC032913 regulation in CRC cells. For example, TNFSF10 was downregulated by BC032913, indicating that BC032913 may influence CRC carcinogenesis and progression by modulating TNFSF10. Because of the complexity of control factors in the gene expression network, further investigations are needed to address these issues.
Collectively, the findings of this study may contribute to a better understanding of the significance of deregulated lncRNAs in CRC. In addition, the lncRNA BC032913 may serve as a new molecular biomarker or a therapeutic target for CRC.
Materials and Methods
Human Tissue Samples and Cell Lines
Snap-frozen tissues were recruited from a total of 141 CRC patients who underwent radical resection at Sun Yat-sen University Cancer Centre between 2004 and 2012. Six samples were used for the lncRNA microarray analysis, whereas the remaining 135 were used for additional evaluations. The diagnosis of all patients was histologically confirmed, and the patients did not receive neoadjuvant therapy. All samples were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction. This study was approved by the Ethics Committee of Sun Yat-sen University Cancer Centre, and written informed consent was obtained from all patients. Detailed information on the six CRC patient samples included in the microarray is summarized in Table S1.
Human CRC cell lines (HCT116 and DLD-1) and a human embryonic kidney cell line (HEK293) were obtained from the American Type Culture Collection. All CRC cell lines and HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). All cells were maintained in a humidified 5% CO2 atmosphere at 37°C.
Microarray
The Human LncRNA Array v2.0 (8 × 60 K, Arraystar) was used to evaluate the differences in lncRNA profile between the six human CRC tissue specimens and the corresponding normal tissues. Information regarding the 33,045 lncRNAs and 30,215 coding transcripts that can be detected by the microarray was collected from the most authoritative databases, including Refseq, UCSC Known Genes, and Ensembl, as well as many related reports in the literature. RNA quantification and quality were determined using a NanoDrop ND-1000. RNA integrity and genome DNA (gDNA) contamination were examined using denaturing agarose gel electrophoresis. The microarray work and data collection were performed by KangChen Bio-tech.
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA from tissues and cultured cells was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA). Reverse transcription reactions were performed using M-MLV reverse transcriptase (Promega, Madison, WI, USA). For real-time qPCR, cDNA was amplified using Platinum SYBR Green real-time qPCR SuperMix UDG reagents (Invitrogen) and the MX3000P system (Stratagene). The coding and noncoding gene expression values were normalized to those of the housekeeping gene GAPDH (ΔCt = Ct [gene] – Ct [GAPDH]). The primer sequences are provided in Table S3.
Plasmid Construction and Transfection
The cDNAs of lncRNA BC032913 (NCBI: NR_036580.1) and TIMP3 were synthesized by Invitrogen and subcloned into the pcDNA3.1 plasmid. For the experiments involving TIMP3 siRNAs, a pool of three siRNAs with the following sequences was used (sc-44331, Santa Cruz Biotechnology): GGUAUCACCUGGGUUGUAAtt, GAACCUGUAUUCCUCUUCUtt, and GAGAGUAGGUGAUAAUGUAtt. To perform transient transfections, 5 × 105 HCT116 or DLD-1 cells were seeded in 6-well plates. Twenty-four hours later, the cells were transfected with 4 μg of plasmid DNA or 100 nM siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. After 48 hr, the cells were harvested for qPCR, western blotting, migration, invasion, and sphere formation assay.
Lentivirus Packaging and Transduction
We used a retrovirus system (Clontech Laboratories) to establish cell lines stably expressing BC032913. The lncRNA BC032913 cDNA was subcloned into the pLNCX2 plasmids. GP293 cells were transfected with pLNCX2-BC032913 or pLNCX2 and the package plasmids according to the manufacturer’s instructions. Virus particles were harvested 48 hr after transfection. HCT116 or DLD-1 cells were infected with virus particles and maintained in DMEM containing 1000 μg/mL or 600 μg/mL G418. After 2 weeks of selection, cells stably transfected with pLNCX2-BC032913 or pLNCX2 were pooled. Quantitative real-time PCR was performed as described to detect BC032913 expression.
In Vitro Migration and Invasion Assay
The migration and invasion abilities of the cells were measured using transwell chambers (8-μm pores, BD Falcon) either coated with Matrigel (BD Falcon) or left uncoated. Cells suspended (1 × 105) in serum-free DMEM were pipetted into the upper insert of a 24-well chamber 24 hr after transfection, and the bottom chamber was filled with DMEM containing 10% FBS. After 24 hr, cells that had migrated to the lower surface of the membrane were stained with crystal violet, and cells in five independent fields were counted under a microscope. All experiments were performed in triplicate.
