Abstract
This prospective study developed an MRI-based method for identification of individual motor neuron disease (MND) patients and test its accuracy at the individual patient level in an independent sample compared with mimic disorders. 123 patients with amyotrophic lateral sclerosis (ALS), 44 patients with predominantly upper motor neuron disease (PUMN), 20 patients with ALS-mimic disorders, and 78 healthy controls were studied. The diagnostic accuracy of precentral cortical thickness and diffusion tensor (DT) MRI metrics of corticospinal and motor callosal tracts were assessed in a training cohort and externally proved in a validation cohort using a random forest analysis. In the training set, precentral cortical thickness showed 0.86 and 0.89 accuracy in differentiating ALS and PUMN patients from controls, while DT MRI distinguished the two groups from controls with 0.78 and 0.92 accuracy. In ALS vs controls, the combination of cortical thickness and DT MRI metrics (combined model) improved the classification pattern (0.91 accuracy). In the validation cohort, the best accuracy was reached by DT MRI (0.87 and 0.95 accuracy in ALS and PUMN vs mimic disorders). The combined model distinguished ALS and PUMN patients from mimic syndromes with 0.87 and 0.94 accuracy. A multimodal MRI approach that incorporates motor cortical and white matter alterations yields statistically significant improvement in accuracy over using each modality separately in the individual MND patient classification. DT MRI represents the most powerful tool to distinguish MND from mimic disorders.
Keywords: Motor neuron disease, Amyotrophic lateral sclerosis, Diagnosis, MRI, Random forest analysis
Highlights
-
•
Motor cortical and white matter alterations yield high accuracy in the individual MND patient classification.
-
•
DT MRI represents the most powerful tool to distinguish MND from mimic disorders.
-
•
The most pronounced damage in MND patients relative to mimic subjects was found in the motor callosal fibers.
1. Introduction
Amyotrophic Lateral Sclerosis (ALS) is an adult-onset neurodegenerative disorder of the motor system characterized by upper (UMN) and lower motor neuron (LMN) degeneration, leading to progressive muscular paralysis and death (Kiernan et al., 2011). The El Escorial criteria for the diagnosis of ALS, which were established 20 years ago, are essentially clinical and rely on the detection of motor neuron signs in multiple body segments (Brooks et al., 2000). Although these guidelines have been repeatedly shown to be useful inclusion criteria for clinical trials, concerns have been raised regarding their use in clinical practice (Agosta et al., 2015, Belsh, 2000). The false-positive rate has been estimated to be as high as eight to 10%, while the false-negative rate approaches 45% (Davenport et al., 1996, Traynor et al., 2000). Furthermore, the average delay from the symptom onset to diagnosis is 12 months (Mitchell et al., 2010).
The use of magnetic resonance imaging (MRI) in patients suspected of having ALS is yet restricted to exclude other causes of signs and symptoms of motor neuron pathology (Filippi et al., 2010). However, the recent growing recognition of ALS as a diffuse central nervous system pathology has been a major driver of the application of advanced neuroimaging techniques to the study of the disease (Filippi et al., 2015). Structural MRI detects in vivo both grey matter (GM) and white matter (WM) alterations associated with ALS, providing potential reliable diagnostic markers of the disease (Chiò et al., 2014, Menke et al., 2017). A pathological hallmark of ALS is the atrophy of the primary motor cortex, and numerous studies have detected bilateral precentral gyrus thinning in ALS patients (Agosta et al., 2012, Schuster et al., 2013, Verstraete et al., 2011). Degeneration of the corticospinal tracts (CST) and body of the corpus callosum (CC) represents another disease-defining signature (Agosta et al., 2014, Agosta et al., 2010, van der Graaff et al., 2011), particularly in patients with a predominant UMN (PUMN) variant (Agosta et al., 2014, Iwata et al., 2011, Unrath et al., 2010).
Increasing attempts have been made to test the diagnostic accuracy of structural MRI measures in ALS. Precentral gyrus thickness showed a good accuracy in distinguishing ALS from healthy controls (Agosta et al., 2012, Verstraete et al., 2011, Walhout et al., 2015). Diffusion Tensor (DT) MRI measures in small subject cohorts provided good discrimination between ALS and controls (Agosta et al., 2014, Graham et al., 2004, Nelles et al., 2008). However, these studies reported results at a group level, raising the question of the applicability of such measures in clinical settings. An individual patient data meta-analysis of CST fractional anisotropy (FA) revealed its diagnostic power to be modest relative to healthy controls (Foerster et al., 2013). This disappointing finding may result from the heterogeneity of both the methodology and patient populations but it may also suggest that a single MR technique lacks sufficient diagnostic power. A multimodal neuroimaging approach may be a strategy to improve accuracy (Douaud et al., 2011, Foerster et al., 2014, Schuster et al., 2016). A model incorporating the cortical thickness of the precentral gyrus and DT MRI measures of the CST and CC was able to discriminate ALS and healthy controls with good sensitivity (85.7%) and accuracy (78.4%) (Schuster et al., 2016). However, almost all previous MRI studies have recruited healthy controls as reference group to identify ALS-specific abnormalities, while a comparison with ALS-mimic disorders is mandatory to test the specificity of these markers.
The aims of this study were: to develop a method for individual identification of motor neuron disease (MND) patients using multimodal structural MRI data of ALS-specific anatomical regions (i.e., precentral cortical thickness and DT MRI metrics of motor WM tracts), and to test the validity of such an approach in an independent patient cohort relative to subjects with ALS-mimic disorders.
