Abstract
Activation of cardiac fibroblasts (CF) into myofibroblasts is considered to play an essential role in cardiac remodeling and fibrosis. A limiting factor in studying this process is the spontaneous activation of CFs when cultured on two-dimensional (2D) culture plates. Here, a simplified 3D hydrogel platform of contractile cardiac tissue, stimulated by transforming growth factor-β1 (TGF-β1), is presented to recapitulate a fibrogenic micro-environment. It was hypothesized that the quiescent state of CFs can be maintained by mimicking the mechanical stiffness of native heart tissue. To test this hypothesis, a 3D cell culture model consisting of cardiomyocytes and CFs encapsulated within mechanically engineered gelatin methacryloyl (GelMA) hydrogel, was developed. The study shows that CFs maintain their quiescent phenotype in mechanically tuned hydrogels. Additionally, treatment with a beta-adrenergic agonist increased beating frequency, demonstrating physiologic-like behavior of the heart constructs. Subsequently, quiescent CFs within the constructs were activated by the exogenous addition of TGF-β1. The expression of fibrotic protein markers (and the functional changes in mechanical stiffness) in the fibrotic-like tissues were analyzed to validate the model. Overall, this 3D engineered culture model of contractile cardiac tissue enabled controlled activation of CFs, demonstrating the usability of this platform to study fibrotic remodeling.
Keywords: cardiac fibrosis, cardiac tissue engineering, in vitro 3D models, hydrogels, myofibroblast
1. Introduction
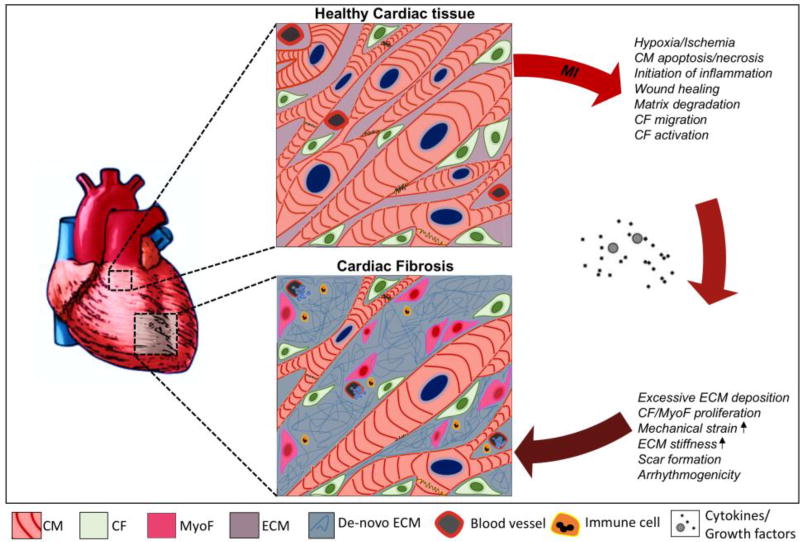
Cardiovascular diseases (CVDs), such as ischemic heart disease and hypertension, have remained in the top 10 major causes of death worldwide.[1] Myocardial infarction (MI), which is responsible for more than 50% of the deaths attributable to CVDs, results in a significant loss of cardiomyocytes.[2, 3] This loss results in the initiation of a reparative wound healing process, which is characterized by an initial inflammatory phase and followed by the proliferation and activation of quiescent cardiac fibroblasts into cardiac myofibroblasts (MyoFs).[4] Cardiac fibrosis results from the excessive synthesis and accumulation of extracellular matrix (ECM) components (eg. collagen, fibronectin), and is caused by the persistent activation and proliferation of both cardiac fibroblasts and MyoFs (Figure 1).[3–5, 6–8] In addition, cardiac MyoFs (hallmarked by the expression of α-smooth muscle actin (α-SMA)) (Figure 1) have contractile properties, which can lead to a sustained contractile stress that is exerted on the infarcted area.[9, 10] In the short term outlook, these pro-fibrotic processes can be beneficial for cardiac function as it prevents dilatation and rupturing of the ventricular wall.[11] However, prolonged and excessive activity of MyoFs results in excessive fibrosis and tissue stiffening, which ultimately impairs cardiac function, increases the risk of arrhythmia, and leads to the progression of end-stage heart failure. [6, 9, 12, 13]
Figure 1. Schematic illustration of the pathophysiological changes during fibrotic cardiac remodeling.
Healthy myocardial tissue consists of a network of cardiomyocytes (CM) and quiescent cardiac fibroblasts (CF) that are interspersed within the extracellular matrix (ECM). After myocardial injury (eg. myocardial infarct (MI)), CMs die and a reparative inflammatory and wound healing process is initiated by the release of various cytokines and growth factors (such as transforming growth factor-β1, angiotensin-II etc.). This results in the activation of cardiac fibroblasts into cardiac myofibroblasts (MyoF). These cells and other resident cardiac fibroblasts are responsible for an excessive and prolonged synthesis and deposition of de-novo ECM proteins (eg. collagen-I, fibronectin, laminin). This results in scarring of the heart tissue and leads to the deterioration of ventricular function followed by diastolic and systolic ventricular dysfunction and may eventually lead to life threatening arrhythmogenicity or heart failure.
Although there are many identified biochemical (eg. transforming growth factor-β1 (TGF-β1), angiotensin II, endothelin-1, platelet derived growth factor),[4, 8, 14] mechanobiological, (eg. tissue stiffness, mechanical strain, and hemodynamic stress)[15, 16] and cellular processes (eg. cardiac fibroblasts migration, MyoF activation)[7] that play a role in cardiac fibrosis, there are a limited number of therapies available that effectively target fibrosis associated heart disease.[8, 9, 17] An important cause of the limited development of improved and more specific therapies against cardiac fibrosis is the lack of biomimetic in vitro platforms to investigate the fibrogenic remodeling after cardiac injury.[4] A suitable in vitro model would preferably maintain cardiac fibroblasts in a quiescent state and enable the integration of more physiological factors, such as contractile tissue activity, cell-cell, cell-ECM, and paracrine and hormonal interactions. Thus there exists a need for a novel, in vitro model system to study the pathological changes in biomimetic and in vivo-like conditions. These systems could not only be used for studying fibrotic changes in heart tissue, but they can potentially contribute to the development of more physiologically relevant assay systems for drug screening.[18]
During the last decade, tissue engineering strategies have shown promise in designing biomimetic in vitro models of cardiac tissue through the use of cardiac cells encapsulated in three-dimensional (3D) hydrogel-based ECM.[19, 20] For instance, the use of a gelatin methacryloyl (GelMA)-based hydrogel in creating a functional and contractile cardiac tissue was demonstrated by the successful encapsulation of cardiomyocytes and cardiac fibroblasts in a gelatin-based hydrogel.[19, 21] In particular, Saini et al have focused on developing and optimizing the co-culture conditions with cardiomyocytes and cardiac fibroblasts within GelMA hydrogels to create healthy and functional cardiac tissue constructs.[19] Despite the significant achievements in this area, the previously engineered 3D cardiac-like tissues have not been used to study the pathological remodeling that occurs during cardiac fibrosis. Furthermore, to date, most of the model systems that have been used to study this, were based on either 2D[22] or mono-cultures of cardiac fibroblasts.[22, 23] However, several 3D culture models using different natural (eg. collagen, hyaluronic acid) and synthetic-derived (eg. polyethylene glycol (PEG)) hydrogels have been developed to control and direct the activation of cardiac fibroblasts and fibroblast-like cells into MyoFs using mechanobiological cues.[22–24] However, there are still remaining challenges in engineering cardiac-like tissues to study MyoF activation and the associated fibrotic remodeling. Consequently, to successfully resemble the fibrotic remodeling process, some of the crucial factors that need to be incorporated within in vitro culture platforms are different cardiac cells in an in vivo-like 3D microenvironment, which can be stimulated (externally) to exhibit a fibrosis phenotype. For example, a recent study found increased MyoF activation when cardiac fibroblasts were encapsulated within a stiff hydrogel condition, as opposed to culturing the cells within softer matrices.[23] Additionally, a better functional and contractile phenotype is observed in cardiomyocytes when cultured on relatively softer hydrogel-based matrices.[25, 26] These studies have suggested that mechanical properties are a key factor in creating native-like heart tissues in vitro. Consequently, physiologically relevant mechanical cues should be incorporated into 3D matrices to fabricate a cardiac tissue model and to maintain cardiac fibroblasts in a quiescent state. Then biochemical components, such as TGF-β1, could be applied to activate the cardiac fibroblasts towards MyoFs to demonstrate an increased synthesis and deposition of ECM components (eg. collagen-I, fibronectin) by TGF-β1 signaling.
In the present study, we developed a 3D hydrogel-platform, composed of primary cardiomyocytes and cardiac fibroblasts, which are used to engineer a physiologically relevant in vitro platform to control the activation of cardiac fibroblasts towards MyoFs. We hypothesized that by mechanically tuning the stiffness of the hydrogels, a native-like ECM environment can be created to enhance the quiescent state of cardiac fibroblasts, and the functional behavior of engineered cardiac tissues. Therefore, the concentration and degree of methacrylation of GelMA macromers, which are important parameters to tune the stiffness of the GelMA hydrogel, was optimized for mimicking the mechanical stiffness of native heart tissue, allowing cardiac fibroblasts to remain in a quiescent state. In addition, the physiological properties of these in vitro cardiac tissues were characterized and the pro-fibrotic consequences of a TGF-β1 induced activation of cardiac fibroblasts were observed. We believe that this disease model of myocardial fibrosis may be a suitable in vitro model to study bio-mechanistic processes of cardiac fibrosis. Moreover, this platform could contribute to the development of better biomimetic pre-clinical drug screening platforms.
