Abstract
Multiple myeloma is a malignancy of plasma cells of the bone marrow. Although the prognosis is variable, no curative therapy has been defined. Vaccinia virus infects cancer cells and kills such cells in a variety of ways. These include direct infection, triggering of immunomediated cell death, and vascular collapse. The potential of the vaccinia virus as an anti-tumor therapy has attracted the attention of oncologists. Interestingly, our preliminary experiments revealed that myeloma cells were particularly susceptible to vaccinia virus. To exploit this susceptibility and to render vaccinia more myeloma specific, we generated thymidine-kinase-deleted microRNA (miRNA)-regulated vaccinia viruses in which the essential viral gene B5R was regulated by miRNAs of normal human cells. Of the miRNAs examined, let-7a was found to be the most reliable in terms of regulating viral transmission. Exposure to unregulated vaccinia virus killed myeloma-transplanted severe combined immunodeficiency (SCID) mice; the animals succumbed to viral toxicity. In contrast, the thymidine-kinase-deleted let-7a-regulated virus remained localized within myeloma cells, triggering tumor regression and improving overall survival. In conclusion, a thymidine-kinase-deleted let-7a-regulated vaccinia virus was safe and effective for mice, warranting clinical trials in humans.
Keywords: oncolytic virotherapy, vaccinia virus, miRNA, multiple myeloma, hematological malignancy
Introduction
Multiple myeloma is a hematological malignancy characterized by excessive production of antibody-producing plasma cells in the bone marrow.1 Although recent advances in myeloma therapy have improved response rates and overall survival, myeloma remains incurable.2 Vaccinia virus (VV) is a member of the pox family of viruses, best known for its use in the eradication of small pox in the 1970s. Recently, VV has generated renewed interest as a potential tool for cancer therapy. VVs have been shown to kill cancer cells in a variety of ways, including direct infection, immune-mediated cell death, and vascular collapse.3 Its high efficiency of infection, wide host range, and high immunogenicity all make VV an attractive option for therapeutic development.3 Unlike most other DNA viruses, entire replication of VV occurs in the cytoplasm, making insertional mutagenesis unlikely.
Recent clinical trials of VV against solid tumors are reporting excellent results.4, 5 Most notably, a phase II clinical trial using a thymidine-kinase (TK)-deleted, human granulocyte-macrophage colony-stimulating factor (hGM-CSF)-armed JX-594, also known as Pexa-Vec, significantly prolonged overall survival of patients with unresectable hepatocellular carcinoma.5 Although previous publications have suggested that hematological malignancies exhibit little to no susceptibility to VV,6 preliminary experiments in our laboratory revealed myeloma to be an exception, exhibiting very high susceptibility to VV. Deng et al.7 also reported that myeloma cells are efficiently infected with a doubly-regulated VV (TK-deleted, vaccinia growth-factor-deleted), and infection with the VV prolongs survival of myeloma-bearing mice. Although excellent infectivity to myeloma relative to other hematological malignancies is attractive, toxicity to normal tissue is still a concern for clinical use. In this study, we attempted to reduce toxicity by introducing another modification to VV.
The highly attenuated, replication-competent VV strain LC16m8 is a well-established viral clone originally isolated from the Lister strain, which has been safely administered to more than 100,000 infants and 3,000 adults with no serious adverse events.8, 9 The LC16m8 strain harbors a single-nucleotide deletion in the B5R gene, which severely impairs viral trafficking and dissemination.10, 11, 12, 13 One method of regulating the expression of genes critical for viral replication or dissemination involves the insertion of microRNA (miRNA) target sequences within the 3′ UTRs of virus transcripts.14 Because recent technological advances have provided greater clarity surrounding miRNA expression profiles, miRNA-mediated regulation of viral gene products constitutes a reasonable option for taming virulence. One such example of this approach can be seen in a study by Hikichi et al.,15 where the authors developed a recombinant VV in which B5R expression was regulated by an miRNA. This miRNA successfully reduced viral pathogenicity while maintaining the desired cytotoxic activity against solid cancer cell lines BxPC-3 and A549. In addition to B5R modifications, another way to regulate the infectivity of VV is by deletion of the TK gene. TK is essential for productive VV infection of non-dividing cells, but it is dispensable in tumor cells,16 with TK-deleted mutant VVs exhibiting tumor-specific replication.17 Although serious adverse events caused by usage of VVs, such as progressive vaccinia (1/1,000,000), eczema vaccinatum (∼8–80/1,000,000), and encephalopathy (∼3–50/1,000,000), are rare,18, 19 additional modifications further restricting viral replication may further reduce the risk of VV use in clinical applications. In that regard, doubly-regulated VV is an attractive way for tumor-specific infection, and it is the field of active research mainly targeting solid tumors.20, 21, 22
In this study, we demonstrate myeloma to be an ideal target for VV, and let-7a-regulated, TK-deleted recombinant VV is shown to specifically infect and kill myeloma cells. These data strongly support the use of VV as a potential therapeutic option for the treatment of myeloma.
Results