2. Methods
2.1. Subjects
All patients were consecutively recruited at three tertiary referral MND Clinics in Northern Italy and underwent a comprehensive evaluation including neurological history, neurophysiological assessment, genetic analysis, and MRI. The main sample consisted of 167 right-handed patients with MND (123 patients with ALS and 44 patients with PUMN), including 17 ALS cases carrying a genetic mutation (11 with hexanucleotide repeat expansions in chromosome 9 open reading frame 72, two with a TARDBP mutation, three with a SOD1 mutation, and one with a FUS mutation) (Table 1). Diagnosis of classic ALS was made according to the revised Escorial criteria (Brooks et al., 2000). Patients with a clinical PUMN phenotype did not have any LMN sign on clinical assessment or any evidence of active denervation on repeated electromyographical examinations (Chio et al., 2011). In the PUMN sample, 33 cases had a disease duration ≥ 3 years and were therefore diagnosed with primary lateral sclerosis (PLS) (Pringle et al., 1992). An additional sample of 20 patients with ALS-mimic disorders (Traynor et al., 2000) was enrolled (Table 1). This group included 13 patients with spinobulbar muscular atrophy (Kennedy's Disease), two patients with multifocal motor neuropathy with conduction blocks, two patients with chronic sensorimotor polyneuropathy, two patients with lumbar radiculopathy, and one case with distal spinal muscular atrophy. Prior to diagnosing a mimic disorder, these patients had been referred to our centers because of a clinical suspicion of ALS. Experienced neurologists blinded to the MRI results performed the clinical assessment. Site of disease onset and disease duration were recorded. Disease severity was assessed using the ALS Functional Rating Scale-revised (ALSFRS-r). The rate of disease progression was calculated as follows: (48–ALSFRS-r score)/time from symptom onset. 72 ALS, 35 PUMN and 14 ALS-mimic patients underwent cognitive and behavioral evaluations following published recommendations (Montuschi et al., 2015, Phukan et al., 2012), as previously described (Agosta et al., 2016).
Table 1.
Demographic and clinical findings of healthy control subjects, and ALS, PUMN and mimic disorder patients.
| Healthy controls | ALS patients | PUMN patients | ALS-mimic patients | p ALS vs HC | p PUMN vs HC | p ALS vs PUMN | p ALS vs ALS-mimic | p PUMN vs ALS-mimic | |
|---|---|---|---|---|---|---|---|---|---|
| Number | 78 | 123 | 44 | 20 | – | – | – | – | |
| Age (years) | 63.23 ± 8.90 | 63.49 ± 10.07 | 62.99 ± 8.22 | 55.85 ± 10.31 | 0.85 | 0.88 | 0.77 | 0.002 | 0.004 |
| Sex (W/M) | 45/33 | 64/59 | 23/21 | 2/18 | 0.43 | 0.56 | 0.98 | < 0.001 | 0.001 |
| Site of onset (bulbar / limb / bulbar + limb) | – | 40/81/2 | 6/38/0 | 0/20/0 | – | – | 0.02 | 0.002 | 0.16 |
| Disease duration (months) | – | 19.28 ± 16.94 | 80.16 ± 56.81 | 118.24 ± 64.54 | – | – | < 0.001 | < 0.001 | < 0.001 |
| ALSFRS-r | – | 38.14 ± 6.88 | 36.67 ± 6.49 | 42.17 ± 1.34 | – | – | 0.23 | 0.046 | 0.01 |
| Rate of disease progression | – | 0.75 ± 0.70 | 0.29 ± 0.48 | 0.05 ± 0.02 | – | – | < 0.001 | < 0.001 | 0.10 |
| No CI or BI/CI or BI/MND-FTD | – | 37/30/5 | 17/18/0 | 7/7/0 | – | – | – | – | – |
Values are means ± standard deviations or number. p values refer to Fisher exact test or ANOVA models, followed by post hoc pairwise comparisons. Abbreviations: ALS = amyotrophic lateral sclerosis; ALSFRS-r = ALS functional rating scale-revised; BI = behavioral impairment; CI = cognitive impairment; HC = healthy controls; M = men; MND = motor neuron disease; W = women.
In addition, data were acquired in 78 right-handed, age-matched healthy controls who were recruited among spouses of patients and by word of mouth (Table 1). Healthy controls were included if the neurological assessment was normal and the Mini-Mental State Examination was ≥ 28. Patients and controls were excluded if they had: significant medical illnesses or substance abuse that could interfere with cognitive functioning; any (other) major systemic, psychiatric, or neurological illnesses; and (other) causes of focal or diffuse brain damage, including lacunae, and extensive cerebrovascular disorders at routine MRI. Approval was obtained from the local ethical standards committee on human experimentation and written informed consent from all subjects (or their legal guardians) before enrolment.
2.2. MRI study
2.2.1. MRI protocol
Using a 3.0 Tesla Philips Intera scanner, the following brain MRI sequences were acquired: T2-weighted spin echo; fluid-attenuated inversion recovery; 3D T1-weighted fast field echo (FFE); and pulsed-gradient spin echo, echo planar with sensitivity encoding and diffusion gradients applied in 32 noncollinear directions (Agosta et al., 2014).
2.2.2. Cortical thickness measurement
Cortical reconstruction and estimation of cortical thickness were performed on the 3D T1-weighted FFE images using the FreeSurfer image analysis suite, version 5.3 (http://surfer.nmr.mgh.harvard.edu/). After registration to Talairach space and intensity normalization, the process involved an automatic skull stripping, which removes extra-cerebral structures, cerebellum and brainstem, by using a hybrid method combining watershed algorithms and deformable surface models. Images were carefully checked for skull stripping errors. After this step, images were segmented into GM, WM, and cerebrospinal fluid (CSF), cerebral hemispheres were separated, and subcortical structures were divided from cortical components. The WM/GM boundary was tessellated and the surface was deformed following intensity gradients to optimally place WM/GM and GM/CSF borders, thus obtaining the WM and the pial surfaces. The results of this segmentation procedure were inspected visually, and if necessary, edited manually by adding control points. Afterwards, surface inflation and registration to a spherical atlas were performed and the cerebral cortex was parcellated into 34 regions per hemisphere, based on gyral and sulcal structures, as described by Desikan et al. (Desikan et al., 2006). Finally, cortical thickness was estimated as the average shortest distance between the WM boundary and the pial surface. Mean cortical thickness of the precentral gyrus bilaterally was obtained.
2.2.3. DT MRI tractography
DT MRI analysis was performed using the FMRIB software library (FSL) tools (http://www.fmrib.ox.ac.uk/fsl/fdt/index.html) and the JIM6 software (Version 6.0, Xinapse Systems, Northants, UK, http://www.xinapse.com). The diffusion-weighted data were skull-stripped using the Brain Extraction Tool implemented in FSL. Using FMRIB's Linear Image Registration Tool (FLIRT), the two diffusion-weighted scans were coregistered by applying the rigid transformation needed to correct for position between the two b0 images (T2-weighted, but not diffusion-weighted). The rotation component was also applied to diffusion-weighted directions. Eddy currents correction was performed using the JIM6 software. Then, the two acquisitions were concatenated. The DT was estimated on a voxel-by-voxel basis using DTIfit provided by the FMRIB Diffusion Toolbox. Maps of mean diffusivity (MD), FA, axial diffusivity (axD) and radial diffusivity (radD) were obtained.