2. Results
2.1 Engineering and Characterization of GelMA Scaffolds
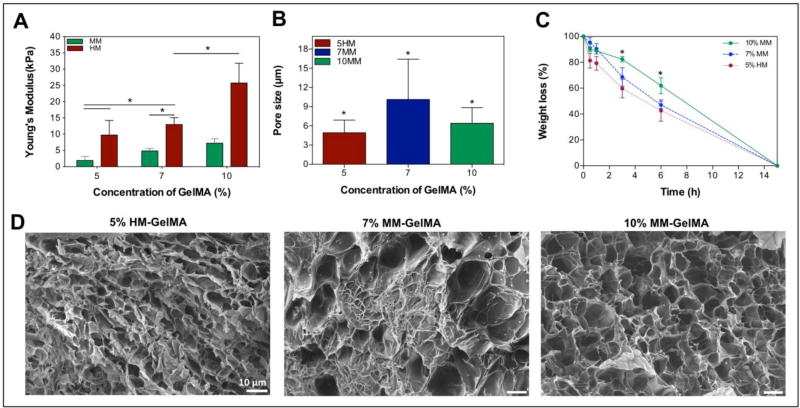
The elastic moduli of a healthy neonatal rat heart ranges from 4 to ~11 kPa.[25] In this study, we encapsulated cells from neonatal rat hearts in a GelMA-based hydrogel to engineer 3D myocardial tissues in vitro. The hydrogels showed an increased stiffness with increasing methacryloyl modification degree and macromer concentration (Figure 2A). As such, 10% HM-GelMA hydrogel exhibited the highest mechanical stiffness (25.76 ± 6.07 kPa) compared to all other hydrogel conditions (p < 0.05). However, 7% HM-GelMA hydrogel (12.97 ± 2.12 kPa) showed a significantly higher compressive modulus than 7% MM-GelMA hydrogel (4.48 ± 0.76 kPa) (p < 0.05) and a significantly lower compressive modulus when compared to 10% HM-GelMA hydrogel. This verifies that the mechanical stiffness can be tuned by varying the methacryloyl modification degree and macromer concentration of GelMA independently.[21] The following three hydrogels 5% HM-GelMA (9.76 ± 4.48 kPa), 7% MM-GelMA (4.48 ± 0.76 kPa), and 10% MM-GelMA (7.25 ± 1.38 kPa), exhibited a mechanical stiffness that was in the range of native neonatal rat hearts (Figure 2A) and were therefore further characterized.
Figure 2. Mechanical, porosity and degradation properties of GelMA hydrogels.
A) pre modulus of GelMA-hydrogels varies with different macromer concentration and degree of methacryloyl modification. B) Pore size analysis of GelMA hydrogels. C) Degradation of GelMA hydrogels with various macromer concentration and methacryloyl modification degree in the presence of collagenase. D) Cross-Sectional scanning electron microscopy images of 5% HM-GelMA, 7% MM-GelMA, and 10% MM-GelMA hydrogels reveal different porosity. Data depict Mean ± Standard deviation. *p<0.05
To access the effect of the GelMA macromer concentration and methacryloyl modification degree on the morphology of hydrogel, all SEM samples were prepared by same cryogenic treatment. SEM images indicated that all three selected hydrogels showed highly microporous structure which were obtained by lyophilization (Figure 2).[27] There is a significant decrease in porosity with an increased macromer concentration. In addition, a significantly lower porosity was observed in the 5% HM-GelMA hydrogel as compared to the 7% and 10% MM-GelMA hydrogels (Figure 2B). This indicates an inverse relationship between porosity and degree of methacryloyl modification and macromer concentration. Although there was a significant decrease in the porosity of 5% HM-GelMA hydrogel, no significant increase was observed in the mechanical properties of 5% HM-GelMA hydrogel when compared to 7% and 10% MM-GelMA hydrogels (Figure 2A).
In native cardiac tissue, MMPs are excreted and activated by cells to induce and promote the cleavage of ECM components.[28] These proteins play an important role in the maintenance and remodeling of the heart ECM. To assess the presence of physiological binding substrates for an MMP-mediated degradation of the GelMA-based scaffold, a degradation assay was performed with collagenase type II (also known as MMP-8). The results revealed significantly faster degradation – described as percentage of weight loss – of 5% HM-GelMA hydrogel after 3 and 6 h when compared to 10% MM-GelMA hydrogel (p < 0.05). Furthermore, a complete degradation of all three hydrogels was observed after 15 h of incubation with MMP-8 (Figure 2C). These results confirm the existence of MMP substrates in the GelMA hydrogel but also indicate the opportunity for use of GelMA based scaffolds for engineered physiological heart ECM tissue.
2.2 GelMA Hydrogel Characteristics Affect Cell Spreading but not Viability
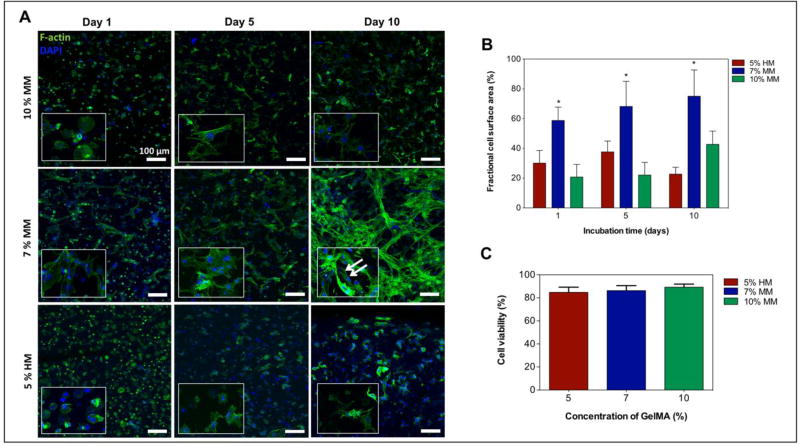
To determine what hydrogel condition enabled the best cellular spreading and networking, we investigated the spreading of encapsulated cells inside the three selected GelMA hydrogels. To visualize this, fluorescent confocal z-stack images were taken after F-actin (cytoskeletal fiber) staining of the cell-laden hydrogels (Figure 3A). After 10 days of culture, the fluorescence images demonstrated that the majority of the cells inside 5% HM-GelMA and 10% MM-GelMA hydrogels had limited spreading, as the cells retained a round shape. Interestingly, the expression of F-actin fibers throughout the 7% MM-GelMA hydrogel clearly demonstrated an increase in cellular spreading and networking (Figure 3A). A higher cellular spreading was expected due to a lower methacryloyl modification degree and macromer concentration, thereby allowing for an increased degradation and spreading throughout the ECM by the cells. Therefore, a higher cellular spreading and networking in 7% MM-GelMA hydrogel could be attributed to the lower macromer concentration and degree of methacryloyl modification as compared to 10% MM-GelMA and 5% HM-GelMA hydrogels, respectively. Additionally, a quantitative analysis of the area covered by F-actin fibers confirmed a significantly higher percentage of fractional coverage in 7% MM-GelMA hydrogel (74.98 ± 17.70 %) compared to 5% HM-GelMA (22.71 ± 4.66 %) and 10% MM-GelMA (42.68 ± 8.98 %) hydrogels at day 10 of culture (p < 0.05) (Figure 3B).
Figure 3. Viability and spreading characteristics of cardiac fibroblasts and cardiomyocytes encapsulated in mechanically tuned GelMA hydrogels.
A) Representative fluorescence images of encapsulated cardiac fibroblasts and cardiomyocytes within various GelMA hydrogels at day 1, 5, and 10 of culture. B) Representation of a quantitative analysis of fractional F-actin coverage within selected windows of 400 µm × 400 µm. C) Quantitative analysis of the viability of cardiomyocytes and cardiac fibroblasts within various GelMA hydrogels conditions on day 1 of culture. Data depict Mean ± Standard deviation. *p<0.05
The viability of encapsulated cells in the different conditions was also assessed after one day of culture. This time point was chosen to evaluate the survival of cells following UV exposure during the fabrication of the constructs. Figure 3C depicts the quantitative analysis of the percentage of live cells at day 1 of culture. Across all experimental conditions, the percentage of live cells was higher than 84% and was not significantly different between groups.
2.3 Functional Properties of the 3D Engineered Cardiac Tissues
For further experiments, we selected the 7% MM-GelMA hydrogel for having the best spreading and networking features for the cells. Additionally, we studied the viability of cells in this condition for 2 weeks of culture. The results revealed that the engineered cardiac tissues remained viable throughout a culture period of 14 days (Figure 4A). Compared to day 1, there was a significant increase in the metabolic activity after 14 days (Figure 4A) (p < 0.05). Given the fact that cardiomyocytes have limited proliferative capacity[19], we believe that cardiac fibroblasts were responsible for the increase in metabolic activity.
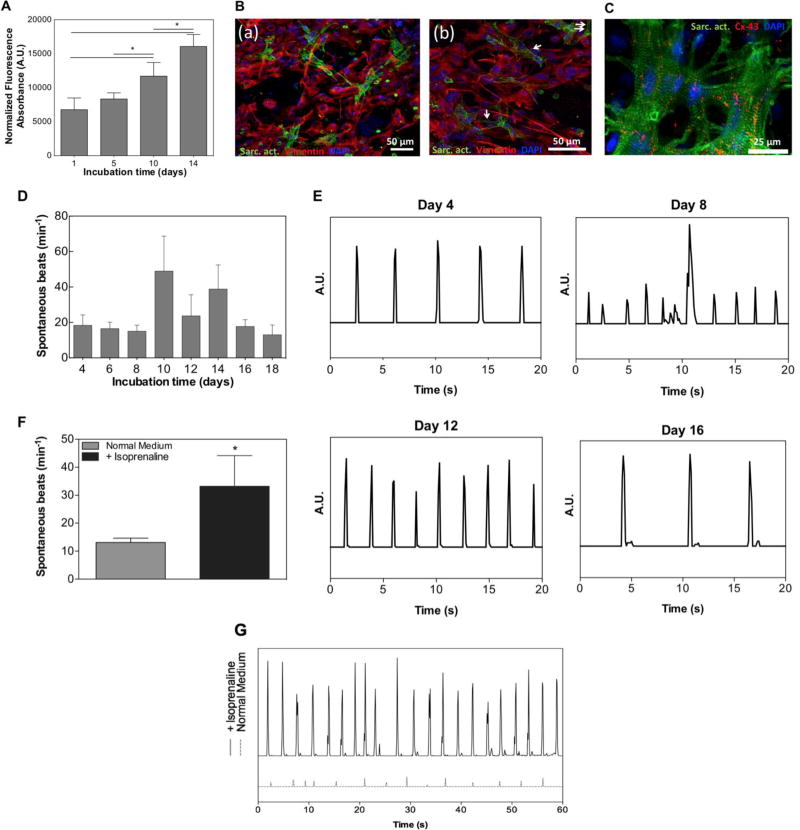
Figure 4. Functional Properties of 3D engineered cardiac tissues.