Multiple Myeloma Cells Have High Susceptibility to VV
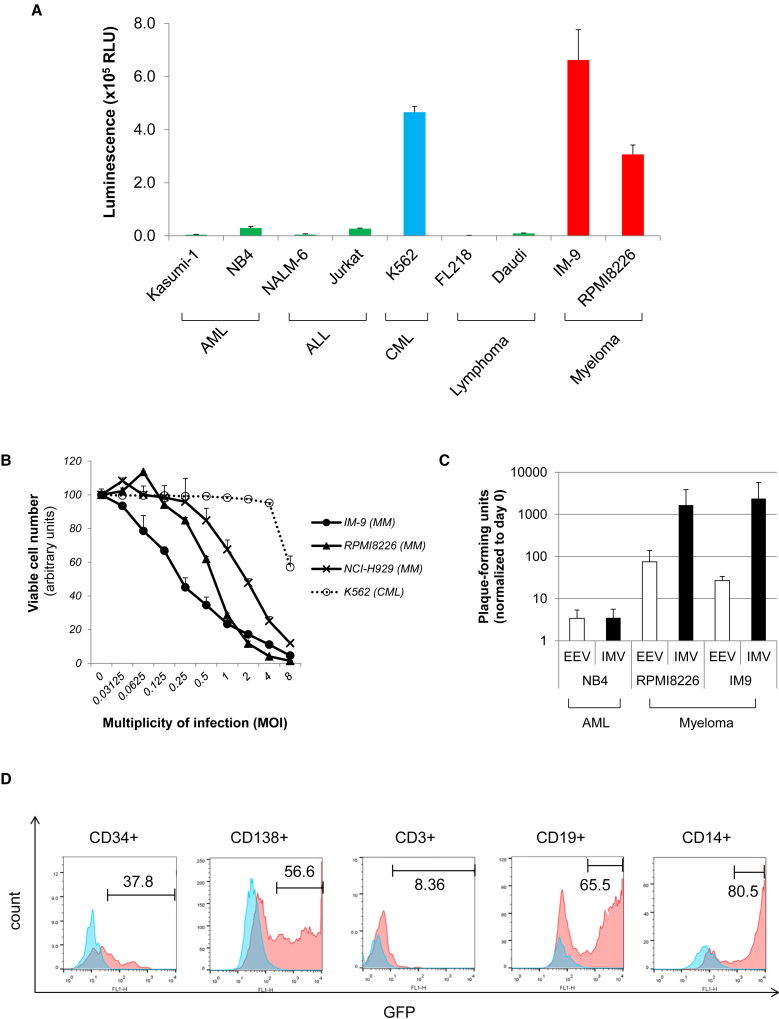
To identify optimal targets for VV therapy, we first infected different cell lines derived from hematological malignancies, including leukemia (Kasumi-1, NB4, NALM6, Jurkat, K562), lymphoma (FL218, Daudi), and myeloma (IM9, RPMI8226), with a luciferase-expressing VV (LC16m8Δ-B5R-FlucIRESgfp) at an MOI of 1. VV levels were determined by luciferase assay at 12 hr after infection. Myeloma-derived cell lines IM-9 and RPMI8226, along with leukemia cell line K562, showed the highest degree of sensitivity relative to other hematological malignancies (Figure 1A). To examine the biological consequence of infection, we infected myeloma-derived cell lines IM-9, RPMI8226, and NCI-H929, as well as K562, with VV strain LC16m8Δ-B5R-FlucIRESgfp at MOIs of 0–8; viable cell numbers were then determined using the WST-8 assay 72 hr after infection. Proliferation of myeloma-derived cell lines was significantly inhibited with a low titer of VV (MOI < 1), whereas K562 cells were resistant to MOIs ≤ 4 (Figure 1B). Given that most oncolytics depend on viral replication, we tested VV replication. Multiple myeloma (MM) cell lines (RPMI8226, IM-9), as well as a leukemia cell line NB4, were infected with VV strain LC16m8Δ-B5Rgfp at an MOI of 0.1. Extracellular enveloped virus (EEV) and intracellular mature virus (IMV) were collected from supernatant and cell pellet at 72 hr after infection and titrated. As expected, myeloma-derived cell lines RPMI8226 and IM-9 showed significant (>1,000-fold increase) VV replication (Figure 1C). To determine whether VV can infect primary cells, we infected bone marrow mononuclear cells from a myeloma patient with LC16m8Δ-B5Rgfp at an MOI of 1 and cultured them for 48 hr in RPMI 1640 medium containing 10% fetal bovine serum. Samples were gated by the expression of CD34 (hematopoietic progenitor), CD138 (myeloma), CD3 (T cell), CD19 (B cell), and CD14 (monocyte), and virus-infected cells (GFP+ cells) were determined by flow cytometry. Myeloma cells (CD138+), B cells (CD19+), and monocytes (CD14+) showed a high degree of sensitivity to VV, even at an MOI of 1, whereas hematopoietic progenitor cells (CD34+) were resistant (Figure 1D). These results suggested that myeloma would be a good target for a VV-based oncolytic therapy.
Figure 1.
Myeloma Cells Are Sensitive to VV
(A) Infectivity of leukemia/myeloma cell lines to VV. Cell lines derived from acute myelogenous leukemia (AML), acute lymphoblastic leukemia (ALL), chronic myeloid leukemia (CML), malignant lymphoma, and multiple myeloma were infected with a firefly-luciferase-expressing VV (LC16m8Δ-B5R-FlucIRESgfp) at an MOI of 1. Luciferase activity was determined at 12 hr after infection. Data represent the means and SD of three independent experiments. (B) Cell viability after infection. Cells (1 × 104/well) were seeded in 96-well plates and cultured for 72 hr in the absence or presence of different concentrations of the VV strain LC16m8Δ-B5R-FlucIRESgfp (MOIs = 0–8). The number of viable cells was determined using the Cell Counting Kit-8 (Dojindo). Data represent the mean and SD of three independent experiments. (C) Cells were infected with LC16m8Δ-B5Rgfp at an MOI of 0.1. Extracellular enveloped virus (EEV) and intracellular mature virus (IMV) were collected from supernatant and cell pellet at 72 hr after infection. Harvested virus was titrated by the infection to RK13 cells. Data represent the means and SD of three independent experiments. (D) Infectivity of myeloma patient bone marrow cells. Bone marrow mononuclear cells derived from a myeloma patient were infected with LC16m8Δ-B5Rgfp at an MOI of 1 and cultured for 48 hr. The frequency of virus-infected cells (GFP+) was compared among CD34+ cells (hematopoietic progenitors), CD138+ cells (myeloma), CD3+ cells (T lymphocytes), CD19+ cells (B lymphocytes), and CD14+ cells (monocytes). Histograms of non-infected cells (blue) are overlaid with infected cells (red).
Identification of miRNAs Differently Expressed in Normal and Myeloma Cells
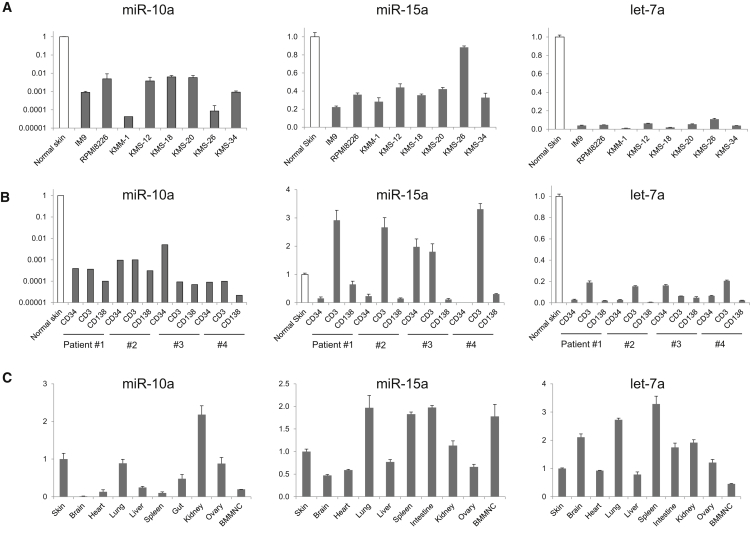
To make VV more tumor specific, we sought to identify miRNAs that could regulate the virus in target cells. We began by looking for miRNAs that are expressed at a high level in normal cells, but not in myeloma cells. Recent publications suggested that miR-10a, miR-15a, and let-7a are downregulated in myeloma or leukemia cells.22, 23, 24 Corthals et al.23 reported that let-7 families are downregulated in myeloma cells using the expression profiling of 365 miRNAs in plasma cells of 45 newly diagnosed MM patients. To validate these observations, we determined the expression of miR-10a, miR-15a, and let-7a in both normal and myeloma cells by qRT-PCR. All three of these miRNAs were downregulated in myeloma cell lines relative to controls (Figure 2A), with similar effects seen in patient-derived myeloma (CD138+) cells (Figure 2B). In contrast, expression of miR-10a was variable in normal tissues, whereas both miR-15a and let-7a were expressed in most normal tissues (Figure 2C).
Figure 2.
qRT-PCR of miRNAs
(A) Expression of miR-10a, miR-15a, and let-7a in myeloma cell lines. (B) Expression in myeloma patient samples. Expression level was compared with human normal skin fibroblasts. (C) Expression in mouse normal tissues. Tissue samples from a C57BL/6 mouse were used. RNA expression data were normalized to endogenous controls by RNU48 (human samples) and by snoRNA202 (mouse samples), respectively. Data represent the means and SD of three independent experiments.