Seeds for tractography of the CST and CC were defined in the Montreal Neurological Institute (MNI) space on the FA template provided by FSL, as previously described (Agosta et al., 2014). Fiber tracking was performed in native DT MRI space using a probabilistic tractography algorithm implemented in FSL (probtrackx), which is based on Bayesian estimation of diffusion parameters (Bedpostx). Fiber tracking was initiated from all voxels within the seed masks in the diffusion space to generate 5000 streamline samples with a step length of 0.5 mm and a curvature threshold of 0.2. Using a “single-seed” approach, the reconstructions of the CC and bilateral CST were obtained. In addition, using a “seed to target” approach, the CC was segmented into three portions to identify the callosal fibers linking the primary motor cortices (CC-precentral), lateral premotor cortices (CC-premotor) and supplementary motor areas (CC-SMA). Tract maps were then normalized taking into consideration the number of voxels in the seed masks. To do so, the number of streamline samples present in the voxels of the tract maps was divided by the way-total, which corresponds to the total number of streamline samples that were not rejected by the exclusion masks. The tract masks obtained were thresholded at a value equal to 40% of the 95th percentile of the distribution of the intensity values of the voxels included in the tract, as previously described. This normalization procedure allowed us to correct for possible differences between tracts due to the different sizes of the starting seeds. In this way, we also excluded the background noise and avoided a too restrictive thresholding when the maximum intensity value was an outlier. Group probability maps of each thresholded tract were produced to visually check their anatomical consistency across study subjects. For each tract, the average MD, FA, axD, and radD were calculated in the native space.
2.3. Statistical analysis
2.3.1. Demographic and clinical data
Normal distribution assumption was checked by means of Q-Q plot and Shapiro-Wilks and Kolmogorov-Smirnov tests. Group comparisons were performed using ANOVA models, followed by post hoc pairwise comparisons. Standardized differences between groups were computed. All analyses were performed using SAS Release 9.3 (SAS Institute, Cary, NC, USA).
2.3.2. MRI data
Precentral gyrus mean cortical thickness and DT MRI measures were compared between groups using ANOVA models, false-discovery rate (FDR)-corrected for multiple comparisons and adjusting for subject's age.
2.3.3. Random forest analysis
The diagnostic accuracy of precentral cortical thickness measures and DT MRI metrics of the CST and motor callosal tracts were assessed in a training cohort (70 randomly selected ALS patients, 22 PUMN patients, and 78 healthy controls) and externally proved in a validation cohort (remaining 53 ALS and 22 PUMN patients, and 20 mimic disorders), using a random forest classifier (Breiman, 2001). This machine learning statistical algorithm, based on an ensemble of classification trees, allowed us to classify subjects according to their clinical status using the set of MRI variables. 100,000 individual classification trees of the forest were built using 0.632 + bootstrap subsampling without replacement. The best split at each node was selected from a random subset of covariates adopting a conditional permutation scheme to avoid possible bias in variable selection. The left-out observations (i.e., “out of bag” observations) were then used to obtain the classification error of each tree considered. The bootstrap re-sampling and the permutation strategy simulate, de facto, the natural variability of measures and provide an internal validation of the results: using a different bootstrap sample of the data and a different subset of predictors, randomly chosen to build each tree of the forest, random forest overcomes the concern of false positive discoveries. Analyses were performed using R (A language and environment for statistical computing).
2.3.4. Power analysis
The analysis was powered on the worst case scenario in terms of our study design (i.e., 20 ALS-mimic vs 22 PUMN patients). In this case, the study had a power of 80%, assuming a type I error of 5%, to detect an accuracy of 0.74. Therefore, for the other comparisons, the power to detect an accuracy of 0.74 was higher or, with a power of 80%, we were able to detect also smaller accuracies.