A) Quantitative analysis of cellular metabolic activity throughout 14 days of culture. B) Representative fluorescence images of immunostained cardiomyocytes (α-sarcomeric actin = green) and cardiac fibroblasts (vimentin = red) on day 14 of culture (a). Higher magnification images of immunostained cardiomyocytes and cardiac fibroblasts showed sarcomeric cross-striations (white arrows) (b). C) Fluorescent image showing well developed sarcomeric striations (= green) and the expression of a gap-junctional protein, connexin-43 (= red). D) Quantitative representation of the spontaneous beats per minute of 3D engineered cardiac tissues from day 4 up until a maximum of 18 days of culture. E) Representative beating pattern of the cardiac tissues at day 4, 8, 12, and 16 of culture. F) Quantitative analysis of the spontaneous beats per minute of cardiac tissues (n=3) in the absence and presence of 1µM isoproterenol (isoprenaline). G) Representation of the beating pattern of cardiac tissues in the absence and presence of isoproterenol at day 6. Data depict Mean ± Standard deviation. *p<0.05
On day 14, we also assessed the phenotype of the encapsulated cardiomyocytes and cardiac fibroblasts by immunostaining with sarcomeric α-actinin and vimentin. As depicted in Figure 4B, a confocal z-stack image of an immunostained cell-laden GelMA hydrogel displayed both sarcomeric α-actinin and vimentin positive cells. In addition, we investigated the expression of connexin-43, a gap junction protein that is important for electrical coupling of cardiomyocytes and is typically found in the cardiac tissue.[29] From the confocal image in Figure 4C, it is clear that cardiomyocytes demonstrated the expression of both connexin-43 and sarcomeric α-actinin. The expression of gap-junctions and functional electro-mechanical coupling was also confirmed by the observation of spontaneous, synchronous, and cardiac tissue-like contraction of the engineered 3D cardiac tissues (Supplemental Video 1, supporting information). Spontaneous individual cell beating activity began after 2 days of culture; however, videos of the beating were recorded and analyzed from when synchronous and tissue-like contraction began (Figure 4D). The engineered cardiac tissues maintained synchronous and tissue-like contraction for as long as 18 days of culture (Supplemental Video 2, supporting information). Additionally, quantitative analysis of synchronous beats per minute (BPM) revealed that the constructs reached a maximum of 48 (± 19.75) BPM on day 10 (Supplemental Video 3, supporting information) and a minimum of 13 (± 5.57) BPM on day 18 of culture. This variable beating behavior is consistent with previously reported studies on engineered GelMA-based cardiac tissues in vitro.[19, 30, 31] The decrease in beating rate with increasing culture time was also shown in previous studies, in which rat neonatal cardiomyocytes were cultured on hydrogel-based tissue-engineered models.[32, 33] In one of these studies, it was hypothesized that this decrease might be attributable to the phenotypical transition of fetal cardiomyocytes towards the neonatal stage.[33] We further went on to analyze the beating pattern of the beating constructs. As depicted in Figure 4E, the cardiac tissues showed a stable and regular beating pattern throughout culture days 4, 8, 12, and 16.
To assess physiological functionality of the engineered cardiac tissues, we also investigated the effect of a beta-adrenergic drug, isoproterenol (isoprenaline), on the beating behavior on day 6. Videos of the beating samples were recorded before and after 45 min incubation with 1 µM isoproterenol. An analysis of the videos revealed that the cardiac tissues exhibited physiological behavior in response to the drugs (Supplemental Video 4 and 5, supporting information). Figure 4F and 4G show the synchronous beating pattern (and amplitude) and beating frequency (in beats/min), respectively, of the cardiac tissues before and after the administration of the drug. In the presence of isoproterenol, the cardiac tissues developed a significant increase in their spontaneous beating frequency (Figure 4F). Furthermore, an increase in the amplitude of all the samples was observed after exposure to the drug (Figure 4G).
2.4 TGF-β1 Induces Proliferation of Cardiac Fibroblasts
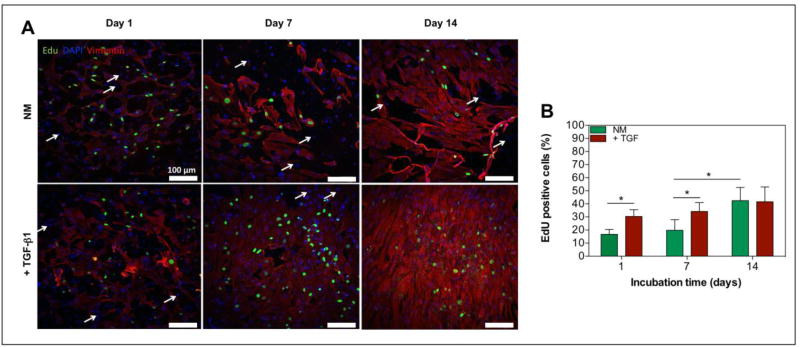
TGF-β1 is a well characterized protein in the pathophysiology of cardiac fibrosis, and is a potent stimulator of cardiac fibroblast proliferation during the course of this disease.[14] In order to assess the effect of TGF-β1 on cell proliferation within our engineered 3D cardiac tissues, we analyzed cell proliferation by EdU labeling. Fluorescent z-stack images of the EdU labeled cardiac tissues were taken on day 1, 7, and 14 of culture (Figure 5A). To identify the cellular phenotype of proliferating cells, we stained the cardiac fibroblasts by positive immunostaining for vimentin (Figure 5A). From Figure 5A, it is clear that the EdU labeled cells (green) were also positively stained by vimentin (red), thus confirming that the increase in proliferation across the two culture conditions was attributed to the proliferation of cardiac fibroblasts. Furthermore, a quantitative analysis of the percentage of EdU stained cells, revealed that the TGF-β1 treated samples showed a significantly higher number of EdU positive cells compared to the non-treated samples on day 1 and 7 (Figure 5B) (P < 0.05). However, there was no significant difference on day 14 between the percentage of EdU positive cells in TGF-β1 treated (41.63 ± 11.31 %) and non-treated samples (42.45 ± 10.11 %). Additionally, the percentage of EdU labeled cells in the non-treated samples had a significant increase by day 14 (42.45 ± 10.11 %) compared to day 1 (16.74 ±3.70 %) (Figure 5B) (p < 0.05). A positive control analysis of the proliferation was also obtained by the EdU labeling of TGF-β1 treated and non-treated cardiac fibroblasts after 24h of 2D–seeding in a well-plate (Supplemental Figure 2, supporting information). As expected, the results were consistent with the results from the engineered cardiac tissues.
Figure 5. The exogenous addition of TGF-β1 affects proliferation of cardiac fibroblasts in 3D engineered cardiac tissues.
A) Confocal images of immunofluorescence staining of a cardiac fibroblast marker, vimentin (= red), and EdU click-iT labeling (= green) of 3D engineered cardiac tissues with and without the addition of TGF-β1 at day 1, 7, and 14. Cardiomyocytes were not stained and showed no positive EdU labeling (white arrows) B) Representative quantification of proliferating cells inside 3D cardiac tissues as determined by the percentage of EdU positive cells at day 1, 7, and 14 of culture (n=3). Data depict Mean ± Standard deviation. *p<0.05
2.5 Characterization of the Expression of Cardiac Fibrosis Markers
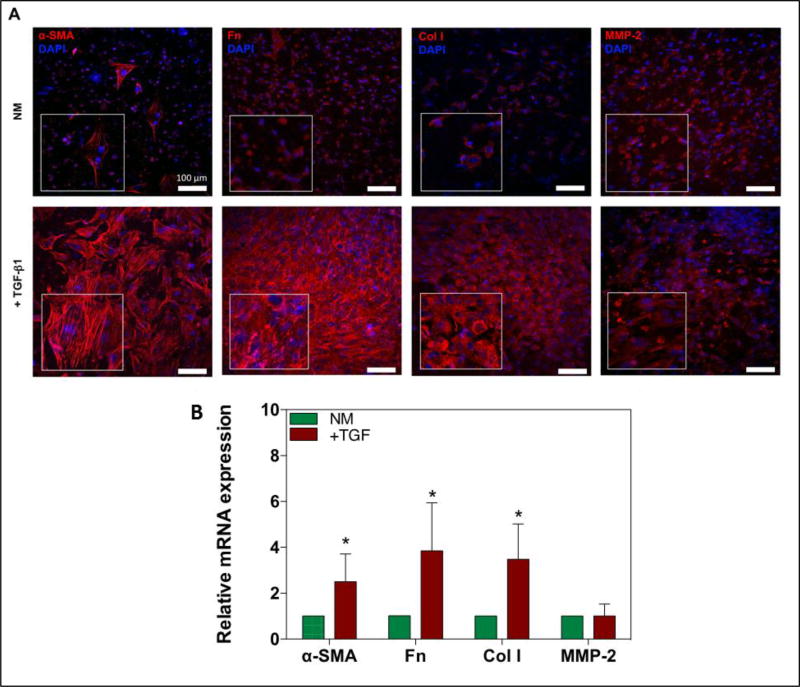
Previous studies have demonstrated that quiescent cardiac fibroblasts spontaneously differentiate into activated MyoFs when cultured in 2D tissue culture polystyrene (TCPS) plates [23, 60]. This is thought to be partly due to the higher mechanical stiffness (GPa range) of TCPS compared to native and even fibrotic myocardium.[15, 34] Positive control of CF activation on TCPS conditions was obtained by culturing CFs in normal and TGF-β1 supplemented medium for 24h. No significant difference was found in the expression of fibrotic markers (α-SMA, collagen-I, fibronectin, and MMP-2) after incubation of these CFs in normal medium compared to the TGF-β1 containing medium for 24h (Supplemental Figure 3A and B, supporting information). To investigate the activation of cardiac fibroblasts in a 3D beating heart-like environment and stiffness, we analyzed the expression of a specific MyoF protein marker, α-SMA after 14 days of culture (Figure 6A). As depicted in Figure 6A, cells in the cell-laden hydrogels showed a minimal expression of α-SMA, indicating that cardiac fibroblasts within the cardiac tissues remained in a quiescent state when cultured in normal culture medium. However, engineered 3D cardiac tissues cultured in the presence of TGF-β1, exhibited a more MyoF-like phenotype after 14 days, as can be seen in Figure 6A. Furthermore, we analyzed the expression of other cardiac fibrosis markers by positive immunofluorescence staining of collagen-I, fibronectin, and MMP-2. Overall, confocal z-stack images clearly demonstrated an increased expression of fibronectin and collagen-I inside the engineered 3D cardiac tissues cultured in TGF-β1 supplemented medium. However, the expression MMP-2 was not clearly increased in the TGF-β1 treated samples. In addition to qualitative protein expression analysis, we also investigated the mRNA expression of α-SMA, collagen-I, fibronectin, and MMP-2 by quantitative real time polymerase chain reaction (RT-PCR) to confirm the upregulation of fibrotic protein expression in the TGF-β1 stimulated samples (Figure 6B). The expression levels of α-SMA, collagen-I, and fibronectin were shown to be elevated for TGF-β1 stimulated samples when compared to normal growth medium. The greatest increase was observed in the fibronectin samples, where the mRNA expression was 3.84 (± 2.33) (p < 0.05) times higher in the TGF-β1 treated samples as compared to the control group. Additionally, mRNA expression levels of α-SMA and collagen-I were shown to be 2.51 (± 1.21) (p < 0.05) and 3.48 (± 1.55) (p < 0.05) times higher, respectively, in TGF-β1 cultured samples as compared to control medium. However, as demonstrated by the protein expression analysis (Figure 6A), the expression of MMP-2 was not significantly changed in TGF-β1 stimulated samples (1.01 ± 0.53). Altogether, these results indicated that a quiescent cardiac fibroblast phenotype could be effectively cultured in a mechanically tuned GelMA-based cardiac tissue construct. Moreover, these results demonstrated that the phenotypic state of cardiac fibroblasts can be directed by designing a 3D cardiac tissue with a physiological co-culture of cells and an in vivo-like dynamic contraction.