Sphere Formation Assay
To examine the sphere formation capability of the CRC cell lines, single-cell suspensions were seeded at 5 × 103 cells/well in ultra-low attachment 6-well plates (Corning Life Sciences). The cells were cultured with DMEM/F12 supplemented with 20 ng/mL epidermal growth factor (R&D Systems), 20 ng/mL basic fibroblast growth factor (R&D Systems), and B-27 supplement (Gibco). After 10 days, the number and size of the spheres that had formed (diameter, >100 μm) were evaluated using microscopy, and the sphere formation ratio was calculated as the sphere formation ratio = sphere number / 5,000. All experiments were performed in triplicate.
Human Tumor Metastasis RT2 Profiler PCR Array
Total RNA was extracted from HCT116-BC032913 and HCT116-control cells, followed by removal of contaminating DNA and use of an RNeasy MinElute cleanup kit (QIAGEN). First-strand cDNA synthesis was achieved using SuperScript III reverse transcriptase (Invitrogen). Real-time PCR was then performed with the commercially available human tumor metastasis RT2 Profiler PCR array (PAHS-028Z, QIAGEN) according to the manufacturer’s instructions. Each 96-well PCR array contained 84 genes related to human tumor metastasis, five different housekeeping genes, one well for a genomic DNA contamination control, three replicate reverse transcription controls, and three replicate positive PCR controls. The data were normalized to GAPDH via the ΔΔCt method. Fold changes and p values were calculated using Student’s t test. p < 0.05 with a fold change greater than 2.0 was considered to indicate significant dysregulation.
Western Blot Analysis
Total cellular proteins were extracted and separated using SDS-PAGE, and western blotting was performed in accordance with standard procedures. GAPDH was employed as a loading control on the same membrane. The primary antibodies used included anti-TIMP3 (sc-9906, Santa Cruz), anti-GAPDH (sc-32233), anti-Histone H3 (sc-8654), anti-β-catenin (8480, Cell Signaling Technology), and anti-E-cadherin (3195, Cell Signaling Technology).
Luciferase Reporter Assay
To assay the activity of the Wnt/β-catenin pathway regulated by BC032913, HEK293, HCT116, and DLD-1 cells were co-transfected, respectively, with 200 ng of TOPFlash /FOPFlash (Addgene) vectors, 20 ng of pRL-TK (Promega) vectors, and 300 ng of pcDNA3.1-BC032913/pcDNA3.1 plasmids according to the manufacturer’s instructions. Twenty-four hours after transfection, the HCT116 and DLD-1 cells were lysed, whereas the HEK293 cells were incubated with 20 mM LiCl for an additional 6 hr followed by cell lysis. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) and normalized to Renilla luciferase activity. All transfection experiments were performed in triplicate and were independently repeated three times.
In Vivo Assays
Female BALB/c nude mice (4–5 weeks old) were purchased from the Animal Centre of the Chinese Academy of Science. All animal experiments were conducted in accordance with current Chinese regulations and standards regarding the use of laboratory animals. All animal procedures were approved by the Sun Yat-sen University Institutional Animal Care and Use Committee.
For the lung metastasis model, HCT116-BC032913 or HCT116-control cells (1 × 106 cells/mouse/100 μL) were injected into the tail vein of each nude mouse, with eight mice in each group. Ten weeks after injection, the animals were sacrificed, and their lungs were dissected and embedded in paraffin. Micrometastases in the lungs were counted under a microscope.58
For the hepatic metastasis model, tumor cell suspensions (HCT116-BC032913 or HCT116-control, 1 × 106 cells/mouse/100 μL) were injected into the distal tip of the spleen. After 8 weeks, the mice were euthanized, and their spleens and livers were removed for pathological examination. The metastases were counted as described above.
Statistical Analysis
Statistical analysis was performed using the SPSS software package (version 17.0). Significant associations between BC032913 expression and clinicopathological parameters were assessed using χ2 tests. A total of 115 CRC patients were included in the survival analysis, which was conducted using the Kaplan-Meier method. A log-rank test was employed to compare differences between survival curves. Significance differences between the in vitro and in vivo data were determined using two-tailed Student’s t tests. The data are presented as the means ± SD, and the threshold for statistically significant differences was set at p < 0.05.
Author Contributions
J.L., J.W., and W.H. contributed to the concept and design of the study. J.L. and X.T. performed the experiments. J.L., L.H., and J.W. contributed to data analysis and interpretation. J.L. wrote the manuscript. All authors contributed to data acquisition and revision of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81272513 and 81272638) and the National High Technology Research and Development Program of China (863 Program, 2012AA02A204).
Footnotes
Supplemental Information includes three tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.07.009.
Contributor Information
Jiangxue Wu, Email: gladysw@163.com.