Endogenous let-7a Effectively Regulates VV Infectivity In Vitro
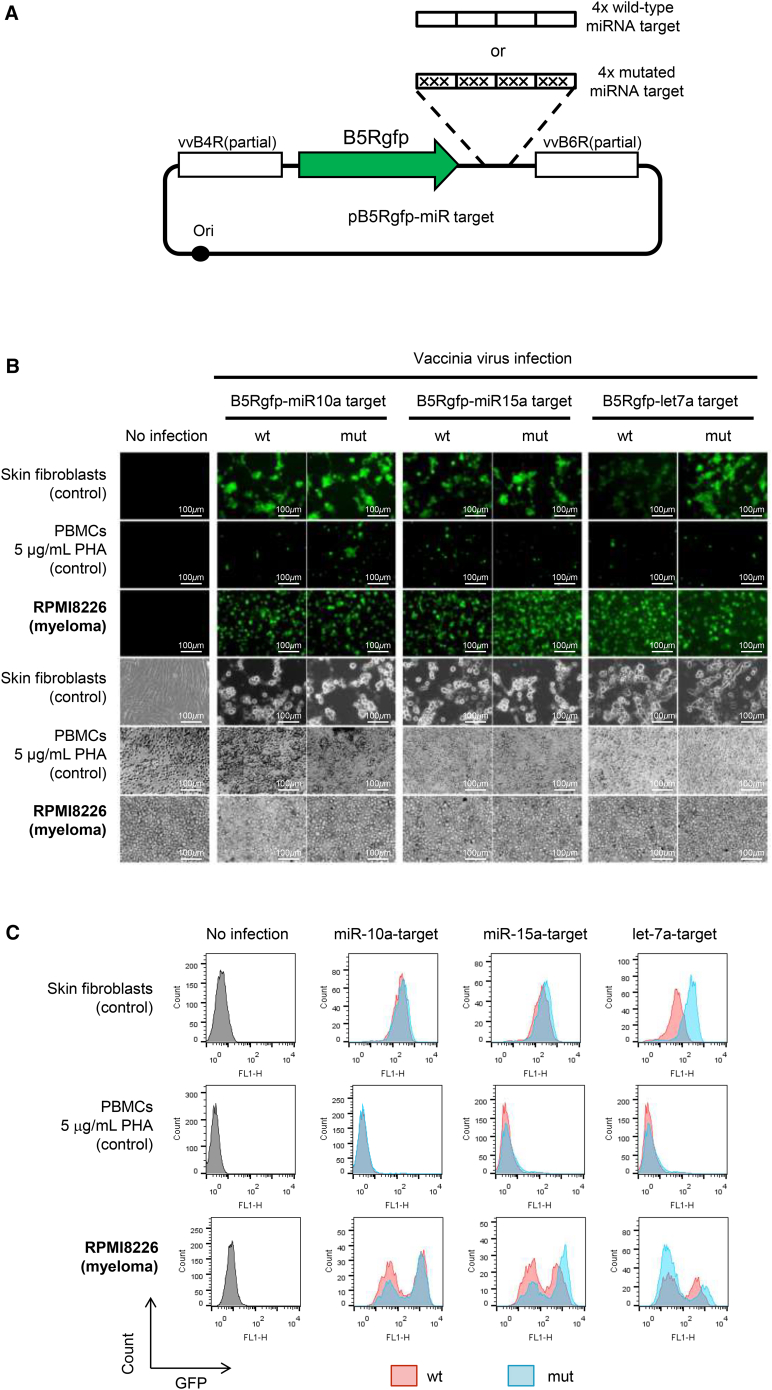
To utilize miRNA expression patterns to improve the tumor specificity of viral infection, we generated recombinant VVs that have four copies of either wild-type or mutated target sequences for miR10a, miR-15a, or let-7a in the 3′ UTR of B5Rgfp (Figure 3A). Infection (MOI = 1, 48 hr) was performed with each of these recombinant VVs to normal skin fibroblasts, normal peripheral blood mononuclear cells (with stimulation with 5 μg/mL phytohemagglutinin), and myeloma cell line RPMI8226 (Figure 3B). The GFP expression level was determined by using flow cytometry (Figure 3C). The myeloma-derived RPMI8226 cells showed a higher infectivity than normal mononuclear cells, and the GFP expression levels were close between viruses with wild-type miRNA target and mutated miRNA target. Normal skin fibroblasts showed variable infectivity to miRNA-regulated VVs, with let-7a-regulated VV exhibiting the most significant reduction. These data confirmed that let-7a-mediated regulation of B5R inhibits viral propagation in normal skin while maintaining infectivity to myeloma cells.
Figure 3.
Infection with miRNA-Regulated VVs In Vitro
(A) Construction of recombinant VV vectors. Four copies of the miRNA target sequence of either wild-type or mutated miR-10a, miR-15a, or let-7a were inserted into the 3′ UTR of B5Rgfp. (B) In vitro infection with miRNA-regulated VVs. Normal human skin fibroblasts (control), peripheral blood mononuclear cells stimulated with 5 μg/mL phytohemagglutinin (control), and RPMI8226 cells (myeloma cell line) were infected with each VV at an MOI of 1 and cultured for 48 hr. (C) Flow cytometric analysis. GFP expression by the VV with wild-type miRNA target is shown in red, and mutated miRNA-target is shown in blue. mut, mutated; PHA, phytohemagglutinin; WT, wild-type.
let-7a Overexpression Inhibits VV Infectivity
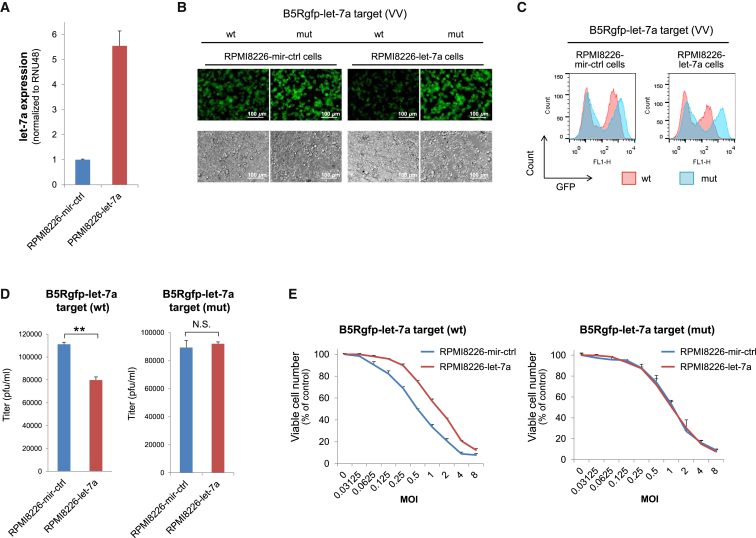
To confirm that let-7a overexpression controls viral replication, we transduced the RPMI8226 cell line with a lentiviral vector expressing either a control miRNA (RPMI8226-miR-control [ctrl] cells) or human let-7a (RPMI8226-let-7a cells) under regulation of the human elongation factor 1α promoter. After transduction, RPMI8226-let-7a cells showed 5.6-fold higher expression of let-7a than that of RPMI8226-miR-ctrl cells (Figure 4A). These cells were infected with VVs that contained either the wild-type or mutated target sequence of let-7a in the 3′ UTR of B5Rgfp (MOI = 1, 48 hr) (Figure 4B). The GFP expression level was determined using flow cytometry. RPMI8226-let-7a cells had much lower B5Rgfp expression than that of RPMI8226-miR-ctrl cells when infected with the VV containing the wild-type let-7a target sequence (Figure 4C). To measure the viral replication in the cell, we infected RPMI8226-miR-ctrl and RPMI8226-let-7a cells with either B5Rgfp-let-7a target (wild-type [WT]) or B5Rgfp-let-7a target (mutated [mut]), and the viruses collected from the infected cells were titrated. A significant reduction in viral replication was observed in RPMI8226-let-7a cells infected with B5Rgfp-let-7a target (wild-type). This reduction was not observed in RPMI8226-let-7a cells infected with the mutated let-7a target (Figure 4D). To observe whether the overexpression of let-7a protects cells from death, we infected RPMI8226-miR-ctrl and RPMI8226-let-7a cells with either B5Rgfp-let-7a target (wild-type) or B5Rgfp-let-7a target (mut) at MOIs of 0–8. Viable cells were then determined using the WST-8 assay 72 hr after infection (Figure 4E). Overexpression of let-7a clearly protected the cells from B5Rgfp-let-7a (wild-type) infection, whereas overexpression of let-7a showed almost no effect on the dose response to B5Rgfp-let-7a (mut). These data confirm that let-7a-mediated regulation of B5R inhibits viral propagation and can be used as an effective method to adjust infectivity in the tissue.
