Abstract
Data obtained by genotyping single nucleotide polymorphisms (SNPs) related to high-density lipoprotein cholesterol (HDL-C) levels were utilized in Genetic Risk Score [unweighted (GRS) and weighted (wGRS)] computation on Hungarian general and Roma populations. The selection process of the SNPs as well as the results obtained are published in our research article (Piko et al., 2017) [1]. Linkage analyses were performed by study groups. Study populations were stratified by quintiles of weighted Genetic Risk Score. Multivariate linear regression analyses were performed using Genetic Risk Scores and HDL-C levels as dependent variables; and ethnicity, sex and age as independent variables. The study subjects were categorized into quintiles according their wGRS values. Associations of Genetic Risk Scores with plasma HDL-C levels (as a continuous variable) were observed in both populations. Finally, the two populations were merged and analyzed together by multivariate logistic regression where reduced plasma HDL-C level was the dependent variable; while ethnicity, age and sex were the independent ones.
Keywords: Single nucleotide polymorphism, Genetic susceptibility, Genetic risk score, High-density lipoprotein cholesterol, Roma population
Specifications Table
| Subject area | Biology |
| More specific subject area | Molecular genetics, Public health genomics |
| Type of data | Figure, Table |
| How data was acquired | Survey, Blood sample collection, MassARRAY platform (Sequenom Inc., San Diego, CA, USA) with iPLEX Gold chemistry |
| Data format | Analyzed |
| Experimental factors | Genomic DNA from peripheral blood was isolated |
| Experimental features | Genotyping method of SNPs was based on MALDI-TOF (Matrix Assisted Laser Desorption-Ionisation-Time Of Flight) analysis, performed on MassARRAY Platform. |
| Data source location | Debrecen, Hungary, Latitude: 47.544062, 21° 38′ 25′′ E & Longitude: 21.64283, 47° 32′ 33′′ N |
| Data accessibility | Data are presented in this article; DNA sample and raw data are available for further analyses in collaborative studies |
Value of the data
-
•
Several studies describe the health status of the Roma, which constitutes the largest ethnic minority in Europe however studies focusing on their genetic predisposition to common chronic diseases are scarce.
-
•
Genetic background of atherosclerosis among Roma as well as the general Hungarian population can be studied separately or in international cohort.
-
•
Genetic risk score assessment can be further utilized to compare susceptibility to reduced HDL-C level among different population groups.
1. Data
Distribution of SNPs related to HDL-C level were analysed in the Hungarian Roma and general populations and weighted Genetic Risk Scores were defined and used to categorize the population into quintiles.
2. Experimental design, materials and methods
2.1. Subjects
Study involved subjects of samples investigated during recent cross sectional surveys [2], [3]. The Roma sample is representative to the Roma population living settlements in North-East Hungary in terms of age and sex and includes 646 individuals (Roma). The “General” sample consisting of 1542 individuals representative for the Hungarian general population in terms of geographic, age and sex distributions.
2.2. DNA extraction
DNA was isolated using a MagNA Pure LC system (Roche Diagnostics, Basel, Switzerland) with a MagNA Pure LC DNA Isolation Kit–Large Volume according to the manufacturer's instructions. Extracted DNA was eluted in 200 µl MagNA Pure LC DNA Isolation Kit-Large Volume elution buffer.
2.3. SNP selection
A systematic literature review on the PubMed, HuGE Navigator and Ensembl databases was conducted to identify SNPs most strongly associated with HDL-C metabolism (Table 1 and Fig. 1). The selection process of the SNPs is demonstrated in detail in our research article [1].
Table 1.