Figure 6. Increased expression of fibrotic makers and increased differentiation of quiescent cardiac fibroblasts into MyoFs by TGF-β1.
A) Confocal images of immunofluorescence stained markers of cardiac fibrosis and MyoF differentiation; α-SMA, collagen-I (Col I), fibronectin (Fn), and MMP-2 after 14 days of culture. B) Data representing RT-PCR of mRNA expression of α-SMA, collagen-I, fibronectin, and MMP-2 in normal culture medium (NM) compared to NM + TGF-β1 after 14 days of culture. Data depict fold-change ± standard deviation. *p<0.05.
2.6 Analysis of the TGF-β1 induced Pro-Fibrotic Changes
During fibrotic remodeling there is a higher risk of arrhythmogenicity as a result of an increased ECM deposition and an altered electrical-coupling between cardiomyocytes and MyoFs.[12, 35] In addition, human fibrotic cardiac tissue is hallmarked by an increased mechanical stiffness (~30–70 kPa) when compared to healthy myocardial tissue (~10 kPa).[36, 37] Furthermore, MyoFs can generate a higher contractile force than cardiac fibroblasts and thereby induce contraction and scarring of the cardiac tissue. We hypothesized that the engineered cardiac tissues would exhibit some of these fibrotic characteristics when the fibrotic response is simulated. To assess the pro-fibrotic changes, we cultured the samples in normal culture medium and added TGF-β1 for 14 days. All TGF-β1 stimulated cardiac tissues showed spontaneous individual cell beating, from day 7 to day 13. Video analysis of the beating behavior revealed that there was a non-synchronous and irregular contraction of all the tissues when stimulated with TGF-β1 (Figure 7A and Supplemental Video 6, supporting information). These results correlated well with previous reports that MyoFs induce changes in the beating behavior of cardiomyocytes.[12, 35]
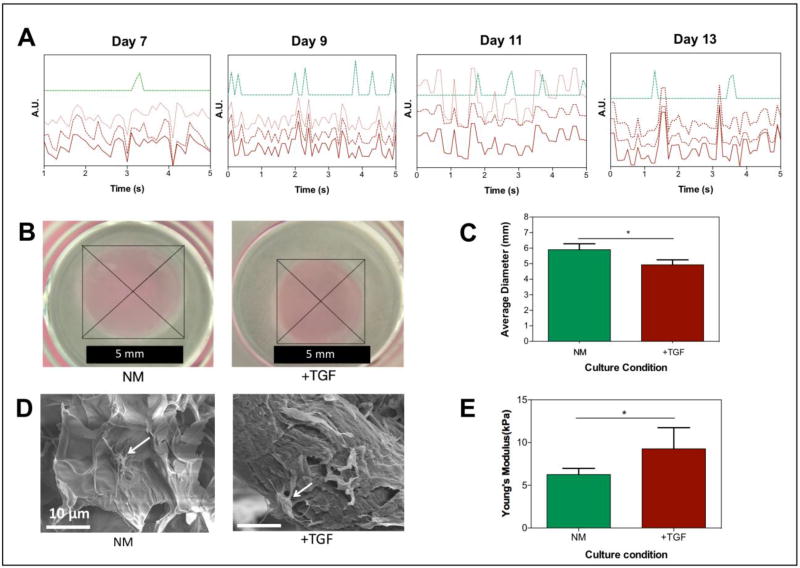
Figure 7. TGF-β1 induces pro-fibrotic changes, such as increased contractility of hydrogels, increased mechanical stiffness, and asynchronous beating, in 3D engineered cardiac tissues.
A) Beating patterns of 3D engineered cardiac tissues cultured in NM (green) and NM + TGF-㬡1 (red) at day 7, 9, 11, and 13. The three red lines (solid and 3dotted) represent three independently areas of beating within the same area of view. B) Hydrogel contraction test. Optical images of TGF-㬡1 treated and non-treated hydrogels with encapsulated cardiomyocytes and cardiac fibroblasts. C) Quantitative analysis of the contraction test of cardiomyocytes/cardiac fibroblast encapsulated GelMA hydrogels in NM compared to NM + TGF-㬡1. D) Representative scanning electron microscopy images of cell (white arrows)-encapsulated GelMA hydrogels cultured in NM and NM + TGF-㬡1 on day 14 of culture. E) Mechanical stiffness of the 3D cardiac tissues in the two different culture conditions (NM and NM+TGF-㬡1) at day 14 of culture. Data depict Mean ± Standard deviation. *p<0.05
Compared with normal culture medium, we also observed a significant decrease of ~17 % in the average diameter of the GelMA-based cardiac tissues, thus indicating an increase in the MyoF-mediated contraction of the hydrogel (Figure 7 B , C) (p < 0.05). In addition, SEM images of both culture conditions showed that cardiac tissues from TGF-β1 stimulated samples clearly had a more fibrous and fibrillar structure than cardiac tissues in the control group (Figure 7D). This is thought to be mainly attributable to the elevated deposition of ECM components (eg. collagen-I, fibronectin) in the fibrotic-like tissues. On day 14, the mechanical stiffness of TGF-β1 treated engineered 3D cardiac tissues was compared to cardiac tissues cultured in normal culture medium. These measurements revealed an increase in the mechanical stiffness of TGF-β1 stimulated encapsulated cells in GelMA hydrogels (Figure 7E).
Our findings highlight the opportunity to use these GelMA-based engineered 3D cardiac tissue constructs to identify fibrotic changes, and to study the pathophysiological cells and factors that play a role in cardiac remodeling and myocardial fibrosis.
3. Discussion
For many decades, conventional TCPS plates have successfully contributed to a better understanding of fibrogenesis in the context of cardiovascular diseases. However, these models lack the in vivo like presence of tissue-level properties (such as cell-ECM interactions). Furthermore, cardiac fibroblasts cultured in a 2D TCPS plate, spontaneously activate into MyoFs, thereby complicating the ability to study phenotypical changes of these cells during disease development. In this work, we designed a simplified 3D in vitro model of cardiac fibrosis by tuning the mechanical ECM of engineered cardiac tissues followed by stimulating these hydrogel-based tissues with TGF-β1; an established and potent mediator in fibrotic remodeling.[14] During cardiac fibrosis, there is an increased synthesis and deposition of ECM components, including collagen (mainly type I and III), laminin, fibronectin, and elastin.[4, 38] An increase in the expression of collagen-I and accumulation of fibrillar collagen deposition directly influences the structural integrity, biochemical, mechanical, and electrical properties of the cardiac tissue, and is considered as a hallmark of cardiac fibrosis.[5b] During this process, the main effector cells are activated MyoFs and proliferating cardiac fibroblasts. In addition, there is an increased (early phase remodeling) and decreased (late phase remodeling) production and activation of MMPs, which play a role in the degradation of the ECM.[28, 39] This is of importance since it can enable and facilitate the migration of cells (eg. cardiac fibroblasts) to the area of injury at the early phases of wound healing.[40] The elevated expression of pro-fibrotic genes such as collagen-I, fibronectin, and α-SMA in our stimulated cardiac tissue was consistent with the pathological changes that occur during cardiac fibrosis. Similarly, we observed an activation of quiescent cardiac fibroblasts by positively immunostaining α-SMA; a widely used marker of MyoFs.[41] Immunostaining images further revealed that the TGF-β1 stimulated tissues also increased the expression of collagen-I and fibronectin. Our results also indicated that there was an induced proliferation of cells in the TGF-β1 treated cardiac tissues, as identified by EdU labeling. It is well established that TGF-β1 is an inducer of cardiac fibroblast activation during pathological fibrotic remodeling.[42] In addition, TGF-β1 is a proliferative stimulator of cardiac fibroblasts.[42, 43] Therefore, we investigated whether the proliferating cells are cardiac fibroblasts or cardiomyocytes by simultaneously labeling with EdU and immunostaining specifically for cardiac fibroblasts. Our results showed that the proliferating cells were cardiac fibroblasts rather than cardiomyocytes, which have a low proliferative capacity.[44]
This disease model also showed that it could recapitulate functional properties of MyoFs and fibrotic cardiac tissue. TGF-β1 induced stimulation of the functional cardiac tissues resulted in an asynchronous and irregular beating behavior. Furthermore, the increased conversion of cardiac fibroblasts into MyoFs, resulted in a higher contraction and shrinkage of the 3D GelMA hydrogels. This however, could also be a result of degradation of GelMA hydrogel due to the production of various MMPs in the TGF-β1 stimulated tissues.