Figure 4.
Let-7a Overexpression Inhibits VV Infectivity
(A) qRT-PCR analysis of let-7a. RNA was extracted from RPMI8226 cells lentivirally transduced with miR-ctrl or let-7a. The RNA expression was normalized to that of the endogenous control RNU48. (B) In vitro infection with let-7a-regulated VVs. Cells were infected with each VV at an MOI of 1 and cultured for 48 hr. (C) Flow cytometric analysis. GFP expression by the VV containing wild-type let-7a-target or the mutated let-s7a-target is shown in red or blue, respectively. (D) Replication of VVs. Cells (1 × 104/well) were infected with VVs with either the wild-type or mutated let-7a-target at an MOI of 0.1 and grown in 96-well plates. Viruses were collected from the infected cells 72 hr after infection. Harvested viruses were titrated via infection of RK13 cells. **p < 0.01 (Student’s t test). (E) Cell viability after infection. Cells (1 × 104/well) were seeded in 96-well plates and cultured for 72 hr in the absence or presence of different concentrations of the VVs containing either the wild-type or mutated let-7a-target (MOI = 0–8). The number of viable cells was determined using the Cell Counting Kit-8. The error bars represent the SDs of three independent experiments. mut, mutated; WT, wild-type.
Let-7a-Regulated VV Inhibits Tumor Growth In Vivo
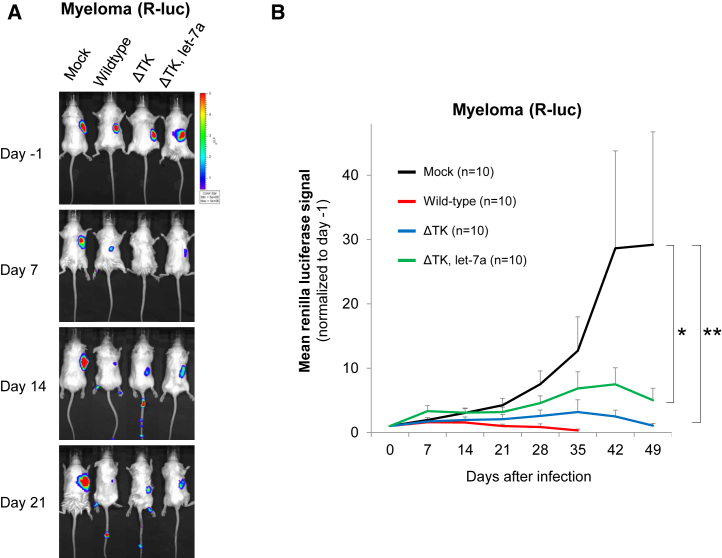
To examine the role of let-7a in vivo, we transduced RPMI8226 myeloma cells with the lentiviral vector expressing Renilla luciferase and the GFP (Rluc-GFP), after which Rluc-GFP-positive cells were collected by fluorescence-activated cell sorting. The sorted cells (1 × 107 cells) were resuspended in Matrigel (BD Biosciences) and injected subcutaneously into the back of immunodeficient severe combined immunodeficiency (SCID) mice. Four weeks later, 1 × 106 plaque-forming units of firefly luciferase (Fluc)-expressing viruses (wild-type, TK-deleted VV [ΔTK], and dual regulation by TK and let-7a [ΔTK let-7a-targeted]) were injected intravenously (n = 10). Tumor volumes and viral titers were determined using an in vivo imaging system to examine Rluc (tumor) and Fluc (virus) expression, respectively. Representative bioimages of infected mice are shown in Figure 5A. All VVs exhibited a robust tumor-inhibitory effect. The tumor-inhibitory effect was attenuated to some extent with ΔTK and ΔTK let-7a, but these VVs significantly reduced tumor burden by 27.5-fold with ΔTK (p < 0.01) and 5.8-fold with ΔTK let-7a (p < 0.05) compared with that of mock-infected mice (Figure 5B).
Figure 5.
TK Depletion, let-7a Regulation, and Anti-tumor Effect In Vivo
A total of 1 × 107 RPMI8226-RlucGFP cells were injected subcutaneously into 6-week-old CB.17-SCID mice. Four weeks later, 1 × 106 plaque-forming units of Fluc-expressing viruses (wild-type, ΔTK, and ΔTK let-7a-targeted) were infected intravenously, and the quantity of virus was determined using an in vivo imaging system. (A) Representative in vivo image of tumor mass detected by Renilla luciferase (Rluc). (B) Quantification of Rluc bioluminescence signals. A statistical analysis (n = 10) was performed using Welch’s t test. *p < 0.05; **p < 0.01. Error bars represent SEM.
Let-7a-Regulated VV and Tumor Specificity
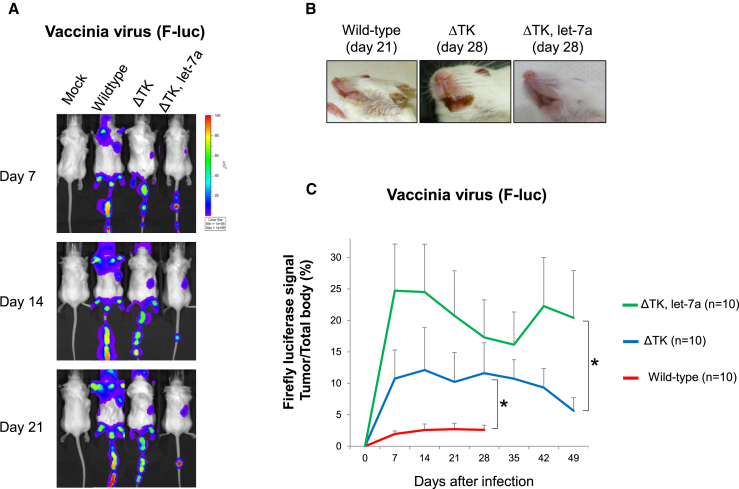
To examine the tropism of VV infection, we quantified VVs in the body using an in vivo imaging system and Fluc (VV) signals. Wild-type VV infected not only the myeloma, but also normal tissues, as shown in Figure 6A. Infected mice developed pock lesions in the skin of the ear, lips, feet, and tail (Figures 6A and 6B). TK deletion and regulation with let-7a alleviated viral toxicity. Although the skin lesions were still observed with these VVs in the tail, ΔTK let-7a-targeted lesions were significantly milder. In particular, lesions in the mouth and ears were less severe than those of the wild-type and ΔTK (Figures 6A and 6B). Fluc signal intensities in the tumor and total body were quantified. The tumor signal percent of the total body was compared among wild-type, ΔTK, and ΔTK let-7a. TK deletion and regulation with let-7a significantly improved tumor specificity (Figure 6C).