List of the SNPs which were involved in the research.
| Nearest Gene | Gene (short) | SNP (rs number) | Chromosome |
|---|---|---|---|
| Apolipoprotein B | APOB | rs693 | 2 |
| ATP-binding cassette transporter ABCA1 | ABCA1 | rs4149268 | 9 |
| Cholesteryl ester transfer protein | CETP | rs1532624 | 16 |
| Cholesteryl ester transfer protein | CETP | rs5882 | 16 |
| Cholesteryl ester transfer protein | CETP | rs708272 | 16 |
| Cholesteryl ester transfer protein | CETP | rs7499892 | 16 |
| Cholesteryl ester transfer protein | CETP | rs9989419 | 16 |
| Endothelial lipase | LIPG | rs2000813 | 18 |
| Endothelial lipase | LIPG | rs4939883 | 18 |
| Hepatic lipase | LIPC | rs10468017 | 15 |
| Hepatic lipase | LIPC | rs1077834 | 15 |
| Hepatic lipase | LIPC | rs1532085 | 15 |
| Hepatic lipase | LIPC | rs1800588 | 15 |
| Hepatic lipase | LIPC | rs2070895 | 15 |
| Hepatic lipase | LIPC | rs4775041 | 15 |
| HMG-CoA Reductase | HMGCR | rs3846662 | 5 |
| Lipoprotein lipase | LPL | rs328 | 8 |
| Polypeptide N-acetylgalactosaminyltransferase 2 | GALNT2 | rs2144300 | 1 |
| Polypeptide N-acetylgalactosaminyltransferase 2 | GALNT2 | rs4846914 | 1 |
| Potassium channel tetramerization domain containing 10 | KCTD10 | rs2338104 | 12 |
| WW Domain Containing Oxidoreductase | WWOX | rs2548861 | 16 |
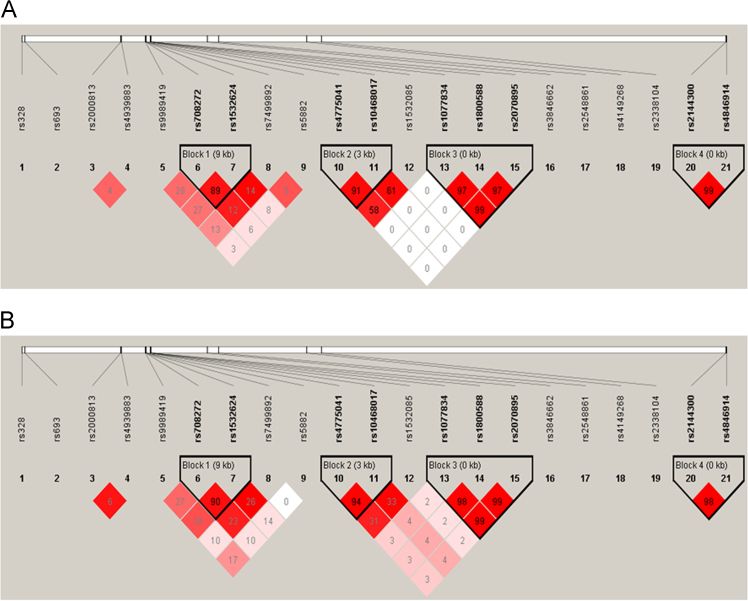
Fig. 1.
Haplotype block organization of SNPs related to high-density lipoprotein cholesterol level on the LD maps for the Hungarian general (A) and Roma (B) populations. Linkage analyses were performed separately in the study populations. According to the LD map generated by Haploview, there are four haplotype blocks (outlined in a bold black line) consisting of variants that are in high LD. The blocks were formed by the SNPs of the CETP, LIPC and GALNT2 genes. The numbers above the map show the rs numbers of SNPs. The colour scheme is a standard Haploview colour scheme (white D′<1 and LOD<2, shades of pink/red: D′<1 and LOD≥2, and bright red D′=1 and LOD≥2). Numbers in squares are D′ values.
2.4. Genotyping
Genotyping was performed on a MassARRAY platform (Sequenom Inc., San Diego, CA, USA) with iPLEX Gold chemistry. Validation, concordance analysis and quality control were conducted by the facility according to their protocols.