3. Results
3.1. MRI variables
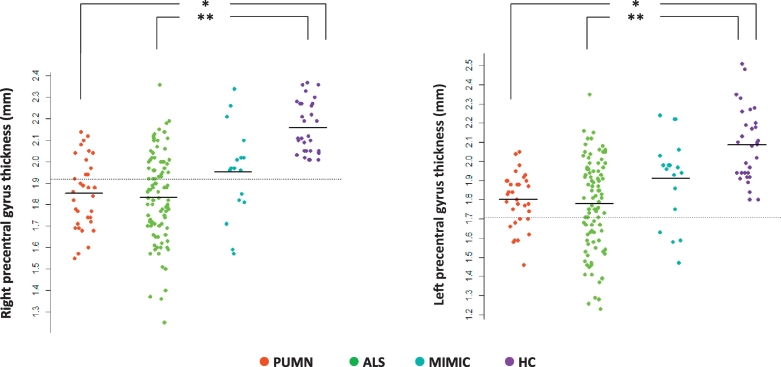
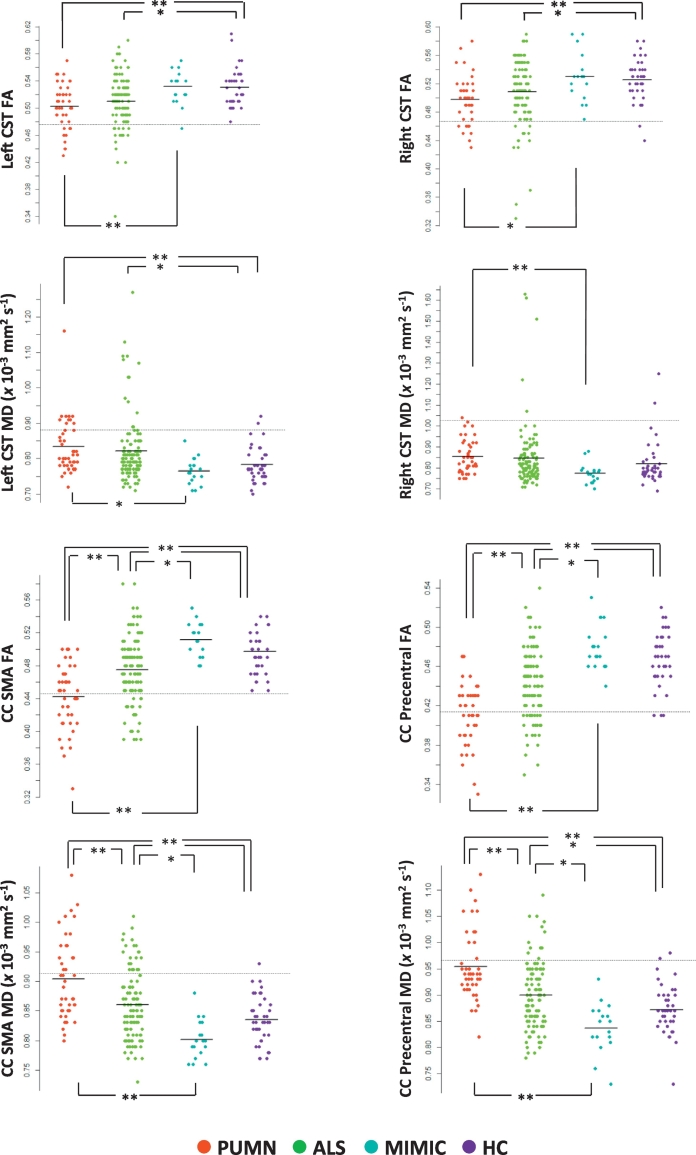
Compared with healthy subjects, ALS and PUMN patients showed a marked cortical thinning of the precentral gyrus bilaterally and altered DT MRI metrics of the CST bilaterally, CC-SMA and CC-precentral fibers (Fig. 1, Fig. 2, E-Table 1, Table 2). Compared to healthy controls, no DT MRI and cortical thickness abnormalities were observed in the whole ALS-mimic patient group (Fig. 2, E-Table 1, Table 2). When patients with Kennedy's disease were considered separately from other cases with mimic disorders relative to healthy controls, they showed no precentral cortical thinning and DT MRI abnormalities (E-Table 1). Relative to ALS-mimic cases, ALS patients showed a greater involvement of the CC-SMA and CC-precentral fibers, and PUMN patients had a more severe damage of the CST bilaterally, CC-SMA and CC-precentral fibers (Fig. 2, E-Table 1, Table 2). When PUMN cases were compared with patients with Kennedy's disease, DT MRI differences were slightly less significant (E-Table 1). PUMN patients showed a greater involvement of the callosal tracts relative to ALS cases (Fig. 2, E-Table 1, Table 2). No cortical thickness difference was found between ALS, PUMN and mimic disorder patients (Fig. 1, E-Table 1, Table 2).
Fig. 1.
Cortical thickness of the precentral gyrus. Mean cortical thickness of the precentral gyrus bilaterally are plotted per study group (healthy controls and patients with ALS, PUMN and ALS-mimic disorders). The dashed horizontal line indicates the mean cortical thickness minus 2 standard deviations of the healthy control group. Cortical thickness measures are in mm. *p < 0.05, **p < 0.001 (ANOVA model followed by pair-wise comparisons, false-discovery rate-corrected for multiple comparisons and adjusted for subject's age). Abbreviations: ALS = amyotrophic lateral sclerosis; HC = healthy controls; PUMN = predominantly upper motor neuron.
Fig. 2.
DT MRI measures of the CST and motor callosal tracts. Mean FA and MD values of the CST bilaterally and motor callosal tracts are plotted per study group (healthy controls and patients with ALS, PUMN and ALS-mimic disorders). The dashed horizontal line indicates the mean value minus 2 standard deviations of the healthy control group. MD values are in × 10− 3 mm2 s− 1. *p < 0.05, **p < 0.001 (ANOVA model followed by pair-wise comparisons, false-discovery rate-corrected for multiple comparisons and adjusted for subject's age). Abbreviations: ALS = amyotrophic lateral sclerosis; CC-precentral = callosal fibers linking the primary motor cortices; CC-SMA = callosal fibers linking the supplementary motor areas; CST = corticospinal tract; FA = fractional anisotropy; HC = healthy controls; MD = mean diffusivity; PUMN = predominantly upper motor neuron.
Table 2.
Diagnostic accuracy values of MRI variables estimated using the random forest analysis.
| Specificity | Sensitivity | Accuracy | |
|---|---|---|---|
| Training sets: | |||
| ALS patients vs healthy controls | |||
| Cortical thickness values | 0.97 | 0.81 | 0.86 |
| DT MRI values | 0.58 | 0.91 | 0.78 |
| Combined MRI model | 0.92 | 0.91 | 0.91 |
| PUMN patients vs healthy controls | |||
| Cortical thickness values | 1.00 | 0.69 | 0.89 |
| DT MRI values | 0.95 | 0.86 | 0.92 |
| Combined MRI model | 1.00 | 0.75 | 0.90 |
| ALS vs PUMN patients | |||
| Cortical thickness values | 0.00 | 1.00 | 0.73 |
| DT MRI values | 0.14 | 1.00 | 0.77 |
| Combined MRI model | 0.00 | 1.00 | 0.72 |
| Validation sets: | |||
| ALS vs PUMN patients | |||
| Cortical thickness values | 0.00 | 1.00 | 0.75 |
| DT MRI values | 0.45 | 0.93 | 0.80 |
| Combined MRI model | 0.00 | 1.00 | 0.74 |
| ALS vs ALS-mimic patients | |||
| Cortical thickness values | 0.56 | 0.82 | 0.73 |
| DT MRI values | 0.72 | 0.92 | 0.87 |
| Combined MRI model | 0.75 | 0.92 | 0.87 |
| PUMN vs ALS-mimic patients | |||
| Cortical thickness values | 0.67 | 0.82 | 0.74 |
| DT MRI values | 0.94 | 0.95 | 0.95 |
| Combined MRI model | 0.94 | 0.94 | 0.94 |
Abbreviations: ALS = amyotrophic lateral sclerosis; DT MRI = diffusion tensor MRI; PUMN = predominantly upper motor neuron.