GelMA hydrogels have been a widely used scaffolding biomaterial in the past few years for applications ranging from tissue engineering[30], organs-on-a-chip,[45] and in vitro disease models.[24] Previous studies have suggested that photocrosslinked cell-laden GelMA hydrogels are a suitable matrix to create functional tissue constructs. It has been reported that UV light alone at low intensity (6 mW/cm2 or 4 mW/cm2) for several minutes does not affect cell viability and cell cycle significantly.[62] Furthermore, previous studies have suggested that photocrosslinked cell-laden GelMA hydrogels are a suitable matrix to create functional tissue constructs for various tissue engineering applications (such as cardiac and cartilage).[4, 21, 24, 47, 63a–c] Moreover, a high cell viability (>80%) after photocrosslinking of the cell-laden GelMA hydrogels has been reported in the literature. [47] Similarly, in this study, we observed a viability of >84% after 1 day of culture throughout all GelMA hydrogel conditions. Therefore, in this study, we do not expect that the exposure times used to adequately crosslink the GelMA (20 sec) had a significant effect on cell viability and behavior. Additionally, most of the previous studies take advantage of the easy fabrication methods, high and easy accessibility, and high biocompatibility of GelMA hydrogels.[21] Moreover, GelMA hydrogels can be tuned mechanically to obtain a physiologically relevant stiffness.[21, 46] This can be obtained by varying the macromer concentration, the degree of methacryloyl modification, the concentration of photoinitiator, the UV intensity, or the UV exposure time.[21] Although GelMA hydrogel is a semi-synthetic, photocrosslinkable scaffolding material, it provides relevant biomimetic cues of the native ECM, such as RGD-binding peptides and MMP degradable sites.[47] In addition, gelatin is a denaturized form of collagen, which is the main constituent of native heart tissue, and therefore exhibits comparable biocompatible and biomimetic properties to collagen. However, a limitation of this in vitro system is the lack of other ECM components that are present in the native heart (eg. collagens, glycosaminoglycans, laminin, and elastin). Interestingly, previous studies have shown that it is possible to engineer functional cardiac tissue by using naturally derived decellularized ECM from native heart tissues.[48] In a recent study, Visser et al. have combined the fabrication methods of GelMA molecules to engineer crosslinkable hydrogels derived from various native tissues.[49] Thus indicating that in future work, it could be possible to combine the advantages of GelMA hydrogels with the native properties of a decellularized heart ECM.
Mechanotransduction is a process in which cells sense their mechanical microenvironment and transmit the physical stresses of their surroundings to biochemical signals that result in various cellular outputs (eg. differentiation, proliferation).[50, 51] These mechano-physical signals are mainly mediated through cell-ECM and cell-cell connections, which are converted to cellular signaling pathways by integrins, focal adhesions, and cadherins.[4, 50, 52] Cardiac fibroblasts and cardiomyocytes can sense their extracellular microenvironment by attaching to their ECM with focal adhesions.[53] Increasing ECM stiffness or ECM-induced strain, can affect both cardiomyocytes and cardiac fibroblasts in a way that alters contractile function and stimulates MyoFs activation.[15, 54] Consequently, the mechanical stiffness of the extracellular microenvironment of cardiac tissue is an important factor in both normal physiology and cardiac fibrosis. In fact, in a previous study by Engler et al. it was reported that cardiomyocytes cultured on stiff, fibrotic-like (34 kPa) substrates, showed a decreased beating activity and lacked the development of well-striated sarcomere structures.[37] In addition, results from other recent studies have demonstrated that the mechanical stiffness of the matrix or substrate can facilitate the transition of cardiac fibroblasts into MyoFs.[22, 23] Zhao and colleagues engineered PEG-substrates with varying degrees of stiffness to study the migration, proliferation, and activation of quiescent cardiac fibroblasts in a 2D in vitro model system.[22] This culture platform suggested that the quiescent state of cardiac fibroblasts could be maintained and directed by mechanically tuning a hydrogel-based substrate. These results highlight the importance of the tunability of matrix stiffness for the engineering of functional and contractile cardiac tissues, while simultaneously maintaining the quiescent-like phenotype of cardiac fibroblasts. In this study, we used 7% MM-GelMA hydrogel (4.48 ± 0.76 kPa) to engineer a physiological stiffness in the same range of native neonatal rat hearts (4–11 kPa).[25] We showed that neonatal rat cardiac fibroblasts remained in a quiescent-state while co-cultured with cardiomyocytes in a mechanically tunable 3D hydrogel model.
Cardiac muscle is a syncytium in which a network of cardiomyocytes and cardiac fibroblasts are connected to each other electrically and mechanically. In order to engineer a physiologic-like heart tissue in vitro, it is essential for cardiomyocytes and cardiac fibroblasts to interact with each other through both direct, and indirect cell-cell and cell-ECM interactions. Here, we hypothesized that the spreading and networking of cells over time would lead to the development of a system which better mimics native cardiac tissue. Consequently, 7% MM-GelMA hydrogel was selected as a 3D substrate for culturing cardiac fibroblasts and cardiomyocytes. The expression of cardiac differentiation markers (sarcomeric α-actinin, connexin-43) demonstrated a well-developed electrical coupling and contractile apparatus inside these hydrogel conditions. Although the cardiomyocytes had an isotropic orientation inside GelMA hydrogel, there was a clear elongation and a well-defined sarcomeric structure visible. In native cardiac tissue however, the cardiomyocytes are elongated and aligned in anisotropic layers of muscle tissue.[55] This is important for the anisotropic propagation of electrical signals, which plays a critical role in synchronous and rhythmic cardiac contraction.[56] One limitation of our engineered cardiac tissue was the lack of cellular anisotropic alignment, which could potentially lead to impaired electrical pulse propagation and contraction in comparison to the native heart muscle. In future studies, we could overcome this through the use of engineering strategies by applying topographical and electrical cues to the tissues to enhance the elongation and alignment of cells.[57]
In this work, we used neonatal rat cardiomyocytes and cardiac fibroblasts to model an in vitro platform of myocardial fibrosis. Previous reports have shown evidence that neonatal cardiomyocytes possess several advantages over adult cardiomyocytes.[59a] In particular, neonatal rat cardiomyocytes seeded on hydrogel-based substrates exhibit spontaneous beating after seeding, whereas adult cardiomyocytes often require external electrical stimulation.[19, 31, 32] By enabling spontaneous contraction of the engineered tissue constructs, a more physiological mechanical cue could be added to the tissue constructs. Additionally, adult cardiomyocytes are terminally differentiated cells and therefore do not have the ability to proliferate in vitro. This, however, would be desirable in a cardiac fibrosis model since activated cardiac fibroblasts may outgrow cardiomyocytes more readily, thereby complicating the ability to analyze the electrical changes of cardiac tissue during fibrogenesis. Furthermore, aging of the heart affects cardiac phenotype by inducing cellular hypertrophy and fibrotic changes. [59b–59c] Consequently, an adult heart or a morphologically more fibrotic heart, may not be a suitable cell source to study the phenotypical changes that take place during fibrogenic remodeling. Moreover, to date, different studies have exploited the use of neonatal rat cardiac fibroblasts as an in vitro model of myofibroblast differentiation, suggesting that neonatal rat heart cells are a suitable source for modeling this process. [59c, 24] Additionally, the use of rats for in vitro studies of heart disease is the standard in both academia and the pharmaceutical and biotechnology industry. Therefore, this indicates that our model may be a suitable and pre-clinically relevant in vitro platform for studies of pathophysiology and drug screening applications.
However, we recognize the simplicity of this in vitro platform, as it lacks the incorporation of other dynamic (eg. blood flow) and physiological factors (eg. blood vessels, inflammatory cells), which are present in the native heart. Recently, microfluidic organs-on-a-chip have emerged and demonstrated the possibility of incorporating several dynamic factors (such as fluid flow and mechanical stretch) within microengineered cardiac tissues.[58] Further development of microfluidic techniques and tissue engineering strategies could therefore aid in the creation of a more physiologically relevant cardiac tissue in the future.
4. Conclusion
In this study, we engineered a simplified but functional and physiologic-like heart tissue to study cardiac fibrosis in a 3D GelMA-based hydrogel platform. By tuning the mechanical stiffness of GelMA hydrogels, we were able to create a well-defined and beating network of cardiomyocytes and quiescent cardiac fibroblasts. Subsequently, we showed that we were able to stimulate the activation of cardiac fibroblasts into MyoFs by adding TGF-β1 to the culture medium. Furthermore, our results demonstrated that the engineered fibrotic-like tissues presented non-synchronous beating behavior and mechanical alterations after incubation with TGF-β1. In particular, the fibrotic-like heart constructs demonstrated an increase in mechanical stiffness after 14 days of culture. In conclusion, our study presents a physiologic-like in vitro model of cardiac fibrosis that could enhance our understanding of this disease, while increasing the potential of these systems to be used for pre-clinical drug screenings.
5. Experimental Section
Synthesis of GelMA
GelMA was synthesized as described in a previous protocol.[21] Briefly, type A gelatin (10% (w/v)) from porcine skin (Sigma-Aldrich) was added to Dulbecco’s phosphate buffer saline (DPBS; Gibco). This mixture was then stirred and heated at 50 °C for 1h to obtain a clear gelatin solution. Subsequently, 1.25% (v/v) or 8% (v/v) methacrylic anhydride (Sigma-Aldrich) was added dropwise to synthesize middle- (MM) and high-degree methacryloyl modification (HM) GelMA. The solution was stirred and remained on a hot plate for 1h (middle methacryloyl modification) or 2h (high methacryloyl modification), after which, DPBS was added to stop the reaction. Following this, the GelMA solution was dialyzed (molecular weight cut off: 12 – 14 kDa) with deionized water for 10 days at 40 °C to remove any salts and unreacted methacrylate anhydride. Finally, the GelMA solution was filtered (0.2 µm), frozen (−80 °C), and lyophilized for 5 days to obtain GelMA foam. The foam was stored at room temperature until further experimental use.
Preparation of Hydrogel Constructs
GelMA pre-polymer solutions were prepared by dissolving 5%, 7%, and 10% (w/v) MM and HM GelMA in DPBS containing 0.25% (w/v) photoinitiator (PI; Igracure 2595 Sigma). The solutions were briefly vortexed, and placed in an oven at 80 °C for 15 min to obtain pre-polymer solutions of GelMA. To prepare disc-shaped hydrogels, 12 µL of the pre-polymer solution was pipetted between two 600 µm tall spacers and covered with a 3-(trimethoxysilyl) propyl methacrylate (TMSPMA) treated glass-slide (Supplemental Figure 1, supporting information). This pre-polymer solution was placed into a customized UV-chamber and exposed to UV light (800 mW, 360–480 nm) for 20 s, resulting in the creation of 600 µm tall hydrogel discs (Supplemental Figure 1, supporting information). After this, the GelMA hydrogels were removed manually from the glass slide and utilized for further experiments.