2.5. Statistical analyses
Two-sided t tests were used to compare the distribution of genetic risk scores in populations. To reveal the association between genetic risk, serum HDL-C level and ethnicity several statistical models were used (Table 1, Table 2, Table 3, Table 4, Table 5, Table 6).
Table 2.
Distribution of study populations by wGRS quintiles.
| Hungarian general population (%) | Hungarian Roma population (%) | p-value | |
|---|---|---|---|
| 1st quintile of wGRS (0.15–≤0.30) | 1.83 | 0.51 | 0.025 |
| 2nd quintile of wGRS (0.31–≤0.45) | 17.18 | 10.45 | <0.001 |
| 3rd quintile of wGRS (0.46–<0.59) | 48.38 | 49.14 | 0.756 |
| 4th quintile of wGRS (0.6–≤ 0.74) | 30 | 34.76 | 0.037 |
| 5th quintile of wGRS (0.75–0.88) | 2.61 | 5.14 | 0.004 |
Table 3.
Output of multiple regression models using unweighted and weighted genetic risk scores as dependent variable and ethnicity, age and sex as independent variables.
| Dependent variable: GRS | R Square=0.009 | ||
| Independent variables | Coefficient | p-value | β |
| Ethnicity (Roma vs. General) | 0.667 | <0.001 | 0.092 |
| Sex women vs. men) | 0.106 | 0.477 | 0.016 |
| Age | −0.0003 | 0.068 | −0.001 |
| β: relative strength of predictors | |||
| Dependent variable: wGRS | R Square=0.017 | ||
| Independent variables | Coefficient | p-value | β |
| Ethnicity (Roma vs. General) | 0.029 | <0.001 | 0.125 |
| Sex (women vs. men) | −0.001 | 0.774 | −0.006 |
| Age | −0.0002 | 0.202 | −0.028 |
| β: relative strength of predictors | |||
Multivariate regression analysis using age, sex as covariates did not change the inference neither for the GRS nor for wGRS.
Table 4.
Proportion of subjects with reduced plasma HDL-C level in the General and Roma populations according to wGRS quintiles.
| 1st quintile of wGRS (0.15–≤0.30) | 2nd quintile of wGRS (0.31–≤0.45) | 3rd quintile of wGRS 0.46–<0.59) | 4th quintile of wGRS | 5th quintile of wGRS (0.75–0.88) | p-valuesfor trend | |
|---|---|---|---|---|---|---|
| (0.6–≤0.74) | ||||||
| General (Men; Women) | N=26 (8;18) | N=241 (115;126) | N=681 (320;361) | N=417 (200;217) | N=36 (21;15) | |
| Average HDL-C level (mmol/l) | 1.56 | 1.47 | 1.41 | 1.38 | 1.33 | 0.021 |
| Reduced plasma HDL-C (%) | 11.54 | 23.77 | 27.8 | 28.5 | 31.43 | 0.083 |
| Roma (Men; Women) | N=3 (1;2) | N=61 (28;33) | N=287 (112;175) | N=203 (76;127) | N=30 (11;19) | |
| Average HDL-C level (mmol/l) | 1.26 | 1.24 | 1.23 | 1.2 | 1.09 | 0.076 |
| Reduced plasma HDL-C (%) | 33.33 | 44.26 | 49.83 | 52.22 | 56.67 | 0.054 |
Table 5.