3.2. Diagnostic accuracy values of MRI metrics
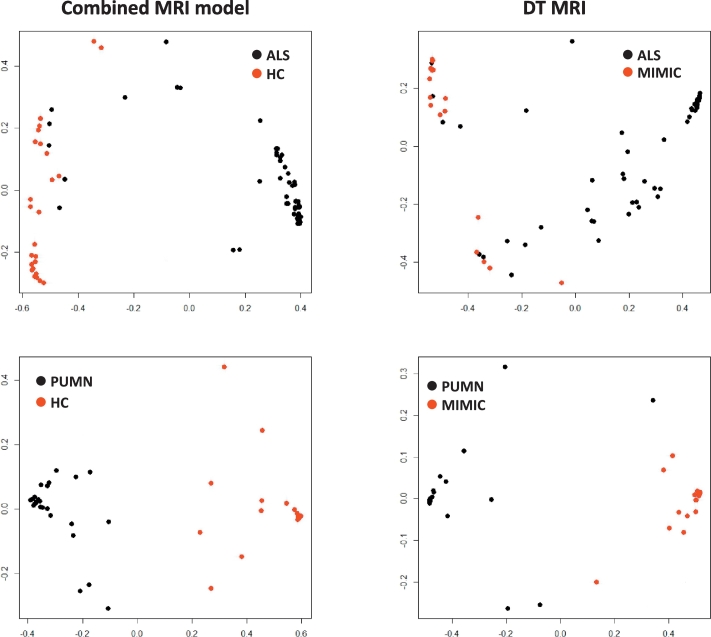
Table 2 reports accuracy, sensitivity and specificity of the MRI metrics in the individual identification of subjects in both training and validation sets. In the training set, precentral cortical thickness showed 0.86 accuracy and DT MRI measures 0.78 accuracy in differentiating ALS patients from healthy controls. The classification pattern considerably improved when the MRI metrics were combined, reaching 0.91 accuracy, 0.91 sensitivity and 0.92 specificity (Fig. 3). In the comparison PUMN patients vs controls, the best diagnostic performance was achieved by the DT MRI which showed 0.92 accuracy (0.86 sensitivity, 0.95 specificity) compared with precentral cortical thickness showing 0.89 accuracy (0.69 sensitivity, 1.00 specificity). The combined model did not improve the ability to differentiate PUMN and healthy subjects (Fig. 3). Both precentral cortical thickness and DT MRI metrics showed a fair accuracy (0.73 and 0.77, respectively) with high sensitivity (1.00) but low specificity (0.00 and 0.14, respectively) in distinguishing ALS from PUMN patients. The analysis of the validation test confirmed a low performance of MRI measures in separating ALS and PUMN groups (Table 2). In the validation cohort, the best accuracy vs mimic disorders was reached by DT MRI (0.87 and 0.95 in ALS and PUMN, respectively), while cortical thickness provided 0.73 and 0.74 accuracy (Table 2). The combined model distinguished ALS and PUMN from mimic syndromes with performance (0.87 and 0.94 accuracy, respectively) similar to that of DT MRI alone (Table 2) (Fig. 3).
Fig. 3.
Diagnostic ability of MRI models. Figure shows the ability of the combined MRI model and the DT MRI measures in differentiating ALS and PUMN patients from healthy controls and ALS-mimic disorders, respectively. Scaling coordinates of the proximity matrix from random forest were used to represent the distance – in terms of MRI values – between subjects with different diagnoses. Abbreviations: ALS = amyotrophic lateral sclerosis; DT = diffusion tensor; HC = healthy controls; PUMN = predominantly upper motor neuron.
4. Discussion
This is a structural MRI study suggesting the potential of multimodal imaging as a diagnostic marker for individual ALS and PUMN cases, and its accuracy in discriminating MND patients from ALS-mimic diseases in an independent patient cohort. We demonstrated that a multiparametric MRI approach significantly improves diagnostic accuracy over using each modality independently in the identification of single patients with ALS and PUMN relative to healthy controls, while DT MRI represents the most powerful tool to distinguish MND from mimic disorders.
Our MRI approach combining cortical thickness and microstructural integrity of MND-specific anatomical regions achieved high sensitivity and moderate to high specificity in distinguishing ALS and PUMN individual cases from healthy controls. Findings in ALS are in keeping with a recent classification study using a cross-validation binary logistic regression model (Schuster et al., 2016), although the specificity of our approach was higher. This can be related to the different methods applied to obtain MRI features, particularly to the greater reliability of cortical thickness measures relative to GM density values in assessing the motor cortex involvement in MND (Chiò et al., 2014, Menke et al., 2017). In a clinical setting, misdiagnosis of MND remains a common challenge (Davenport et al., 1996, Traynor et al., 2000). Therefore, a high degree of both sensitivity and specificity is paramount to prevent patients with mimic conditions to be misclassified as MND.
To prove further the specificity of our approach, we tested the model in an independent validation sample of MND patients relative to subjects with ALS-mimic conditions. Previous MRI-based classification studies of MND did not include patients with mimic disorders (Ben Bashat et al., 2011, Foerster et al., 2013, Schuster et al., 2016). We found significant thinning of the primary motor cortex and altered DT MRI metrics of motor WM tracts in patients with ALS and PUMN but not in the whole group of ALS-mimic cases relative to healthy controls. Furthermore, when we compared MND with mimic patients, the former group showed greater WM damage involving the CST and motor callosal fibers. To the best of our knowledge, only a few studies compared brain MRI features of ALS patients to those of mimic subjects. The lack of significant precentral cortical atrophy in ALS-mimics is in agreement with a single previous study (Walhout et al., 2015). Higher apparent diffusion coefficients were observed in the precentral gyrus and CST in patients with ALS relative to subjects with cervical spondylotic myelopathy (Koike et al., 2015). It is noteworthy that the majority of our mimic cases were patients with Kennedy's disease. Previous studies reported some extent of brain damage in this LMN variant, such as frontal GM atrophy (Kassubek et al., 2007) and glucose hypometabolism (Lai et al., 2013), CST degeneration (Pieper et al., 2013, Unrath et al., 2010), and cerebellar involvement (Kassubek et al., 2007, Pieper et al., 2013), although findings were not replicated by all studies. The small group of patients with Kennedy's disease included in the present study did not show MRI abnormalities relative to healthy controls. Methodological differences (e.g., region-of-interest-based cortical thickness measurement vs voxel-based morphometry; tractography vs voxel-wise analysis) may explain inconsistent findings among studies. Nevertheless, we cannot exclude that the subtle cerebral pathology in Kennedy's disease cases could at least partially explain the spread of MRI values in the mimic group in Fig. 2.
Our classification model demonstrated that DT MRI measures alone are able to discriminate classical ALS and PUMN patients from mimic syndromes with the highest accuracy. These findings are likely to be associated with the elevated sensitivity of DT MRI to UMN involvement and the inclusion of mimic patients with a clinically prominent LMN impairment. Future studies should test the validity of MRI-based models in distinguishing MND patients from cases with prominent UMN impairment.