Cardiac Cell Isolation and Culture
Primary ventricular cardiomyocytes and cardiac fibroblasts were isolated from two-day-old neonatal Sprague Dawley rats. These procedures were based on a previously well-defined protocol approved by the Institution’s Committee on Animal Care.[33] Briefly, the hearts of neonatal pups were surgically removed from the thoracic cavity after euthanasia. Upon removing the atria, the ventricular tissues were cut into multiple small pieces and incubated overnight (at 4 °C) on a shaker in a 0.05% (w/v) trypsin solution prepared in Hank’s Balanced Salt Solution (HBSS, Gibco, USA). The heart tissues were subjected to four collagenase type II (LS004176, Worthington, Lakewood, NJ) digestions (10 minutes, 37 °C, 80 rpm) to further digest the heart tissues. The cell suspension was then collected, centrifuged (1000 rpm) for 5 min, and pre-plated for 1 h to enrich the cardiomyocytes for immediate experimental use. The attached cardiac fibroblasts were cultured for a maximum of three passages for future experimental use. The cardiac fibroblasts were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco USA) with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin/streptomycin (P/S; Gibco, USA). To identify the purity of cardiomyocytes, we have performed fluorescence-activated cell sorting (FACS) using Troponin-T as a CM specific marker immediately after cell isolation. Our data indicated that we have successfully obtained 77.3% Troponin positive cells, which we further used to adjust the concentration of CFs to achieve a final CM:CF ratio of 1:1 (Supplemental Figure 4, supporting information). In our previous study, we have demonstrated that a co-culture of CM:CF (1:1) exhibit cardiac-like tissue functions such as spontaneous beating, gap-junction expression and sarcomeric structure. [61]
Engineering Cell-Laden Hydrogel Constructs
To fabricate cell-laden hydrogel constructs, a pre-polymer solution was prepared with minor modifications to the described protocol above. In brief, 5%, 7%, and 10% (w/v) MM and HM GelMA was dissolved in DMEM containing 0.25% PI, 50% FBS, 1% P/S, and 2% (w/v) L-glutamine (Gibco USA). Pre-polymer solutions were removed from the 80 °C oven, and placed in a water bath at 37 °C until cell encapsulation. Cultured cardiac fibroblasts (passage 1–3) were trypsinized and mixed at a 1:1 ratio with the freshly isolated cardiomyocytes to obtain a final concentration of 25 × 106 cells/mL. The cells were centrifuged at 1200 rpm for 5 min, and the pellet was resuspended in the GelMA pre-polymer solution. Gels were created following the preparation of hydrogel constructs (above) and placed in culture medium containing DMEM, supplemented with 10% FBS, 1% P/S, and 2% L-glutamine. In some conditions, media was additionally supplemented with TGF-β1 at a concentration of 2 ng/mL (100-21C, PeproTech, USA) Medium was replaced consistently every 24 h throughout all experimental conditions.
Characterization of Hydrogels and Engineered Cardiac Tissues
Hydrogels were fabricated according to the described methods above to determine the compressive modulus of the constructed (cell-laden) hydrogels. After fabrication, non-cell-laden hydrogels were detached from the glass slide and allowed to swell overnight in DPBS at 4 °C. Engineered cardiac tissues, however, were cultured in normal and TGF-β1 containing medium for 14 days before mechanical testing (n=5). Hydrogels were cut with a 5 mm biopsy punch, and excess liquid was removed from the hydrogel. Gels were compressed with a uniaxial tensile loading machine (Instron, 5542, USA) at a rate of 1mm/min with a 10 N cell load capacity. The compressive modulus was calculated as the slope from 0–15% strain (n=4).
Scanning electron microscopy (SEM; Zeiss Ultra 55 SEM; Carl Zeiss, Thornwood, NY, USA) was performed to characterize the hydrogel porosity. Cell-laden hydrogels were fixed at day 1 and day 14 in 4% (v/v) paraformaldehyde (PF, 15700, Electron Microscopy Sciences, Hatfield, PA) for 30 min at room temperature. Following fixation, the cell-laden hydrogels were washed with DPBS and incubated at 4 °C overnight. Hydrated hydrogels were placed in liquid nitrogen for 20 min and stored at −80 °C overnight. After freezing, the hydrogels were lyophilized for 2 days to obtain a porous and foam-like GelMA hydrogel. The foams were broken in half and coated with Pt/Pd to allow for cross-sectional imaging by SEM. Quantification of the pore-size was performed by measuring pore-size diameter (n=150) from SEM images (n=3) made from 5% HM, 7% MM, and 10% MM GelMA foams.
Hydrogel degradation was assessed by fabricating hydrogels and subjecting them to collagenase-induced degradation. Hydrogels were fabricated and allowed to swell in DPBS overnight at 4 °C. Hydrogels were then placed in a 0.5 U/mL collagenase type II solution (in DPBS) at 37 °C. Excess liquid was removed, and the hydrogels were weighed before and after incubation with collagenase. The weight loss percent was determined after 0.5, 1, 3 and 6 h (n=3).
Characterization of Cell Spreading and Cell Viability
Cell spreading within the 3D engineered cardiac tissues was determined by visualizing the organization of F-actin fibers within the cells. The cell-laden hydrogels were fixed with 4% PF solution for 30 min. Subsequently, the 3D encapsulated cells were permeabilized with 0.1% X-100 Triton (Sigma-Aldrich) for 40 min at room temperature. This was followed by 45 min incubation with Alexa Fluor 488 Phalloidin (Invitrogen) with a 1:40 dilution in DPBS. Cell nuclei were counterstained with 4’,6-diamidino-2-phenyl indole dihydrochloride (DAPI; Vector Laboratories) for 20 min at room temperature. Hydrogels were then washed three times in DPBS for 5 min. 3D imaging was performed by confocal microscopy (Leica SP5 X MP, Germany) to visualize the fluorescently stained F-actin fibers and to determine the degree of cell spreading within the hydrogels. Z-stack (100 µm each) images were taken of each hydrogel per condition and four areas (400 µm × 400 µm) were selected for further quantification of cell spreading. Fractional area coverage by F-actin was determined within the four selected windows using ImageJ software.
Cell viability was examined with a Live/Dead fluorescent labeling kit (Invitrogen) on day 1 of culture according to the manufacturer’s protocol. Hydrogels were first washed with DPBS followed by an incubation with calcein-AM (0.5 µL/mL) and ethidium homodimer-1 (2 µL/mL) in DPBS for 15 min at 37 °C. After washing with DPBS, fluorescent images were taken from 4 selected areas using an inverted microscope (Nikon TE 2000-U, Nikon instruments Inc., USA). To quantify viability, images were taken at 4 different focal planes within the hydrogel by adjusting the height of the objective manually. Three cell-laden hydrogels were used to determine the cell viability in each condition, and ImageJ software was used to quantify the number of viable cells. Data depicted represents the percentage of live cells within the engineered constructs. Cell metabolic activity was assessed throughout culture with PrestoBlue® Cell Viability Reagent (PB; Life Technologies). The cell-laden hydrogels were incubated with PrestoBlue for 2 h at 37 °C in a 1:10 dilution in normal culture medium (n=4). The fluorescence was determined (560 nm – 590 nm) using a fluorescence reader (Synergy HT-Reader, BioTek, Winooski, VT). The data was normalized to hydrogel samples without encapsulated cells. The data represent the normalized fluorescence absorbance at day 1, 5, 10, and 14 of culture.
Cell Proliferation Analysis
Click-iT Plus EdU Alexa Fluor® 488 Imaging Kit (Life Technologies) was used to specifically and quantifiably assess the number of proliferating cells within 3D cardiac tissues. Proliferating cells were labeled following the manufacturer’s guidelines. Briefly, cell-encapsulated hydrogels (n=3) were incubated with 10 × 10−6 M EdU in normal culture medium at 37 °C. After 24 h of incubation, the samples were fixed with 4% PF (30 min) and permeabilized with 0.1% X-100 Triton (40 min) at room temperature. The samples were blocked with 3% (w/v) bovine serum albumin (BSA; Sigma-Aldrich) solution and subsequently incubated for 30 min with the Click-iT solution at room temperature. Additionally, cell-specific proliferation was assessed by immunostaining with vimentin, a mesenchymal cell specific marker. Subsequently, the samples were washed twice with DPBS and counterstained for 20 min with DAPI at room temperature. 3D z-stack (50 µm each) images were taken by confocal microscopy. To quantify proliferation, fluorescence images were taken with an inverted microscope at 3 different focal planes within the hydrogel by adjusting the height of the objective manually (n=3). ImageJ software was used to count the number of EdU positive cells. Positive control for EdU labeling was determined by staining cardiac fibroblasts cultured for 24 h in normal, and TGF-β1 supplemented culture medium (n=3). Fluorescence images were taken from each sample by using an inverted microscope (n=10). The percentage of proliferating cells was calculated by counting the EdU labeled cells using ImageJ software. Cell proliferation was calculated by dividing the EdU positive cells by the total number of DAPI positive cells.
Immunofluorescence Staining for Cardiac (fibrosis) Specific Markers
The 3D engineered cardiac tissues were immunostained for cardiac tissue (sarcomeric α-actinin, connexin-43) and cardiac fibrosis (α-SMA, collagen type I, fibronectin, and matrix-metalloproteinase-2 (MMP-2)) markers. Samples were fixed with a 4% PF solution for 30 min, followed by three washing steps (5 min each) with DPBS. Subsequently, the cell-hydrogels were permeabilized by incubation with 0.1% X-100 triton for 45 min, after which the samples were washed with DPBS and blocked for 30 min with a 10% goat serum solution in DPBS. After blocking, the hydrogels were incubated with a monoclonal mouse anti-sarcomeric α-actinin (Abcam, catalogue #9465), polyclonal rabbit anti-connexin-43 (Abcam, catalogue #11370), monocloncal rabbit anti-vimentin (Abcam, catalogue #92547), monoclonal rabbit anti-α-SMA (Abcam, catalogue #32575), polyclonal rabbit anti-collagen I (Abcam, catalogue #292), polycloncal rabbit anti-fibronectin (Abcam, catalogue #23751), or a polycloncal rabbit anti-MMP-2 (Abcam, catalogue #37150) for 16 h at 4 °C. After incubation with the primary antibody (diluted 1:200 in 10% goat serum), the samples were washed three times (10 min each) in DPBS at room temperature. The secondary antibodies (goat anti-rabbit Alexa Fluor 594 or goat anti-mouse Alexa Fluor 488 (Abcam)) were diluted 1:200 in 10% goat serum, followed by incubation with the samples for 2 h at room temperature. The nuclei were counterstained with DAPI. Immunofluorescence double staining was performed by incubating two primary antibodies (eg. sarcomeric α-actinin and connexin-43) simultaneously. After washing with DPBS three times, the secondary antibodies were incubated separately for 2 h each. 3D confocal z-stack images (150 µm each) were taken and processed with ImageJ software.