Wenlin Huang, Email: hwenl@mail.sysu.edu.cn.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Zhang S., Zhao P., Zeng H., Zou X. Report of cancer incidence and mortality in China, 2010. Ann. Transl. Med. 2014;2:61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 5.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 9.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., Wu Z., Fu X., Han W. Long Noncoding RNAs: Insights from Biological Features and Functions to Diseases. Med. Res. Rev. 2013;33:517–553. doi: 10.1002/med.21254. [DOI] [PubMed] [Google Scholar]
- 12.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Chang H.Y. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z., Zhou L., Wu L.M., Lai M.C., Xie H.Y., Zhang F., Zheng S.S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 16.Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., Burghardt R., Kim S., Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 18.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin R., Maeda S., Liu C., Karin M., Edgington T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 20.Lai M.C., Yang Z., Zhou L., Zhu Q.Q., Xie H.Y., Zhang F., Wu L.M., Chen L.M., Zheng S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 21.Ren S., Wang F., Shen J., Sun Y., Xu W., Lu J., Wei M., Xu C., Wu C., Zhang Z. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer. 2013;49:2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 22.Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 23.Shi D., Zheng H., Zhuo C., Peng J., Li D., Xu Y., Li X., Cai G., Cai S. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med. Oncol. 2014;31:31. doi: 10.1007/s12032-014-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J., Rong L.F., Shi C.B., Dong X.G., Wang J., Wang B.L., Wen H., He Z.Y. Screening of lymph nodes metastasis associated lncRNAs in colorectal cancer patients. World J. Gastroenterol. 2014;20:8139–8150. doi: 10.3748/wjg.v20.i25.8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y., Ma G., Gu D., Zhu L., Hua Q., Du M., Chu H., Tong N., Chen J., Zhang Z., Wang M. Genome-wide analysis of long noncoding RNA signature in human colorectal cancer. Gene. 2015;556:227–234. doi: 10.1016/j.gene.2014.11.060. [DOI] [PubMed] [Google Scholar]
- 26.Wood L.D., Parsons D.W., Jones S., Lin J., Sjöblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 27.Visse R., Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ. Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 28.Lindsey J.C., Lusher M.E., Anderton J.A., Bailey S., Gilbertson R.J., Pearson A.D., Ellison D.W., Clifford S.C. Identification of tumour-specific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis. 2004;25:661–668. doi: 10.1093/carcin/bgh055. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y., Goldberg I.D., Shi Y.E. Complex roles of tissue inhibitors of metalloproteinases in cancer. Oncogene. 2002;21:2245–2252. doi: 10.1038/sj.onc.1205291. [DOI] [PubMed] [Google Scholar]
- 30.Chirco R., Liu X.W., Jung K.K., Kim H.R. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006;25:99–113. doi: 10.1007/s10555-006-7893-x. [DOI] [PubMed] [Google Scholar]
- 31.Hilska M., Roberts P.J., Collan Y.U., Laine V.J., Kössi J., Hirsimäki P., Rahkonen O., Laato M. Prognostic significance of matrix metalloproteinases-1, -2, -7 and -13 and tissue inhibitors of metalloproteinases-1, -2, -3 and -4 in colorectal cancer. Int. J. Cancer. 2007;121:714–723. doi: 10.1002/ijc.22747. [DOI] [PubMed] [Google Scholar]
- 32.Lin H., Zhang Y., Wang H., Xu D., Meng X., Shao Y., Lin C., Ye Y., Qian H., Wang S. Tissue inhibitor of metalloproteinases-3 transfer suppresses malignant behaviors of colorectal cancer cells. Cancer Gene Ther. 2012;19:845–851. doi: 10.1038/cgt.2012.70. [DOI] [PubMed] [Google Scholar]
- 33.Bachman K.E., Herman J.G., Corn P.G., Merlo A., Costello J.F., Cavenee W.K., Baylin S.B., Graff J.R. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 34.Qin S., Zhu Y., Ai F., Li Y., Bai B., Yao W., Dong L. MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma. 2014;61:27–34. [PubMed] [Google Scholar]
- 35.Li F., Chong Z.Z., Maiese K. Winding through the WNT pathway during cellular development and demise. Histol. Histopathol. 2006;21:103–124. doi: 10.14670/hh-21.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polakis P. Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 2012;4:a008052. doi: 10.1101/cshperspect.a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 39.Kikuchi A. Regulation of beta-catenin signaling in the Wnt pathway. Biochem. Biophys. Res. Commun. 2000;268:243–248. doi: 10.1006/bbrc.1999.1860. [DOI] [PubMed] [Google Scholar]