Notably, the most pronounced damage in MND patients relative to ALS-mimic subjects was detected in the motor callosal fibers. Damage of the body of the CC is a well-known feature in patients with UMN impairment. A greater CC involvement has been consistently reported in PLS patients relative to classic ALS cases (Agosta et al., 2014, Iwata et al., 2011, Unrath et al., 2010). Previous DT MRI studies have demonstrated that motor callosal fibers are impaired in ALS but not in predominant LMN variants (Ben Bashat et al., 2011, Spinelli et al., 2016). In the present study, we have widened these findings, demonstrating that CC damage also represents a powerful tool in the differential diagnosis with ALS-mimic syndromes including (non-ALS) pure LMN and peripheral nervous system disorders. The assessment of additional WM tracts according to the staging hypothesis of ALS (Brettschneider et al., 2013), such as the corticorubral and corticopontine tracts, the corticostriatal pathway and the perforant path (Kassubek et al., 2014), may further improve the classification model.
The study is not without limitations. The first shortcoming deals with the relatively small number of ALS-mimic cases, although power analysis showed that 20 ALS-mimic patients resulted in 80% power to detect MRI differences. Second, the heterogeneity of our ALS-mimic sample, which included a majority of patients with Kennedy's disease and a minority of subjects with peripheral nervous system pathology, prevented the possibility to perform separate MRI analyses. Third, the moderately low number of genetic patients did not allow us to test the accuracy of the models separately in sporadic and genetic MND. These limitations can be overcome in the framework of multisite collaborations. NiSALS (www.nisals.org) is a thriving multinational, academic-led research consortium, which has established a large MND data repository of quality-controlled MRI scans from countries all over the world providing an ideal resource to test classification models (Filippi et al., 2015). Fourth, our classification model included advanced multimodal structural analyses, which can be time consuming. More effort should be made in order to allow MRI-based machine learning models to be better included in clinical neuroimaging/clinical practice. It is also noteworthy that both cortical thickness measurement and DT MRI tractography present some technical limitations. In particular, although Freesurfer has the potential to generate accurate surface representations, some manual intervention can be necessary when segmentation errors occur. Probabilistic tractography is extremely powerful in WM damage detection; however, this approach might lack sensitivity in the identification of some important tracts including the lateral portions of the motor pathways, due to difficulties in estimating multiple diffusion orientations, and recent studies have therefore proposed important extensions of this model. Finally, the role of functional MRI (fMRI) metrics in the differential diagnosis of MND still need to be explored. A number of fMRI studies investigating differences between MND patients and controls at rest have revealed functional connectivity abnormalities within the sensorimotor network (Chiò et al., 2014, Menke et al., 2017). Thus, the inclusion of resting state fMRI analyses is likely to considerably increase the overall accuracy in future classification models of multimodal datasets (Welsh et al., 2013). Despite these limitations, the present study provides a validated diagnostic MRI model to differentiate MND variants from healthy controls and mimic disorders with clinically prominent LMN impairment, providing a roadmap for translation of MRI markers into daily clinical practice.
Funding
The study was funded by AriSLA - Fondazione Italiana di Ricerca per la SLA (MacLearnALS Project).
Disclosure statement
P.M. Ferraro, N. Riva, E.G. Spinelli, Y. Falzone have nothing to disclose.
F. Agosta is Section Editor of NeuroImage: Clinical and serves as a member of the editorial board of Journal of Neurology; has received speaker honoraria from ExceMED – Excellence in Medical Education and Biogen Idec; and receives research supports from the Italian Ministry of Health, AriSLA (Fondazione Italiana di Ricerca per la SLA), and the European Research Council.
M. Copetti has received compensation for consulting and/or serving on advisory boards from Teva Pharmaceuticals and Biogen Idec.
G. Sorarù receives research support from the Italian Ministry of Health, Telethon, and the French Muscular Dystrophy Association.
G. Comi has received consulting fees for participating on advisory boards from Novartis, Teva Pharmaceutical Ind. Ltd., Sanofi, Genzyme, Merck Serono, Bayer, Actelion and honorarium for speaking activities for Novartis, Teva Pharmaceutical Ind. Ltd., Sanofi, Genzyme, Merck Serono, Bayer, Biogen, ExceMED.
A. Chiò serves as a member of the editorial board of Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration; has served on scientific advisory boards for Biogen Idec, Cytokinetics, Italfarmaco, Neuraltus, Mitsubishi Tanabe; and has received research support from the Italian Ministry of Health, Italian Ministry of Education University and Research, European Commission, Compagnia di San Paolo, AriSLA (Fondazione Italiana di Ricerca per la SLA), Fondazione Vialli e Mauro Onlus, and Associazione Piemontese per l'Assistenza alla SLA (APASLA).
M. Filippi is Editor-in-Chief of the Journal of Neurology; serves on a scientific advisory board for Teva Pharmaceutical Industries; has received compensation for consulting services and/or speaking activities from Biogen Idec, ExceMED, Novartis, and Teva Pharmaceutical Industries; and receives research support from Biogen Idec, Teva Pharmaceutical Industries, Novartis, Italian Ministry of Health, Fondazione Italiana Sclerosi Multipla, Cure PSP, Alzheimer's Drug Discovery Foundation (ADDF), the Jacques and Gloria Gossweiler Foundation (Switzerland), and ARiSLA (Fondazione Italiana di Ricerca per la SLA).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nicl.2017.08.002.