Figure 6.
TK-Depletion, let-7a Regulation, and Viral Toxicity In Vivo
(A) Distribution of VVs detected by Fluc. (B) Skin lesions of infected mice. (C) Quantification of Fluc bioluminescence signals. Fluc signals in the tumor region and total body were measured, and tumor signal percent of the total body was determined. A statistical analysis (n = 10) was performed using Welch’s t test. *p < 0.05. Error bars represent SEM.
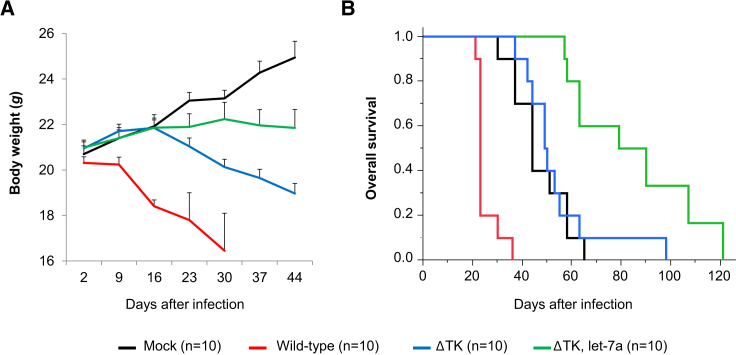
Let-7a-Regulated VV Prolongs Overall Survival of Tumor-Bearing Mice
Although mice infected with wild-type VV exhibited significantly fewer tumors, these mice developed skin pock lesions, quickly lost weight (Figure 7A), and died between days 21 and 36 (median, day 23) (Figure 7B). Pock lesions in the skin of the ears, lips, feet, and tail were prominent, but the histological analysis showed viral lesions only in the skin and tumor; critical organs such as the brain, lung, liver, and kidney were spared regardless of VV type (Figure S1). The most likely cause of death was insufficient food intake due to oral skin lesions rather than tumor progression in wild-type-infected mice. ΔTK-infected mice showed less severe lesions but still showed a continuous decline in body weight and died at a median time of 49.5 days. The probable cause of death of the ΔTK-infected mice was viral toxicity, but together with the reduction in tumor mass, ΔTK-infected mice lived as long as control mice. Control tumor mice died within 65 days because of tumor progression (median, 44 days), whereas mice infected with ΔTK let-7a-targeted VV showed limited skin lesions on the tail and survived for up to 121 days (median, 84.5 days) (Figure 7B). Survival was significantly longer in ΔTK let-7a-targeted-infected than in mock-infected mice (p < 0.05, log rank test), suggesting successful stabilization of the disease (Figure 7B).
Figure 7.
Effect of VVs on Survival
(A) Body weight of the mice after infection (n = 10). Error bars represent SEM. (B) Kaplan-Meier survival curves for control and infected mice.
Discussion
Recent advances in gene recombination technology have greatly accelerated the development of oncolytic virotherapy. JX-594, a Wyeth vaccine strain with disruption of TK, along with an insertion of hGM-CSF, showed significant improvement in overall survival of patients with hepatocellular carcinoma.5 However, a previous in vitro study suggested significantly lower infectivity to VV among hematological malignancies, thereby limiting the use of VV as a potential therapy in these diseases.6 Here, we observed exceptionally high sensitivity of myeloma cells to VV, as compared with other hematological malignancies, leading us to focus on myeloma as a potential target of VV therapy. Parato et al.6 assessed the median effective dose of VV (JX-594) on a panel of cancer cell lines of different tissue origins (NCI60 cell line panel), in which a myeloma cell line RPMI8226 showed similar sensitivity (MOI < 1) to cancer cell lines of the lung and colon; the data presented here are broadly consistent with these results. An important case report in the 1980s using a VV against myeloma showed a significant decrease of serum M protein levels; however, no confirmation of these results has been provided in the decades since.24
VVs bind to target cells via heparan sulfate, a ubiquitous cell-surface glycoprotein,25 and are taken into the cell through a ubiquitous micropinocytosis process.26 Therefore, tissue selectivity of VV infection is solely mediated within the cytoplasm of the target cell. The mechanism of high selectivity of VVs for myeloma remains unclear, although abnormal signal transduction through RAS/mitogen-activated protein kinase (MAPK) pathways in the myeloma is suspected to contribute to this effect, because RAS/MAPK pathways are necessary for the transmission, proliferation, and survival of virus in the host.27, 28 It is noteworthy that activating mutations of RAS, RAF, and MAPK are the most common (nearly 50%) genetic abnormalities in myeloma patients.29
miRNA-mediated regulation of viruses for the purpose of tissue-specific infection is a well-designed strategy.14 Hikichi et al.15 reported that miRNA-mediated regulation of B5R successfully reduces toxicity in normal tissues while maintaining oncolytic effects toward cancer cells of the lung, breast, and pancreas. Recent publications have demonstrated downregulation of miRNAs such as miR-10a,30 miR-15a,31 and let-7a32 in leukemia and myeloma cells. Using a series of in vitro infection experiments with different miRNA target sequences, we demonstrated that let-7a was the most effective miRNA for distinguishing between normal and myeloma cells. let-7a functions as a developmental regulator and is one of the first two miRNAs ever described.33 Because let-7a is highly expressed in differentiated cells, but only weakly expressed or absent in human and mouse stem cells,34, 35 let-7a in normal tissues would be a very effective tool for regulating critical viral genes. Recent studies have shown a strong correlation between decreases in let-7a expression in cancer cells and poor prognosis.36, 37, 38 In addition, K-RAS, N-RAS, and H-RAS all have let-7 binding sites in their 3′ UTRs, and inhibition of let-7 upregulates RAS expression.39 Taken together, these data provide a strong basis for let-7a-mediated regulation of VV as a means of promoting tumor specificity.