- 40.Giles R.H., van Es J.H., Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 41.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han D., Wang M., Ma N., Xu Y., Jiang Y., Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361:13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Nissan A., Stojadinovic A., Mitrani-Rosenbaum S., Halle D., Grinbaum R., Roistacher M., Bochem A., Dayanc B.E., Ritter G., Gomceli I. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int. J. Cancer. 2012;130:1598–1606. doi: 10.1002/ijc.26170. [DOI] [PubMed] [Google Scholar]
- 45.Graham L.D., Pedersen S.K., Brown G.S., Ho T., Kassir Z., Moynihan A.T., Vizgoft E.K., Dunne R., Pimlott L., Young G.P. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes Cancer. 2011;2:829–840. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling H., Spizzo R., Atlasi Y., Nicoloso M., Shimizu M., Redis R.S., Nishida N., Gafà R., Song J., Guo Z. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shulha H.P., Crisci J.L., Reshetov D., Tushir J.S., Cheung I., Bharadwaj R., Chou H.J., Houston I.B., Peter C.J., Mitchell A.C. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 2012;10:e1001427. doi: 10.1371/journal.pbio.1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park H.S., Yeo H.Y., Chang H.J., Kim K.H., Park J.W., Kim B.C., Baek J.Y., Kim S.Y., Kim D.Y. Dipeptidyl peptidase 10, a novel prognostic marker in colorectal cancer. Yonsei Med. J. 2013;54:1362–1369. doi: 10.3349/ymj.2013.54.6.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egea V., Zahler S., Rieth N., Neth P., Popp T., Kehe K., Jochum M., Ries C. Tissue inhibitor of metalloproteinase-1 (TIMP-1) regulates mesenchymal stem cells through let-7f microRNA and Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E309–E316. doi: 10.1073/pnas.1115083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourboulia D., Han H., Jensen-Taubman S., Gavil N., Isaac B., Wei B., Neckers L., Stetler-Stevenson W.G. TIMP-2 modulates cancer cell transcriptional profile and enhances E-cadherin/beta-catenin complex expression in A549 lung cancer cells. Oncotarget. 2013;4:166–176. doi: 10.18632/oncotarget.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia Y., Wu S. Tissue inhibitor of metalloproteinase 2 inhibits activation of the β-catenin signaling in melanoma cells. Cell Cycle. 2015;14:1666–1674. doi: 10.1080/15384101.2015.1030557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hojilla C.V., Kim I., Kassiri Z., Fata J.E., Fang H., Khokha R. Metalloproteinase axes increase beta-catenin signaling in primary mouse mammary epithelial cells lacking TIMP3. J. Cell Sci. 2007;120:1050–1060. doi: 10.1242/jcs.003335. [DOI] [PubMed] [Google Scholar]
- 53.Garouniatis A., Zizi-Sermpetzoglou A., Rizos S., Kostakis A., Nikiteas N., Papavassiliou A.G. FAK, CD44v6, c-Met and EGFR in colorectal cancer parameters: tumour progression, metastasis, patient survival and receptor crosstalk. Int. J. Colorectal Dis. 2013;28:9–18. doi: 10.1007/s00384-012-1520-9. [DOI] [PubMed] [Google Scholar]
- 54.Mashita N., Yamada S., Nakayama G., Tanaka C., Iwata N., Kanda M., Kobayashi D., Fujii T., Sugimoto H., Koike M. Epithelial to mesenchymal transition might be induced via CD44 isoform switching in colorectal cancer. J. Surg. Oncol. 2014;110:745–751. doi: 10.1002/jso.23705. [DOI] [PubMed] [Google Scholar]
- 55.Cho S.H., Park Y.S., Kim H.J., Kim C.H., Lim S.W., Huh J.W., Lee J.H., Kim H.R. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int. J. Oncol. 2012;41:211–218. doi: 10.3892/ijo.2012.1453. [DOI] [PubMed] [Google Scholar]
- 56.Gao Y., Ruan B., Liu W., Wang J., Yang X., Zhang Z., Li X., Duan J., Zhang F., Ding R. Knockdown of CD44 inhibits the invasion and metastasis of hepatocellular carcinoma both in vitro and in vivo by reversing epithelial-mesenchymal transition. Oncotarget. 2015;6:7828–7837. doi: 10.18632/oncotarget.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orian-Rousseau V., Schmitt M. CD44 regulates Wnt signaling at the level of LRP6. Mol. Cell. Oncol. 2015;2:e995046. doi: 10.4161/23723556.2014.995046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang S., Jean D., Luca M., Tainsky M.A., Bar-Eli M. Loss of AP-2 results in downregulation of c-KIT and enhancement of melanoma tumorigenicity and metastasis. EMBO J. 1998;17:4358–4369. doi: 10.1093/emboj/17.15.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.