Association of GRSs with plasma HDL-Ca level by study groups.
|
Hungarian General |
Hungarian Roma |
|||
|---|---|---|---|---|
| β (95% CI) | p-value | β (95% CI) | p-value | |
| GRS | ||||
| Model I | −0.01 (−0.018 to −0.003) | 0.004 | −0.013 (−0.023 to −0.003) | 0.011 |
| Model II | −0.011 (−0.018 to −0.004) | 0.003 | −0.013 (−0.023 to −0.003) | 0.009 |
| wGRS | ||||
| Model III | −0.243 (−0.466 to −0.020) | 0.033 | −0.318 (−0.633 to −0.002) | 0.049 |
| Model IV | −0.205 (−0.420 to 0.101) | 0.062 | −0.336 (−0.651 to −0.21) | 0.036 |
The association of GRS and wGRS with plasma HDL-C level were evaluated under unadjusted regression models (Model I and III) and under regression models adjusted for age and sex (Model II and IV) separately in Roma and general subjects. In all models the HDL-C was the dependent variable, the GRS/wGRS were the independent variables.
95% CI: 95% confidence interval
HDL-C values were non-normally distributed and were transformed using a two-step approach suggested by Templeton [4].
Table 6.
The association of HDL-C level with genetic risk scores adjusted by ethnicity, sex and age.
| Dependent variable: reduced plasma HDL-C level | R Square=0.046 | |
| Independent variables | OR (95% CI) | p-value |
| Genetic constitution defined by GRS | 1.07 (1.04–3.31) | <0.001 |
| Ethnicity (Roma vs. General) | 2.70 (2.19–3.31) | <0.001 |
| Sex (women vs. men) | 0.99 (0.81–1.20) | 0.942 |
| Age | 1.00 (0.99–1.01) | 0.393 |
| Dependent variable: reduced plasma HDL-C level | R Square=0.042 | |
| Independent variables | OR (95% CI) | p-value |
| Genetic constitution defined by wGRS | 3.89 (1.56–9.69) | 0.004 |
| Ethnicity (Roma vs. General) | 2.69 (2.19–3.31) | <0.001 |
| Sex (women vs. men) | 1.00 (0.83–1.21) | 0.993 |
| Age | 1.00 (0.99–1.01) | 0.353 |
OR: odds ratio.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
This study was approved by the Ethical Committee of the University of Debrecen, Medical Health Sciences Centre (Reference no. 2462-2006) and by the Ethical Committee of the Hungarian Scientific Council on Health (Reference nos. NKFP/1/0003/2005 and 8907-O/2011-EKU).
This article does not contain any studies with animals performed by any of the authors.
Acknowledgements
The project was co-financed by the European Union under the European Social Fund (TÁMOP 4.2.1. B-09/1/KONV-2010-0007 and TÁMOP 4.2.2.A-11/1/KONV-2012-0031) and European Regional Development Fund (GINOP-2.3.2-15-2016-00005), as well as by the Hungarian Academy of Sciences (MTA11010 and TK2016-78).
Footnotes
Transparency data document associated with this article can be found in the online version at doi: 10.1016/j.atherosclerosis.2017.05.028.
Supplementary data associated with this article can be found in the online version at doi: 10.1016/j.atherosclerosis.2017.05.028.
Transparency document. Supplementary material
Transparency document
.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Piko P., Fiatal S., Kosa Z., Sandor J., Adany R. Genetic factors exist behind the high prevalence of reduced high-density lipoprotein cholesterol levels in the Roma population. Atherosclerosis. 2017;263:119–126. doi: 10.1016/j.atherosclerosis.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 2.Szigethy E., Szeles G., Horvath A., Hidvegi T., Jermendy G., Paragh G., Voko Z. Epidemiology of the metabolic syndrome in Hungary. Public Health. 2012;126:143–149. doi: 10.1016/j.puhe.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Kosa Z., Moravcsik-Kornyicki A., Dioszegi J., Roberts B., Szabo Z., Sandor J., Adany R. Prevalence of metabolic syndrome among Roma: a comparative health examination survey in Hungary. Eur. J. Public Health. 2015;25:299–304. doi: 10.1093/eurpub/cku157. [DOI] [PubMed] [Google Scholar]
- 4.Templeton G.F. A two-step approach for transforming continuous variables to normal: implications and recommendations for is research. Commun. Assoc. Inf. Syst. 2011:28. (Article 4) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document
Supplementary material