In regard to the safety of VV in vivo, the wild-type VV strain used here (LC16m8Δ-B5R-FlucIRESgfp) caused severe skin pock lesions in SCID mice, leading to death 21–36 days after infection. Histological analysis of these mice did not indicate any involvement in the vital organs, such as the brain, heart, lung, liver, or kidney, whereas the most likely cause of death was either insufficient food intake due to oral skin lesions or physical debilitation resulting from chronic inflammation. TK-deleted VV (ΔTK) significantly delayed the development of skin lesions, but still led to death within 98 days after infection. Dual regulation by TK and let-7a (ΔTK let-7a-targeted) significantly protected normal tissues, with mice surviving for up to 121 days after infection. In wild-type VV-infected mice, viral lesions were seen in the skin of the ear, lips, feet, and tail, where the skin was not coated with fur. Because the LC16m8 strain was originally established by in vitro culture at low temperature,40, 41 LC16m8-derived strains may prefer hypothermic areas in the body. Severity of the host’s immunodeficiency is another important factor. SCID mice have a deficiency that affects both B and T lymphocytes;42, 43 human patients with a similar disease (X-linked SCID) present in the first few months of life with recurrent severe bacterial, viral, or fungal infections, and the condition is always fatal without definitive treatment such as bone marrow transplantation.44 As compared with the immunological condition of myeloma patients, our SCID mouse model likely represents a far more severe form of immune deficiency. Although Hikichi et al.15 showed that let-7a-mediated regulation of VV reduces viral toxicity without impairing anti-tumor efficacy, we generated a doubly-regulated VV with a TK deletion in addition to the let-7a-mediated regulation and demonstrated its safety using a more stringent immunodeficiency model to observe viral pathogenicity. The SCID mice that we injected intravenously with virus were more susceptible to systemic viral infection than were nude mice injected intratumorally with virus by Hikichi et al.15 Therefore, the safety profile of our doubly-regulated VV was further confirmed. Adverse effects of VV similar to those seen in our SCID mice are unlikely to occur in patients given the long and extensive history of use in humans for small pox eradication. Because the double regulation of VV by ΔTK and let-7a is so robust in terms of cell tropism, the challenge now becomes finding a way to intensify the anti-myeloma effects. Not surprisingly, this specificity and selective toxicity has made VV a promising model for cancer vaccine development.45, 46 As the name “vaccinia” implies, VV is highly immunogenic and elicits strong T cell47 and B cell48 responses. VV can be easily manipulated by inserting large or multiple transgenes up to 25 kb in size,49 enabling insertion of potential cancer antigens, which may further enhance the host’s anti-cancer activity; in combination with ΔTK and let-7a-mediated regulation, this may represent a promising approach to anti-cancer therapy.
In summary, we were able to demonstrate that: (1) myeloma cells can be infected with VV at a very low titer (MOI < 1) in contrast to other hematological malignancies; and (2) let-7a-medited regulation of VV is effective at inhibiting the spread of VV into normal tissues, allowing for targeted anti-tumor activity. Taken together, these results strongly support the use of VV as a potential treatment for myeloma.
Materials and Methods
Cell Culture
The following cell lines (Kasumi-1, NB4, NALM-6, Jurkat, K562, Daudi, IM-9, RPMI8226, NCI-H929, HeLa, and RK13) were obtained from ATCC between 2004 and 2013. FL-218 was obtained from the Japanese Collection of Research Bioresources Cell Bank in 2011. Normal human skin fibroblasts (NB1RGB) were obtained from the RIKEN BioResource Center in 2014. All cell lines were passaged for less than 6 months. No further authentication was done for these cell lines in the past 6 months. Bone marrow samples were collected from patients admitted to the Japanese Red Cross Medical Center between February 2014 and December 2014; written informed consent was obtained from all patients prior to collection. All relevant study-related protocols were approved by the Institutional Review Board of the Japanese Red Cross Medical Center and the Institute of Medical Science, The University of Tokyo. Cells were grown in appropriate culture medium supplemented with 10% fetal bovine serum (JRH Biosciences), 100 U/mL penicillin (Wako), and 100 μg/mL streptomycin (Wako). Cell lines derived from hematological malignancies and patient bone marrow mononuclear cells were grown in RPMI 1640 medium (Wako). HeLa cells and NB1RGB cells were grown in minimum essential medium (MEM), alpha modification (Wako). RK13 cells were grown in standard MEM (Wako).
In Vitro Luciferase Assay
Cells (1 × 104/well) were seeded in 96-well plates and cultured for 12 hr in the presence of VV expressing Fluc (LC16m8Δ-B5R-FlucIRESgfp) (MOI = 1). Luciferase activity was determined by the ONE-Glo luciferase assay system (Promega).
Flow Cytometry
Allophycocyanin (APC)-labeled antibodies against human CD3, CD14, CD19, CD34, and CD138 were purchased from BioLegend. Labeled cells were quantified using the FACSCalibur flow cytometer (BD Biosciences).
Quantification of miRNAs
MicroRNAs were extracted using the miRVana miRNA isolation kit (Life Technologies). qRT-PCR was performed with TaqMan small RNA assays (Life Technologies) using a CFX connect real-time PCR detection system (Bio-Rad). Relative expression levels were determined by the comparative cycle threshold (Ct) method.
Plasmid Construction
Oligonucleotide pairs containing four copies of completed or mutated complementary sequences for miR-10a, miR-15a, or let-7a were inserted into the 3′ UTR of the pTN-B5Rgfp vector. Sequence information and other details can be found in the Supplemental Materials and Methods.
Recombination of VV
RK13 cells were infected with LC16m8Δ virus at an MOI of 0.01, then transfected with either pTN-B5Rgfp-miR-10a × 4, pTN-B5Rgfp-miR-10a-mut × 4, pTN-B5Rgfp-miR-15a × 4, pTN-B5Rgfp-miR-15a-mut × 4, pTN-B5Rgfp-let-7a × 4, or B5Rgfp-let-7a-mut × 4. After harvesting progeny viruses 2–5 days later, recombinant LC16m8Δ-B5Rgfp with miRNA targets was selected on the basis of larger plaque size and GFP expression, by three serial plaque purifications. Likewise, LC16m8Δ-B5R viruses expressing Fluc and GFP were generated by infecting RK13 cells with LC16m8Δ-B5R, then transfecting with pSFJvnc110-FlucIRESgfp; the resulting strain was designated as the “wild-type” in this study. TK-deleted, Fluc-, and GFP-expressing viruses were generated by infecting 143B cells with either LC16m8Δ-B5R or LC16m8Δ-B5R-let-7a × 4 and transfecting with the pTK-FlucIRESgfp. Recombinant virus was purified by 5-bromodeoxyuridine (BrdU) selection. These strains were termed ΔTK and ΔTK-let-7a target, respectively. All viruses were propagated and titrated in RK13 cells and stored at −80°C.
Production of the Lentiviral Vector
To generate lentiviral vectors expressing human let-7a driven by the human elongation factor 1α promoter, we purchased the vector plasmids pLV-hsa-miR-ctrl (MIR-P000) and pLV-hsa-let-7a-1 (MIR-P001) from Biosettia. To generate the plasmid-expressing Renilla luciferase (Rluc) and GFP, we amplified Rluc cDNA by PCR with primers 5′-GAATTCGCCACCATGGCTTCCAAGGT-3′ and 5′-GGATCCCTGCTCGTTCTTCAGCAC-3′ using the pmirGLO plasmid (Promega) as a template. The PCR product was then digested with EcoRI and BamHI, and cloned into CSII-EF-MCS-2A-EGFP, resulting in CSII-EF-Rluc-2A-EGFP. Lentiviral vector particles were produced by co-transfection of Lenti-X293T cells (Clontech) with a transfer plasmid, pMDLg/p.RRE, pRSV-rev, and pMD.G, then titrated in HeLa cells as described previously.50
Mice
CB.17-SCID and C57BL/6J mice were purchased from CLEA Japan. All animal experimentation protocols were approved by the Animal Experiment Committee of the Institute of Medical Science, University of Tokyo. Animals were sacrificed if they had lost >20% of their body weight or had a tumor burden >10% body weight.
In Vivo Imaging
To detect Fluc activity, we dissolved D-luciferin (Promega) in PBS at 15 mg/mL and administered intraperitoneally at a dose of 100 μL (150 μg) per animal. To detect Renilla luciferase (Rluc) activity, we dissolved ViviRen in vivo Renilla luciferase substrate (Promega) in dimethyl sulfoxide at 10 mg/mL, and the stock solution was further diluted 1,000-fold in PBS containing 0.1% bovine serum albumin, after which 100 μL (1 μg) was administered by intravenous injection. The mice were anesthetized with isoflurane before imaging with the in vivo imaging system (IVIS) 100 bioluminescence imaging system (Xenogen). Bioluminescence signals were quantified 10 min after the injection of D-luciferin or immediately after the injection of ViviRen according to the manufacturer’s protocol.
Statistical Analysis
Statistical analyses between two groups were performed by the two-tailed Student’s t test or the Welch’s t test, depending on homogeneity of variances. Kaplan-Meier survival curves were generated using JMP pro 11.2.0 (SAS Institute). Statistical significance between groups was determined using the log rank test.
Author Contributions
Conceptualization, M.F., T.N., and A.T.; Methodology, M.F., K. Sato, and T.N.; Investigation, M.F., K. Sato, and T.N.; Writing, M.F.; Funding Acquisition, M.F.; Resources, K.M. and K. Suzuki; Supervision, T.N., K. Suzuki, and A.T.
Conflicts of Interest
No author has any competing financial interest.
Acknowledgments
M.F. received the Grant-in-Aid for Young Scientists Program B grant 26830103 from the Japan Society for the Promotion of Science. We would like to thank Dr. I.M. Verma (Salk Institute) and Cell Genesys for providing lentiviral vector constructs. The English in this document has been checked by at least two professional editors, both native speakers of English; for a certificate, please see: http://www.textcheck.com/certificate/ult3b9.
Footnotes
Supplemental Information includes Supplemental Materials and Methods and one figure and can be found with this article online at http://dx.doi.org/10.1016/j.omto.2017.07.001.
Supplemental Information
References
- 1.Palumbo A., Anderson K. Multiple myeloma. N. Engl. J. Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S.K., Dispenzieri A., Lacy M.Q., Gertz M.A., Buadi F.K., Pandey S., Kapoor P., Dingli D., Hayman S.R., Leung N. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. 2014;28:1122–1128. doi: 10.1038/leu.2013.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guse K., Cerullo V., Hemminki A. Oncolytic vaccinia virus for the treatment of cancer. Expert Opin. Biol. Ther. 2011;11:595–608. doi: 10.1517/14712598.2011.558838. [DOI] [PubMed] [Google Scholar]
- 4.Park B.H., Hwang T., Liu T.C., Sze D.Y., Kim J.S., Kwon H.C., Oh S.Y., Han S.Y., Yoon J.H., Hong S.H. Use of a targeted oncolytic poxvirus, JX-594, in patients with refractory primary or metastatic liver cancer: a phase I trial. Lancet Oncol. 2008;9:533–542. doi: 10.1016/S1470-2045(08)70107-4. [DOI] [PubMed] [Google Scholar]
- 5.Heo J., Reid T., Ruo L., Breitbach C.J., Rose S., Bloomston M., Cho M., Lim H.Y., Chung H.C., Kim C.W. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat. Med. 2013;19:329–336. doi: 10.1038/nm.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parato K.A., Breitbach C.J., Le Boeuf F., Wang J., Storbeck C., Ilkow C., Diallo J.S., Falls T., Burns J., Garcia V. The oncolytic poxvirus JX-594 selectively replicates in and destroys cancer cells driven by genetic pathways commonly activated in cancers. Mol. Ther. 2012;20:749–758. doi: 10.1038/mt.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng H., Tang N., Stief A.E., Mehta N., Baig E., Head R., Sleep G., Yang X.Z., McKerlie C., Trudel S. Oncolytic virotherapy for multiple myeloma using a tumour-specific double-deleted vaccinia virus. Leukemia. 2008;22:2261–2264. doi: 10.1038/leu.2008.120. [DOI] [PubMed] [Google Scholar]
- 8.Saito T., Fujii T., Kanatani Y., Saijo M., Morikawa S., Yokote H., Takeuchi T., Kuwabara N. Clinical and immunological response to attenuated tissue-cultured smallpox vaccine LC16m8. JAMA. 2009;301:1025–1033. doi: 10.1001/jama.2009.289. [DOI] [PubMed] [Google Scholar]
- 9.Kidokoro M., Shida H. Vaccinia virus LC16m8Δ as a vaccine vector for clinical applications. Vaccines (Basel) 2014;2:755–771. doi: 10.3390/vaccines2040755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi-Nishimaki F., Funahashi S., Miki K., Hashizume S., Sugimoto M. Regulation of plaque size and host range by a vaccinia virus gene related to complement system proteins. Virology. 1991;181:158–164. doi: 10.1016/0042-6822(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 11.Smith G.L., Vanderplasschen A., Law M. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 2002;83:2915–2931. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- 12.Schmelz M., Sodeik B., Ericsson M., Wolffe E.J., Shida H., Hiller G., Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollinshead M., Rodger G., Van Eijl H., Law M., Hollinshead R., Vaux D.J., Smith G.L. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 2001;154:389–402. doi: 10.1083/jcb.200104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly E.J., Hadac E.M., Greiner S., Russell S.J. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat. Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 15.Hikichi M., Kidokoro M., Haraguchi T., Iba H., Shida H., Tahara H., Nakamura T. MicroRNA regulation of glycoprotein B5R in oncolytic vaccinia virus reduces viral pathogenicity without impairing its antitumor efficacy. Mol. Ther. 2011;19:1107–1115. doi: 10.1038/mt.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buller R.M., Smith G.L., Cremer K., Notkins A.L., Moss B. Decreased virulence of recombinant vaccinia virus expression vectors is associated with a thymidine kinase-negative phenotype. Nature. 1985;317:813–815. doi: 10.1038/317813a0. [DOI] [PubMed] [Google Scholar]
- 17.Puhlmann M., Brown C.K., Gnant M., Huang J., Libutti S.K., Alexander H.R., Bartlett D.L. Vaccinia as a vector for tumor-directed gene therapy: biodistribution of a thymidine kinase-deleted mutant. Cancer Gene Ther. 2000;7:66–73. doi: 10.1038/sj.cgt.7700075. [DOI] [PubMed] [Google Scholar]
- 18.Poland G.A., Grabenstein J.D., Neff J.M. The US smallpox vaccination program: a review of a large modern era smallpox vaccination implementation program. Vaccine. 2005;23:2078–2081. doi: 10.1016/j.vaccine.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Casey C.G., Iskander J.K., Roper M.H., Mast E.E., Wen X.J., Török T.J., Chapman L.E., Swerdlow D.L., Morgan J., Heffelfinger J.D. Adverse events associated with smallpox vaccination in the United States, January-October 2003. JAMA. 2005;294:2734–2743. doi: 10.1001/jama.294.21.2734. [DOI] [PubMed] [Google Scholar]