Appendix A. Supplementary data
Supplementary tables
References
- Agosta F., Pagani E., Petrolini M., Caputo D., Perini M., Prelle A., Salvi F., Filippi M. Assessment of white matter tract damage in patients with amyotrophic lateral sclerosis: a diffusion tensor MR imaging tractography study. AJNR Am. J. Neuroradiol. 2010;31:1457–1461. doi: 10.3174/ajnr.A2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta F., Valsasina P., Riva N., Copetti M., Messina M.J., Prelle A., Comi G., Filippi M. The cortical signature of amyotrophic lateral sclerosis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta F., Galantucci S., Riva N., Chio A., Messina S., Iannaccone S., Calvo A., Silani V., Copetti M., Falini A., Comi G., Filippi M. Intrahemispheric and interhemispheric structural network abnormalities in PLS and ALS. Hum. Brain Mapp. 2014;35:1710–1722. doi: 10.1002/hbm.22286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta F., Al-Chalabi A., Filippi M., Hardiman O., Kaji R., Meininger V., Nakano I., Shaw P., Shefner J., van den Berg L.H., Ludolph A. The El Escorial criteria: strengths and weaknesses. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16:1–7. doi: 10.3109/21678421.2014.964258. [DOI] [PubMed] [Google Scholar]
- Agosta F., Ferraro P.M., Riva N., Spinelli E.G., Chio A., Canu E., Valsasina P., Lunetta C., Iannaccone S., Copetti M., Prudente E., Comi G., Falini A., Filippi M. Structural brain correlates of cognitive and behavioral impairment in MND. Hum. Brain Mapp. 2016;37:1614–1626. doi: 10.1002/hbm.23124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsh J.M. ALS diagnostic criteria of El Escorial revisited: do they meet the needs of clinicians as well as researchers? Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2000;1(Suppl. 1):S57–60. doi: 10.1080/14660820052415925. [DOI] [PubMed] [Google Scholar]
- Ben Bashat D., Artzi M., Tarrasch R., Nefussy B., Drory V.E., Aizenstein O. A potential tool for the diagnosis of ALS based on diffusion tensor imaging. Amyotroph. Lateral Scler. 2011;12:398–405. doi: 10.3109/17482968.2011.582646. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Brettschneider J., Del Tredici K., Toledo J.B., Robinson J.L., Irwin D.J., Grossman M., Suh E., Van Deerlin V.M., Wood E.M., Baek Y., Kwong L., Lee E.B., Elman L., McCluskey L., Fang L., Feldengut S., Ludolph A.C., Lee V.M., Braak H., Trojanowski J.Q. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann. Neurol. 2013;74:20–38. doi: 10.1002/ana.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., Miller R.G., Swash M., Munsat T.L. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- Chio A., Calvo A., Moglia C., Mazzini L., Mora G. Phenotypic heterogeneity of amyotrophic lateral sclerosis: a population based study. J. Neurol. Neurosurg. Psychiatry. 2011;82:740–746. doi: 10.1136/jnnp.2010.235952. [DOI] [PubMed] [Google Scholar]
- Chiò A., Pagani M., Agosta F., Calvo A., Cistaro A., Filippi M. Neuroimaging in amyotrophic lateral sclerosis: systematic insight into structural and functional changes. Lancet Neurol. 2014;13:1228–1240. doi: 10.1016/S1474-4422(14)70167-X. [DOI] [PubMed] [Google Scholar]
- Davenport R.J., Swingler R.J., Chancellor A.M., Warlow C.P. Avoiding false positive diagnoses of motor neuron disease: lessons from the Scottish motor neuron disease register. J. Neurol. Neurosurg. Psychiatry. 1996;60:147–151. doi: 10.1136/jnnp.60.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R.S., Segonne F., Fischl B., Quinn B.T., Dickerson B.C., Blacker D., Buckner R.L., Dale A.M., Maguire R.P., Hyman B.T., Albert M.S., Killiany R.J. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Douaud G., Filippini N., Knight S., Talbot K., Turner M.R. Integration of structural and functional magnetic resonance imaging in amyotrophic lateral sclerosis. Brain. 2011;134:3470–3479. doi: 10.1093/brain/awr279. [DOI] [PubMed] [Google Scholar]
- Filippi M., Agosta F., Abrahams S., Fazekas F., Grosskreutz J., Kalra S., Kassubek J., Silani V., Turner M.R., Masdeu J.C. EFNS guidelines on the use of neuroimaging in the management of motor neuron diseases. Eur. J. Neurol. 2010;17 doi: 10.1111/j.1468-1331.2010.02951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M., Agosta F., Grosskreutz J., Benatar M., Kassubek J., Verstraete E., Turner M.R. Progress towards a neuroimaging biomarker for amyotrophic lateral sclerosis. Lancet Neurol. 2015;14:786–788. doi: 10.1016/S1474-4422(15)00134-9. [DOI] [PubMed] [Google Scholar]
- Foerster B.R., Dwamena B.A., Petrou M., Carlos R.C., Callaghan B.C., Churchill C.L., Mohamed M.A., Bartels C., Benatar M., Bonzano L., Ciccarelli O., Cosottini M., Ellis C.M., Ehrenreich H., Filippini N., Ito M., Kalra S., Melhem E.R., Pyra T., Roccatagliata L., Senda J., Sobue G., Turner M.R., Feldman E.L., Pomper M.G. Diagnostic accuracy of diffusion tensor imaging in amyotrophic lateral sclerosis: a systematic review and individual patient data meta-analysis. Acad. Radiol. 2013;20:1099–1106. doi: 10.1016/j.acra.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster B.R., Carlos R.C., Dwamena B.A., Callaghan B.C., Petrou M., Edden R.A., Mohamed M.A., Welsh R.C., Barker P.B., Feldman E.L., Pomper M.G. Multimodal MRI as a diagnostic biomarker for amyotrophic lateral sclerosis. Ann. Clin. Transl. Neurol. 2014;1:107–114. doi: 10.1002/acn3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff M.M., Sage C.A., Caan M.W., Akkerman E.M., Lavini C., Majoie C.B., Nederveen A.J., Zwinderman A.H., Vos F., Brugman F., van den Berg L.H., de Rijk M.C., van Doorn P.A., Van Hecke W., Peeters R.R., Robberecht W., Sunaert S., de Visser M. Upper and extra-motoneuron involvement in early motoneuron disease: a diffusion tensor imaging study. Brain. 2011;134:1211–1228. doi: 10.1093/brain/awr016. [DOI] [PubMed] [Google Scholar]
- Graham J.M., Papadakis N., Evans J., Widjaja E., Romanowski C.A., Paley M.N., Wallis L.I., Wilkinson I.D., Shaw P.J., Griffiths P.D. Diffusion tensor imaging for the assessment of upper motor neuron integrity in ALS. Neurology. 2004;63:2111–2119. doi: 10.1212/01.wnl.0000145766.03057.e7. [DOI] [PubMed] [Google Scholar]