Characterization of the Beating Behavior of Engineered Cardiac Tissue
The beating behavior of 3D engineered cardiac tissues was characterized quantitatively by using a temperature controlled chamber (at 37 °C) and real time video recording with a camera (Sony XCD-X710) attached to an inverted optical microscope. Videos of the beating constructs (n=3) were recorded every day from day 3 of culture onwards in three different constructs at three different spots. The beating pattern and frequency of the constructs was determined by a custom written MATLAB program.[59] The single cell beating characteristics of the engineered tissues, cultured in growth medium supplemented with TGF-β1, were also assessed with a modified custom written MATLAB program.
GelMA Hydrogel Contraction Assay
A GelMA hydrogel contraction assay was performed to assess the contractile manifestation of MyoFs inside the engineered fibrotic-like cardiac tissues. The GelMA hydrogel contraction assay was performed in a similar manner as a previously described collagen contraction assay.[22] Briefly, 3D cell-laden hydrogels were fabricated as described above and were cultured in TGF-β1 containing culture medium for 14 days according to the protocol. After 14 days, the culture medium was aspirated and optical images were taken, followed by a quantitative analysis of the gel diameters using ImageJ software. Depicted data represents mean ± SD of gel diameter in each condition (n=5).
Real-Time Polymerase Chain Reaction for Expression of Cardiac Fibrosis Markers
Cell-laden hydrogels were used to examine the expression of cardiac fibrosis markers. First, 3D cardiac tissues were mechanically disrupted and total RNA was extracted from all samples using TRIzol reagent (Life Technologies) and total RNA yield was measured with a NanoDrop (Thermo Scientific). 1 µg of total RNA from each sample was reverse transcribed according to the manufacturer’s instructions using the QuantiTect ® Reverse Transcription kit (Qiagen). All RT-PCR was performed using the iTaq™ Universal SYBR® Green supermix (Bio-Rad, USA). The 20 µL volume reaction component included 10 µL supermix, 1 µL of primer mix (5 µM forward/reverse primer), 100 ng template and nuclease free water (variable). Predesigned KiCqStart® SYBR® Green primers (Sigma-aldrich) were obtained for the following target genes: Collagen1A1 (catalogue #KSPQ12012G), Fibronectin (catalogue #KSPQ12012G), α-SMA (catalogue #KSPQ12012G), and MMP-2 (catalogue #KSPQ12012G). Relative expressions were calculated using ΔΔCt method and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression.
Statistical Analysis
The quantitative results on all sample conditions were plotted by mean ± standard deviation (error bars). Investigators blinded to the various treatment conditions assessed and analyzed the measurements related to mechanical stiffness, RT-PCR, and all quantitative analysis. To perform statistical analysis, a student’s t-test or one-way ANOVA was used. For multiple comparisons, we used a Tukey’s test. Graphpad Prism (v.6, GraphPad, USA) software was used to perform all statistical analyses and results were considered to be significantly different with a p < 0.05.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge funding by the Defense Threat Reduction Agency (DTRA) under Space and Naval Warfare Systems Center Pacific (SSC PACIFIC) Contract No. N66001-13-C-2027. The authors also acknowledge funding from the Office of Naval Research Young National Investigator Award, the National Institutes of Health (EB012597, AR057837, DE021468, HL099073, R56AI105024), the Presidential Early Career Award for Scientists and Engineers (PECASE) and Air Force Office of Sponsored Research under award # FA9550-15-1-0273. We acknowledge the support from Innovation and the Netherlands CardioVascular Research Initiative (CVON): The Dutch Heart Foundation, Dutch Federation of University Medical Centers, the Netherlands Organization for Health Research and Development and the Royal Netherlands Academy of Science. The publication of this material does not constitute approval by the government of the findings or conclusions herein. S.R.S. would like to recognize and thank Brigham and Women’s Hospital President Betsy Nabel, MD, and the Reny family, for the Stepping Strong Innovator Award through their generous funding. I.K.Y. was supported by a NIH Organ Design and Engineering Training fellowship (T32 EB16652).
Footnotes
Supporting information is available from the Wiley Online Library or from the author.
The authors declare no conflict of interests in this work.
Contributor Information
Amir Hossein Sadeghi, Biomaterials Innovation Research Center, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, 65 Landsdowne street, Cambridge, MA 02139, USA.; Harvard-MIT Division of Health Sciences & Technology, Massachusetts Institute of Technology, 65 Landsdowne street, Cambridge, MA 02139, USA. Department of Cardiology, University Medical Center Utrecht, 3584 CX Utrecht, The Netherlands. Department of Cardiothoracic Surgery, University Medical Center Utrecht, 3584 CX, The Netherlands.
Dr. Su Ryon Shin, Biomaterials Innovation Research Center, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, 65 Landsdowne street, Cambridge, MA 02139, USA. Harvard-MIT Division of Health Sciences & Technology, Massachusetts Institute of Technology, 65 Landsdowne street, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA.
Janine C. Deddens, Department of Cardiology, University Medical Center Utrecht, 3584 CX Utrecht, The Netherlands. Netherlands Heart Institute (ICIN), 3584CX Utrecht, The Netherlands.
Giuseppe Fratta, Biomaterials Innovation Research Center, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, 65 Landsdowne street, Cambridge, MA 02139, USA.; Harvard-MIT Division of Health Sciences & Technology, Massachusetts Institute of Technology, 65 Landsdowne street, Cambridge, MA 02139, USA. Department of Electronics and Telecommunications, Politecnico di Torino, 10129 Torino, Italy.
Serena Mandla, Biomaterials Innovation Research Center, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, 65 Landsdowne street, Cambridge, MA 02139, USA.; Harvard-MIT Division of Health Sciences & Technology, Massachusetts Institute of Technology, 65 Landsdowne street, Cambridge, MA 02139, USA.
Dr. Iman K. Yazdi, Biomaterials Innovation Research Center, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, 65 Landsdowne street, Cambridge, MA 02139, USA. Harvard-MIT Division of Health Sciences & Technology, Massachusetts Institute of Technology, 65 Landsdowne street, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA.
Gyan Prakash, Biomaterials Innovation Research Center, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, 65 Landsdowne street, Cambridge, MA 02139, USA.; Harvard-MIT Division of Health Sciences & Technology, Massachusetts Institute of Technology, 65 Landsdowne street, Cambridge, MA 02139, USA.
Silvia Antona, Biomaterials Innovation Research Center, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, 65 Landsdowne street, Cambridge, MA 02139, USA.; Harvard-MIT Division of Health Sciences & Technology, Massachusetts Institute of Technology, 65 Landsdowne street, Cambridge, MA 02139, USA. Department of Electronics and Telecommunications, Politecnico di Torino, 10129 Torino, Italy.
Danilo Demarchi, Department of Electronics and Telecommunications, Politecnico di Torino, 10129 Torino, Italy.
Dr. Marc P. Buijsrogge, Department of Cardiothoracic Surgery, University Medical Center Utrecht, 3584 CX, The Netherlands.
Joost P.G. Sluijter, Department of Cardiology, University Medical Center Utrecht, 3584 CX Utrecht, The Netherlands. Netherlands Heart Institute (ICIN), 3584CX Utrecht, The Netherlands. UMC Utrecht Regenerative Medicine Center, University Medical Center Utrecht, 3584 CX Utrecht, The Netherlands.
Dr. Jesper Hjortnaes, Department of Cardiothoracic Surgery, University Medical Center Utrecht, 3584 CX, The Netherlands. UMC Utrecht Regenerative Medicine Center, University Medical Center Utrecht, 3584 CX Utrecht, The Netherlands.
Ali Khademhosseini, Biomaterials Innovation Research Center, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, 65 Landsdowne street, Cambridge, MA 02139, USA. Harvard-MIT Division of Health Sciences & Technology, Massachusetts Institute of Technology, 65 Landsdowne street, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA. Department of Physics, King Abdulaziz University, Jeddah 21569, Saudi Arabia. Department of Bioindustrial Technologies, College of Animal Bioscience and Technology, Konkuk University, Hwayang-dong, Kwangjin-gu, Seoul, Republic of Korea.
References
- 1.WHO. [accessed: April, 2016];Cardiovascular diseases. http://www.who.int/mediacentre/factsheets/fs317/en/
- 2.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER, 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB C. American Heart Association Statistics, S. Stroke Statistics. Circulation. 2015;131:e29. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 3.Sutton MG, Sharpe N. Circulation. 2000;101:2981. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 4.Deddens JC, Sadeghi AH, Hjortnaes J, van Laake LW, Doevendans BMPA, Khademhosseini A, SLuijter JPG. Adv. Healthcare Mater. 2016;0:1. doi: 10.1002/adhm.201600571. [DOI] [PubMed] [Google Scholar]
- 5.a) Heusch G, Libby P, Gersh B, Yellon D, Bohm M, Lopaschuk G, Opie L. Lancet. 2014;383:1933. doi: 10.1016/S0140-6736(14)60107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kong P, Christia P, Frangogiannis NG. Cell Mol Life Sci. 2014;71:549. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber KT, Sun Y, Bhattacharya SK, Ahokas RA, Gerling IC. Nat Rev Cardiol. 2013;10:15. doi: 10.1038/nrcardio.2012.158. [DOI] [PubMed] [Google Scholar]
- 7.Hermans KC, Daskalopoulos EP, Blankesteijn WM. J Mol Cell Cardiol. 2016;91:35. doi: 10.1016/j.yjmcc.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Leask A. Circ Res. 2010;106:1675. doi: 10.1161/CIRCRESAHA.110.217737. [DOI] [PubMed] [Google Scholar]
- 9.Rockey DC, Bell PD, Hill JA. N Engl J Med. 2015;373:96. doi: 10.1056/NEJMc1504848. [DOI] [PubMed] [Google Scholar]
- 10.Deb A, Ubil E. J Mol Cell Cardiol. 2014;70:47. doi: 10.1016/j.yjmcc.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creemers EE, Cleutjens JP, Smits JF, Daemen MJ. Circ Res. 2001;89:201. doi: 10.1161/hh1501.094396. [DOI] [PubMed] [Google Scholar]
- 12.Rohr S. Heart Rhythm. 2009;6:848. doi: 10.1016/j.hrthm.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 13.Hill JA, Olson EN. N Engl J Med. 2008;358:1370. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 14.Bujak M, Frangogiannis NG. Cardiovasc Res. 2007;74:184. doi: 10.1016/j.cardiores.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroer AK, Merryman WD. J Cell Sci. 2015;128:1865. doi: 10.1242/jcs.162891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yong KW, Li Y, Huang G, Lu TJ, Safwani WK, Pingguan-Murphy B, Xu F. Am J Physiol Heart Circ Physiol. 2015;309:H532. doi: 10.1152/ajpheart.00299.2015. [DOI] [PubMed] [Google Scholar]
- 17.Brown RD, Ambler SK, Mitchell MD, Long CS. Annu Rev Pharmacol Toxicol. 2005;45:657. doi: 10.1146/annurev.pharmtox.45.120403.095802. [DOI] [PubMed] [Google Scholar]
- 18.Benam KH, Dauth S, Hassell B, Herland A, Jain A, Jang KJ, Karalis K, Kim HJ, MacQueen L, Mahmoodian R, Musah S, Torisawa YS, van der Meer AD, Villenave R, Yadid M, Parker KK, Ingber DE. Annu Rev Pathol. 2015;10:195. doi: 10.1146/annurev-pathol-012414-040418. [DOI] [PubMed] [Google Scholar]
- 19.Saini H, Navaei A, Van Putten A, Nikkhah M. Adv Healthc Mater. 2015;4:1961. doi: 10.1002/adhm.201500331. [DOI] [PubMed] [Google Scholar]
- 20.a) Zimmermann WH, Schneiderbanger K, Schubert P, Didie M, Munzel F, Heubach JF, Kostin S, Neuhuber WL, Eschenhagen T. Circ Res. 2002;90:223. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]; b) Hirt MN, Sorensen NA, Bartholdt LM, Boeddinghaus J, Schaaf S, Eder A, Vollert I, Stohr A, Schulze T, Witten A, Stoll M, Hansen A, Eschenhagen T. Basic Res Cardiol. 2012;107:307. doi: 10.1007/s00395-012-0307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H, Li X, Zhao S, Zeng Y, Zhao L, Ding H, Sun W, Du Y. Biofabrication. 2014;6:045009. doi: 10.1088/1758-5082/6/4/045009. [DOI] [PubMed] [Google Scholar]
- 23.Hjortnaes J, Camci-Unal G, Hutcheson JD, Jung SM, Schoen FJ, Kluin J, Aikawa E, Khademhosseini A. Adv Healthc Mater. 2015;4:121. doi: 10.1002/adhm.201400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galie PA, Westfall MV, Stegemann JP. Cardiovasc Pathol. 2011;20:325. doi: 10.1016/j.carpath.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khademhosseini A, Eng G, Yeh J, Kucharczyk PA, Langer R, Vunjak-Novakovic G, Radisic M. Biomed Microdevices. 2007;9:149. doi: 10.1007/s10544-006-9013-7. [DOI] [PubMed] [Google Scholar]
- 26.Kim SB, Bae H, Cha JM, Moon SJ, Dokmeci MR, Cropek DM, Khademhosseini A. Lab Chip. 2011;11:1801. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhana B, Iyer RK, Chen WL, Zhao R, Sider KL, Likhitpanichkul M, Simmons CA, Radisic M. Biotechnol Bioeng. 2010;105:1148. doi: 10.1002/bit.22647. [DOI] [PubMed] [Google Scholar]
- 28.Vlierberghe SV, Cnudde V, Dubruel P, Masschaele B, Cosijns A, Paepe ID, Jacobs PJ, Hoorebeke LV, Remon JP, Schacht E. Biomacromolecules. 2007;8:331. doi: 10.1021/bm060684o. [DOI] [PubMed] [Google Scholar]
- 29.Spinale FG. Circ Res. 2002;90:520. doi: 10.1161/01.res.0000013290.12884.a3. [DOI] [PubMed] [Google Scholar]
- 30.Kreuzberg MM, Willecke K, Bukauskas FF. Trends Cardiovasc Med. 2006;16:266. doi: 10.1016/j.tcm.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim SB, Nikkhah M, Khabiry M, Azize M, Kong J, Wan KT, Palacios T, Dokmeci MR, Bae H, Tang XS, Khademhosseini A. ACS Nano. 2013;7:2369. doi: 10.1021/nn305559j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, Weiss AS. Adv Funct Matery. 2013:23. doi: 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radisic M, Yang L, Boublik J, Cohen RJ, Langer R, Freed LE, Vunjak-Novakovic G. Am J Physiol Heart Circ Physiol. 2004;286:H507. doi: 10.1152/ajpheart.00171.2003. [DOI] [PubMed] [Google Scholar]
- 34.Hinz B. J Biomech. 2010;43:146. doi: 10.1016/j.jbiomech.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 35.Thompson SA, Copeland CR, Reich DH, Tung L. Circulation. 2011;123:2083. doi: 10.1161/CIRCULATIONAHA.110.015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janmey PA, Miller RT. J Cell Sci. 2011;124:9. doi: 10.1242/jcs.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang HY, Speicher DW, Sanger JW, Sanger JM, Discher DE. J Cell Sci. 2008;121:3794. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Y, Halade GV, Lindsey ML. J Cardiovasc Transl Res. 2012;5:848. doi: 10.1007/s12265-012-9398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stawowy P, Margeta C, Kallisch H, Seidah NG, Chretien M, Fleck E, Graf K. Cardiovasc Res. 2004;63:87. doi: 10.1016/j.cardiores.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Shamhart PE, Meszaros JG. J Mol Cell Cardiol. 2010;48:530. doi: 10.1016/j.yjmcc.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 41.van den Borne SW, Diez J, Blankesteijn WM, Verjans J, Hofstra L, Narula J. Nat Rev Cardiol. 2010;7:30. doi: 10.1038/nrcardio.2009.199. [DOI] [PubMed] [Google Scholar]
- 42.Rosenkranz S. Cardiovasc Res. 2004;63:423. doi: 10.1016/j.cardiores.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 43.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Nat Rev Mol Cell Biol. 2002;3:349. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 44.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Science. 2009;324:98. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen MB, Srigunapalan S, Wheeler AR, Simmons CA. Lab Chip. 2013;13:2591. doi: 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- 46.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 47.Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A. Biomaterials. 2015;73:254. doi: 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.a) Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Nat Med. 2008;14:213. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]; b) Williams C, Budina E, Stoppel WL, Sullivan KE, Emani S, Emani SM, Black LD., 3rd Acta Biomater. 2015;14:84. doi: 10.1016/j.actbio.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visser J, Levett PA, te Moller NC, Besems J, Boere KW, van Rijen MH, de Grauw JC, Dhert WJ, van Weeren PR, Malda J. Tissue Eng Part A. 2015;21:1195. doi: 10.1089/ten.tea.2014.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingber DE. FASEB J. 2006;20:811. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 51.a) Bokel C, Brown NH. Dev Cell. 2002;3:311. doi: 10.1016/s1534-5807(02)00265-4. [DOI] [PubMed] [Google Scholar]; b) Israeli-Rosenberg S, Manso AM, Okada H, Ross RS. Circ Res. 2014;114:572. doi: 10.1161/CIRCRESAHA.114.301275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leckband DE, le Duc Q, Wang N, de Rooij J. Curr Opin Cell Biol. 2011;23:523. doi: 10.1016/j.ceb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Chiquet M, Gelman L, Lutz R, Maier S. Biochim Biophys Acta. 2009;1793:911. doi: 10.1016/j.bbamcr.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 54.Sheehy SP, Grosberg A, Parker KK. Biomech Model Mechanobiol. 2012;11:1227. doi: 10.1007/s10237-012-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Proc Natl Acad Sci U S A. 2010;107:565. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.a) Pijnappels DA, Gregoire S, Wu SM. Ann N Y Acad Sci. 2010;1188:7. doi: 10.1111/j.1749-6632.2009.05077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Papadaki M, Bursac N, Langer R, Merok J, Vunjak-Novakovic G, Freed LE. Am J Physiol Heart Circ Physiol. 2001;280:H168. doi: 10.1152/ajpheart.2001.280.1.H168. [DOI] [PubMed] [Google Scholar]
- 57.a) Au HT, Cheng I, Chowdhury MF, Radisic M. Biomaterials. 2007;28:4277. doi: 10.1016/j.biomaterials.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Aubin H, Nichol JW, Hutson CB, Bae H, Sieminski AL, Cropek DM, Akhyari P, Khademhosseini A. Biomaterials. 2010;31:6941. doi: 10.1016/j.biomaterials.2010.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.a) Bhatia SN, Ingber DE. Nat Biotechnol. 2014;32:760. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]; b) Ribas J, Sadeghi H, Manbachi A, Leijten J, Brinegar K, Zhang YS, Ferreira L, Khademhosseini A. Applied In Vitro Toxicology Jun. 2016;2:82. doi: 10.1089/aivt.2016.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.a) Ehler E, Moore-Morris T, Lange S. J Vis Exp. 2013:79. doi: 10.3791/50154. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Biernacka A, Frangogiannis NG. Aging Dis. 2011;2:158. [PMC free article] [PubMed] [Google Scholar]; c) Santiago JJ, Dangerfield AL, Rattan SG, Bathe KL, Cunnington RH, Raizman JE, Bedosky KM, Freed DH, Kardami E, Dixon IM. Dev Dyn. 2010;239:1573. doi: 10.1002/dvdy.22280. [DOI] [PubMed] [Google Scholar]
- 60.Driesen RB, Nagaraju CK, Char JA, Coenen T, Lijnen PJ, Fagard RH, Sipido KR, Petrov VV. Cadiovasc Res. 2014;101:411. doi: 10.1093/cvr/cvt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu K, Shin SR, Van Kempen T, Li Y-C, Ponraj V, Najaspour A, Mandla S, Hu N, Liu X, Leijten J, Li Y-D, Hussain MA, Zhang YS, Tamayol A, Khademhosseini A. Adv Funct Mater. 2017:1605352. doi: 10.1002/adfm.201605352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fedorovich NE, Oudshoorn MH, van Geemen D, Hennink WE, Albas J, Dhert WJ. Biomaterials. 2009;30:344. doi: 10.1016/j.biomaterials.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 63.a) Visser J, Gawlitta D, Benders KE, Toma SM, van Pouran B, Weeren PR, Dhert WJ, Malda J. Biomaterials. 2015;37:174. doi: 10.1016/j.biomaterials.2014.10.020. [DOI] [PubMed] [Google Scholar]; b) Shin SR, Zihlmann C, Akbari M, Assawes P, Cheung L, Zhang K, Manoharan V, Zhang YS, Yuksekkaya M, Wan KT, Nikkhah M, Dokmeci MR, Tang XS, Khademhosseini A. Small. 2016;12:3677. doi: 10.1002/smll.201600178. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Camci-Unal G, Cuttica D, Annabi N, Demarchi D, Khademhosseini A. Biomacromolecules. 2013;14:1085. doi: 10.1021/bm3019856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.