- 20.Lun X., Ruan Y., Jayanthan A., Liu D.J., Singh A., Trippett T., Bell J., Forsyth P., Johnston R.N., Narendran A. Double-deleted vaccinia virus in virotherapy for refractory and metastatic pediatric solid tumors. Mol. Oncol. 2013;7:944–954. doi: 10.1016/j.molonc.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lun X.Q., Jang J.H., Tang N., Deng H., Head R., Bell J.C., Stojdl D.F., Nutt C.L., Senger D.L., Forsyth P.A., McCart J.A. Efficacy of systemically administered oncolytic vaccinia virotherapy for malignant gliomas is enhanced by combination therapy with rapamycin or cyclophosphamide. Clin. Cancer Res. 2009;15:2777–2788. doi: 10.1158/1078-0432.CCR-08-2342. [DOI] [PubMed] [Google Scholar]
- 22.McCart J.A., Ward J.M., Lee J., Hu Y., Alexander H.R., Libutti S.K., Moss B., Bartlett D.L. Systemic cancer therapy with a tumor-selective vaccinia virus mutant lacking thymidine kinase and vaccinia growth factor genes. Cancer Res. 2001;61:8751–8757. [PubMed] [Google Scholar]
- 23.Corthals S.L., Sun S.M., Kuiper R., de Knegt Y., Broyl A., van der Holt B., Beverloo H.B., Peeters J.K., el Jarari L., Lokhorst H.M. MicroRNA signatures characterize multiple myeloma patients. Leukemia. 2011;25:1784–1789. doi: 10.1038/leu.2011.147. [DOI] [PubMed] [Google Scholar]
- 24.Kawa A., Arakawa S. The effect of attenuated vaccinia virus AS strain on multiple myeloma; a case report. Jpn. J. Exp. Med. 1987;57:79–81. [PubMed] [Google Scholar]
- 25.Hsiao J.C., Chung C.S., Chang W. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 1998;72:8374–8379. doi: 10.1128/jvi.72.10.8374-8379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laliberte J.P., Moss B. Appraising the apoptotic mimicry model and the role of phospholipids for poxvirus entry. Proc. Natl. Acad. Sci. USA. 2009;106:17517–17521. doi: 10.1073/pnas.0909376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Magalhães J.C., Andrade A.A., Silva P.N., Sousa L.P., Ropert C., Ferreira P.C., Kroon E.G., Gazzinelli R.T., Bonjardim C.A. A mitogenic signal triggered at an early stage of vaccinia virus infection: implication of MEK/ERK and protein kinase A in virus multiplication. J. Biol. Chem. 2001;276:38353–38360. doi: 10.1074/jbc.M100183200. [DOI] [PubMed] [Google Scholar]
- 28.Andrade A.A., Silva P.N., Pereira A.C., De Sousa L.P., Ferreira P.C., Gazzinelli R.T., Kroon E.G., Ropert C., Bonjardim C.A. The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. Biochem. J. 2004;381:437–446. doi: 10.1042/BJ20031375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohr J.G., Stojanov P., Carter S.L., Cruz-Gordillo P., Lawrence M.S., Auclair D., Sougnez C., Knoechel B., Gould J., Saksena G., Multiple Myeloma Research Consortium Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agirre X., Jiménez-Velasco A., San José-Enériz E., Garate L., Bandrés E., Cordeu L., Aparicio O., Saez B., Navarro G., Vilas-Zornoza A. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol. Cancer Res. 2008;6:1830–1840. doi: 10.1158/1541-7786.MCR-08-0167. [DOI] [PubMed] [Google Scholar]
- 31.Roccaro A.M., Sacco A., Thompson B., Leleu X., Azab A.K., Azab F., Runnels J., Jia X., Ngo H.T., Melhem M.R. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Handa H., Hattori H., Takahashi N., Sasaki Y., Saitoh T., Osaki Y., Tahara K., Koiso H., Mitsui T., Shimizu H. Association between micro-RNA and epigenetic modifiers DNA methyltransferases (DNMTs), histone deacetylases (HDACs) in multiple myeloma (MM) and monoclonal gammmopathy with undetermined significance (MGUS) Blood. 2012;120:3942. [Google Scholar]
- 33.Büssing I., Slack F.J., Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Thomson J.M., Newman M., Parker J.S., Morin-Kensicki E.M., Wright T., Hammond S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair V.S., Maeda L.S., Ioannidis J.P. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J. Natl. Cancer Inst. 2012;104:528–540. doi: 10.1093/jnci/djs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 38.Nana-Sinkam S.P., Croce C.M. MicroRNAs as therapeutic targets in cancer. Transl. Res. 2011;157:216–225. doi: 10.1016/j.trsl.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Hashizume S., Yoshizawa H., Morita M., Suzuki K. Elsevier Science Publishing Co.; 1985. Properties of attenuated mutant of vaccinia virus, LC16m8, derived from Lister strain. [Google Scholar]
- 41.Kenner J., Cameron F., Empig C., Jobes D.V., Gurwith M. LC16m8: an attenuated smallpox vaccine. Vaccine. 2006;24:7009–7022. doi: 10.1016/j.vaccine.2006.03.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bosma G.C., Custer R.P., Bosma M.J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 43.Okazaki K., Nishikawa S., Sakano H. Aberrant immunoglobulin gene rearrangement in scid mouse bone marrow cells. J. Immunol. 1988;141:1348–1352. [PubMed] [Google Scholar]
- 44.Aloj G., Giardino G., Valentino L., Maio F., Gallo V., Esposito T., Naddei R., Cirillo E., Pignata C. Severe combined immunodeficiences: new and old scenarios. Int. Rev. Immunol. 2012;31:43–65. doi: 10.3109/08830185.2011.644607. [DOI] [PubMed] [Google Scholar]
- 45.Odunsi K., Matsuzaki J., Karbach J., Neumann A., Mhawech-Fauceglia P., Miller A., Beck A., Morrison C.D., Ritter G., Godoy H. Efficacy of vaccination with recombinant vaccinia and fowlpox vectors expressing NY-ESO-1 antigen in ovarian cancer and melanoma patients. Proc. Natl. Acad. Sci. USA. 2012;109:5797–5802. doi: 10.1073/pnas.1117208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amato R.J., Stepankiw M. Evaluation of MVA-5T4 as a novel immunotherapeutic vaccine in colorectal, renal and prostate cancer. Future Oncol. 2012;8:231–237. doi: 10.2217/fon.12.7. [DOI] [PubMed] [Google Scholar]
- 47.Walker J.M., Slifka M.K. The immunostimulatory power of acute viral infection. Immunity. 2008;28:604–606. doi: 10.1016/j.immuni.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 48.Pütz M.M., Midgley C.M., Law M., Smith G.L. Quantification of antibody responses against multiple antigens of the two infectious forms of Vaccinia virus provides a benchmark for smallpox vaccination. Nat. Med. 2006;12:1310–1315. doi: 10.1038/nm1457. [DOI] [PubMed] [Google Scholar]
- 49.Smith G.L., Moss B. Infectious poxvirus vectors have capacity for at least 25 000 base pairs of foreign DNA. Gene. 1983;25:21–28. doi: 10.1016/0378-1119(83)90163-4. [DOI] [PubMed] [Google Scholar]
- 50.Bai Y., Soda Y., Izawa K., Tanabe T., Kang X., Tojo A., Hoshino H., Miyoshi H., Asano S., Tani K. Effective transduction and stable transgene expression in human blood cells by a third-generation lentiviral vector. Gene Ther. 2003;10:1446–1457. doi: 10.1038/sj.gt.3302026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.