- Iwata N.K., Kwan J.Y., Danielian L.E., Butman J.A., Tovar-Moll F., Bayat E., Floeter M.K. White matter alterations differ in primary lateral sclerosis and amyotrophic lateral sclerosis. Brain. 2011;134:2642–2655. doi: 10.1093/brain/awr178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassubek J., Juengling F.D., Sperfeld A.D. Widespread white matter changes in Kennedy disease: a voxel based morphometry study. J. Neurol. Neurosurg. Psychiatry. 2007;78:1209–1212. doi: 10.1136/jnnp.2006.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassubek J., Muller H.P., Del Tredici K., Brettschneider J., Pinkhardt E.H., Lule D., Bohm S., Braak H., Ludolph A.C. Diffusion tensor imaging analysis of sequential spreading of disease in amyotrophic lateral sclerosis confirms patterns of TDP-43 pathology. Brain. 2014;137:1733–1740. doi: 10.1093/brain/awu090. [DOI] [PubMed] [Google Scholar]
- Kiernan M.C., Vucic S., Cheah B.C., Turner M.R., Eisen A., Hardiman O., Burrell J.R., Zoing M.C. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- Koike Y., Kanazawa M., Terajima K., Watanabe K., Ohashi M., Endo N., Shimohata T., Nishizawa M. Apparent diffusion coefficients distinguish amyotrophic lateral sclerosis from cervical spondylotic myelopathy. Clin. Neurol. Neurosurg. 2015;132:33–36. doi: 10.1016/j.clineuro.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Lai T.H., Liu R.S., Yang B.H., Wang P.S., Lin K.P., Lee Y.C., Soong B.W. Cerebral involvement in spinal and bulbar muscular atrophy (Kennedy's disease): a pilot study of PET. J. Neurol. Sci. 2013;335:139–144. doi: 10.1016/j.jns.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Menke R.A., Agosta F., Grosskreutz J., Filippi M., Turner M.R. Neuroimaging endpoints in amyotrophic lateral sclerosis. Neurotherapeutics. 2017;14:11–23. doi: 10.1007/s13311-016-0484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.D., Callagher P., Gardham J., Mitchell C., Dixon M., Addison-Jones R., Bennett W., O'Brien M.R. Timelines in the diagnostic evaluation of people with suspected amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND)—a 20-year review: can we do better? Amyotroph. Lateral Scler. 2010;11:537–541. doi: 10.3109/17482968.2010.495158. [DOI] [PubMed] [Google Scholar]
- Montuschi A., Iazzolino B., Calvo A., Moglia C., Lopiano L., Restagno G., Brunetti M., Ossola I., Lo Presti A., Cammarosano S., Canosa A., Chio A. Cognitive correlates in amyotrophic lateral sclerosis: a population-based study in Italy. J. Neurol. Neurosurg. Psychiatry. 2015;86:168–173. doi: 10.1136/jnnp-2013-307223. [DOI] [PubMed] [Google Scholar]
- Nelles M., Block W., Traber F., Wullner U., Schild H.H., Urbach H. Combined 3T diffusion tensor tractography and 1H-MR spectroscopy in motor neuron disease. AJNR Am. J. Neuroradiol. 2008;29:1708–1714. doi: 10.3174/ajnr.A1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukan J., Elamin M., Bede P., Jordan N., Gallagher L., Byrne S., Lynch C., Pender N., Hardiman O. The syndrome of cognitive impairment in amyotrophic lateral sclerosis: a population-based study. J. Neurol. Neurosurg. Psychiatry. 2012;83:102–108. doi: 10.1136/jnnp-2011-300188. [DOI] [PubMed] [Google Scholar]
- Pieper C.C., Konrad C., Sommer J., Teismann I., Schiffbauer H. Structural changes of central white matter tracts in Kennedy's disease - a diffusion tensor imaging and voxel-based morphometry study. Acta Neurol. Scand. 2013;127:323–328. doi: 10.1111/ane.12018. [DOI] [PubMed] [Google Scholar]
- Pringle C.E., Hudson A.J., Munoz D.G., Kiernan J.A., Brown W.F., Ebers G.C. Primary lateral sclerosis. Clinical features, neuropathology and diagnostic criteria. Brain. 1992;115(Pt 2):495–520. doi: 10.1093/brain/115.2.495. [DOI] [PubMed] [Google Scholar]
- Schuster C., Kasper E., Machts J., Bittner D., Kaufmann J., Benecke R., Teipel S., Vielhaber S., Prudlo J. Focal thinning of the motor cortex mirrors clinical features of amyotrophic lateral sclerosis and their phenotypes: a neuroimaging study. J. Neurol. 2013;260:2856–2864. doi: 10.1007/s00415-013-7083-z. [DOI] [PubMed] [Google Scholar]
- Schuster C., Hardiman O., Bede P. Development of an automated MRI-based diagnostic protocol for amyotrophic lateral sclerosis using disease-specific pathognomonic features: a quantitative disease-state classification study. PLoS One. 2016;11 doi: 10.1371/journal.pone.0167331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli E.G., Agosta F., Ferraro P.M., Riva N., Lunetta C., Falzone Y.M., Comi G., Falini A., Filippi M. Brain MR imaging in patients with lower motor neuron-predominant disease. Radiology. 2016;280:545–556. doi: 10.1148/radiol.2016151846. [DOI] [PubMed] [Google Scholar]
- Traynor B.J., Codd M.B., Corr B., Forde C., Frost E., Hardiman O. Amyotrophic lateral sclerosis mimic syndromes: a population-based study. Arch. Neurol. 2000;57:109–113. doi: 10.1001/archneur.57.1.109. [DOI] [PubMed] [Google Scholar]
- Unrath A., Muller H.P., Riecker A., Ludolph A.C., Sperfeld A.D., Kassubek J. Whole brain-based analysis of regional white matter tract alterations in rare motor neuron diseases by diffusion tensor imaging. Hum. Brain Mapp. 2010;31:1727–1740. doi: 10.1002/hbm.20971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraete E., Veldink J.H., Hendrikse J., Schelhaas H.J., van den Heuvel M.P., van den Berg L.H. Structural MRI reveals cortical thinning in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry. 2011;83:383–388. doi: 10.1136/jnnp-2011-300909. [DOI] [PubMed] [Google Scholar]
- Walhout R., Westeneng H.J., Verstraete E., Hendrikse J., Veldink J.H., van den Heuvel M.P., van den Berg L.H. Cortical thickness in ALS: towards a marker for upper motor neuron involvement. J. Neurol. Neurosurg. Psychiatry. 2015;86:288–294. doi: 10.1136/jnnp-2013-306839. [DOI] [PubMed] [Google Scholar]
- Welsh R.C., Jelsone-Swain L.M., Foerster B.R. The utility of independent component analysis and machine learning in the identification of the amyotrophic lateral sclerosis diseased brain. Front. Hum. Neurosci. 2013;7:251. doi: 10.3389/fnhum.2013.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables