Abstract
Gene inactivation is essential for forward and reverse genetic approaches to establish protein function. Techniques such as insertion or chemical mutagenesis have been developed to mutagenize chlamydiae via targeted or random mutagenesis, respectively. Both of these approaches require transformation of chlamydiae to either introduce insertion elements or complement mutants. We have recently developed a targeted mutagenesis strategy, fluorescence-reported allelic exchange mutagenesis (FRAEM), to delete Chlamydia trachomatis L2 genes. This approach overcomes several barriers for genetically manipulating intracellular bacteria. Perhaps most significantly, FRAEM employs fluorescence reporting to indicate successful transformation and subsequent recombination events. Three protocols are provided that detail methods to construct gene-specific suicide vectors, transform C. trachomatis L2 to select for recombinants, and isolate clonal populations via limiting dilution. In aggregate, these protocols will allow investigators to engineer C. trachomatis L2 strains carrying complete deletions of desired gene(s).
Keywords: Transformation, Chlamydia, Mutagenesis
INTRODUCTION
Genetic manipulation of many bacterial species has been feasible for decades, and has helped elucidate how prokaryotic cells grow, divide, utilize nutrients, respond to the environment, communicate with each other, and in the case of pathogenic bacteria, how they interact with their hosts to cause disease. However, for numerous bacterial genera, any kind of targeted genetic manipulation remains elusive. Members of the genus Chlamydia are obligate intracellular parasites that undergo a biphasic developmental cycle. The bacterium is either in the form of an infectious elementary body that is spore-like and specialized for transmission, or in the form of a vegetative, but non-infectious reticulate body. This lifestyle posed significant barriers to achievement of specific inactivation of genes within the chlamydial genome. Reproducible transformation of Chlamydia was achieved via CaCl2 treatment and an expression plasmid containing the eight genes encoded on the 7.5 kb cryptic chlamydial plasmid (Wang, et al. 2011). Allelic exchange by homologous recombination in C. trachomatis has been successfully established only very recently (Mueller, Wolf, & Fields, 2016). This approach relies on a conditionally replicating plasmid generated by re-engineering the portion of the construct containing the 8 chlamydial genes. The chlamydial plasmid encodes eight putative ORFs designated as pgp1–8 (reviewed in (Zhong, 2016). Although the precise function of these genes is unknown, the expression of pgp6 appears to be crucial for maintenance of the plasmid by the bacterium. Deletion of pgp6 leads to loss of the cryptic plasmid after a few rounds of chlamydial infection (Gong, Yang, Lei, Shen, & Zhong, 2013) (Song et al., 2013). Initially, the pL2 plasmid isolated from C. trachomatis, serovar L2/434Bu was utilized to produce the chlamydial cloning plasmid pBOMB4-Tet-mCherry (Bauler & Hackstadt, 2014). This cloning plasmid was then used as a backbone to generate a chlamydial suicide vector, pSUmC, where the expression of pgp6 was placed under control of the tetracycline (Tet) inducible promoter. A bla coding region is located downstream of pgp6, followed by the pUC derivative of the pMB1 origin of replication and the gene encoding the red-fluorescent protein, mCherry. The presence of anhydrotetracycline (ATc) inducer in the culture medium permits expression of pgp6, and confers the ability of C. trachomatis to maintain the pSUmC vector. When ATc is removed, the Tet repressor binds to the operator upstream of pgp6, and expression of this gene is inhibited, resulting in loss of pSUmC (Mueller et al., 2016).
Basic Protocol 1 describes the generation of a construct where a target chlamydial gene flanked by 3 kb sequences is cloned into the vector pUC18A. pUC18A expresses β-lactam and spectinomycin resistance genes bla and aadA, respectively. Chlamydial sequences that are inserted into this plasmid replace the bla gene. Once the construct is propagated and purified from E.coli, the chlamydial target gene in pUC18A is substituted with a bla-gfp cassette. Finally, the bla-gfp cassette surrounded by 3 kb sequences from the chlamydial genome is mobilized into pSUmC. Basic Protocol 2 focuses on transformation of C. trachomatis L2 with the above generated suicide vector, maintenance of infected cultures in the presence of penicillin G and ATc, as well as isolation of transformants. As mentioned above, when transformed chlamydiae are then cultivated in the absence of ATc, the maintenance of the suicide vector becomes impaired. Thus, addition of penicillin G selects for chlamydiae which have spontaneously exchanged the bla-gfp cassette of the suicide vector with the target gene on the chlamydial chromosome through homologous recombination. Generation of chlamydial mutants and isolation of isogenic population of mutant C. trachomatis are the topic of Basic Protocol 3.
CAUTION: Chlamydia trachomatis is a Biosafety Level 2 (BSL2) pathogen. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms. See UNIT 1A.1 and other pertinent resources (APPENDIX 1B) for more information.
BASIC PROTOCOL 1
Assembly of deletion construct
Chlamydial mutants have been successfully generated through the use of deletion constructs based on the suicide vector, pSUmC. However, assembly of these constructs requires multiple modifications of pSUmC in order to introduce the bla-gfp cassette flanked by genomic targeting sequences (Fig 1). Initially, bla in the cloning vector, pUC18A will be replaced with the gene of interest surrounded by the ~3.0 kb flanking sequences of the C. trachomatis genome by insertion/ deletion PCR (Geiser, et al. 2001). Next, the chlamydial gene in this construct will be removed by divergent PCR, and replaced with the bla-gfp cassette amplified from pUC19G by blunt-end ligation. Lastly, the entire region required for allelic exchange by homologous recombination, the ~3.0 kb arms flanking the bla-gfp cassette, will be mobilized into pSUmC by insertion/deletion PCR. E. coli transformed with the suicide vector will be resistant to carbenicillin and display green and red fluorescence. The final pSUmC construct is then propagated in and purified from methyltransferase-deficient E.coli prior to transforming chlamydiae and initiation of the FRAEM approach.
Figure 1.
Schematic of overall cloning strategy (Basic Protocol 1) used for constructing suicide plasmids. 1. The gene of interest (ct00X) is amplified from C. trachomatis L2 genomic DNA with 3 kb of up (us)- and down (ds)-stream homology arm (HR) DNA (steps 1–3). 2. The PCR product is used in an insertion/deletion PCR reaction using pUC18A as template such that the chlamydial DNA sequence replaces plasmid-encoded bla. (steps 4–7). 3. The target gene (ct00X) is deleted using Away primers, annealing immediately outside the ct00X coding sequence, in a divergent PCR reaction to amplify the remaining pUC18@ct00X construct. The divergent PCR product is then ligated to the bla and gfp cassette amplified from pUC19G to generate a pUC18 plasmid where ct00X has been replaced by bla-gfp (steps 8–13). 4. The chlamydial DNA required for homologous recombination plus the bla-gfp selection marker is PCR amplified using engineered primers (Step 14). 5. An insertion PCR is used to replace the bla gene in pSuMc with the homologous-recombination construct (steps 15–18). 6. The completed suicide plasmid is propagated in dam−/dcm− E. coli prior to the transformation of chlamydiae outlined in protocol 2 (step 19).
Materials
pSUmC plasmid DNA
pUC18A plasmid DNA
pUC19G plasmid DNA
Primer pair to amplify the gene targeted for deletion surrounded by approximately 3-kilobase (kb) flanking sequences from chlamydial genomic DNA: X@pUC F and R
Primer pair surrounding, but directed away from, the target gene: X-away F and R
-
Primer pair for amplifying the bla-gfp cassette from pUC19G:
bla-gfp F: GGAAATGTGCGCGGAACCC
bla-gfp R: TTACTTGTATAGTTCATCCATGCCATGTG
-
Primer pair for amplifying the homologous recombination sequence for insertion into pSUmC:
HomRR@pSUmC F: CTGCAGGTACCGGTCGACCATTC GGTCTGACGCTCAGTGGAACG
HomRR@pSUmC R: GATCTTTCTACGGGGTCTGACGCTC CTGGCGTTACCCAACTTAATCGCC
Q5® High-Fidelity DNA Polymerase (M0491S, NEB)
DpnI restriction enzyme (R0176S, NEB)
Cutsmart buffer (B7204S, NEB)
T4 Polynucleotide Kinase (M0201S, NEB)
Quick Ligation™ Kit (M2200S, NEB)
NEB® 10-beta Competent E. coli (C3019H, NEB)
dam−/dcm− Competent E. coli (C2925I, NEB)
Zyppy™ Plasmid Miniprep Kit (D4036, Zymo Research)
Zymoclean™ Gel DNA Recovery Kit (D4001, Zymo Research)
QIAfilter Plasmid Maxi Kit (12262, Qiagen)
Spectinomycin dihydrochloride pentahydrate (J61820, Alfa Aesar)
Carbenicillin disodium salt (J61949, Alfa Aesar)
LB Media (J106, VWR)
Agar Powder (A10752, Alfa Aesar)
Agarose LE (BMK-A1705, KSE Scientific)
Phenol–Chloroform (6805, EMD Millipore)
3M sodium acetate
-
100% ethanol
-
Design primers for amplifying gene × flanked upstream and downstream (±) by 3 kb arms for insertion into pUC18A.
Sequences should be designed with melting temperatures of approximately 60°C which hairpin at temperatures no higher than 40°C, as determined by OligoAnalyzer 3.1 (Integrated DNA Technologies). Forward and reverse sequences should then be added to the 3’ end of X@pUC F and X@pUC R, respectively, to produce the complete primer sequences:X@pUC F: GGTCTGACGCTCAGTGGAACG-XX@pUC R: CTGGCGTTACCCAACTTAATCGCC-X -
Design primers for amplifying away from the target gene.
Sequences should be designed with melting temperatures of approximately 60°C which hairpin at temperatures no higher than 40°C, as determined by OligoAnalyzer 3.1 (Integrated DNA Technologies).X-away FX-away R Amplify the gene targeted for deletion with 3-kb flanking sequences from chlamydial genomic DNA using X@pUC F and X@pUC R primers with Q5® High-Fidelity DNA Polymerase according to manufacturer’s instructions. Remove protein from PCR reactions by phenol-chloroform extraction, and precipitate DNA by ethanol precipitation with sodium acetate. Resuspend the DNA pellet and separate the sample in 0.8%-agarose gel. Extract the correct PCR product with the Zymoclean™ Gel DNA Recovery Kit.
-
Insert the gene × ± 3-kb arms-PCR product into pUC18A by insertion/deletion PCR.
Set up a 50-µl Q5® High-Fidelity DNA Polymerase reaction with 150 ng of pUC18A, 2.8 µg of gene × ± 3-kb arms, and no additional primers, with an extension time of 5 minutes and a melting temperature of 60°C. Remove protein from the insertion/deletion PCR reaction by phenol-chloroform extraction, and precipitate DNA by ethanol precipitation with sodium acetate. Resuspend the DNA pellet in 8 µl water, and add 1 µl of Cutsmart buffer (NEB) and 1 µl of DpnI. Incubate at 37°C for 5 hours. Heat-inactivate DpnI by incubating the sample at 80°C for 20 minutes.
Transform the sample into NEB® 10-beta Competent E. coli according to manufacturer’s instructions. Spread the transformation on LB-agar plates with 100 µg/ml spectinomycin, and incubate overnight at 37°C.
Inoculate 3 ml LB with 100 µg/ml spectinomycin with positive colonies as determined by PCR screening. Allow cultures to grow to saturation at 37°C while shaking at 250 rpm, and extract the plasmid DNA with the Zyppy™ Plasmid Miniprep Kit. Confirm the plasmid is the correct size by separating on a 0.8%-agarose gel.
Amplify the plasmid without the target gene by divergent PCR through use of a 50-µl Q5® High-Fidelity DNA Polymerase reaction with pUC18A (now carrying gene × ± 3-kb arms) as template, and primers X-away F and X-away R. Remove protein from the PCR reaction by phenol-chloroform extraction, and precipitate DNA by ethanol precipitation with sodium acetate. Resuspend the DNA pellet and separate the sample in 0.8%-agarose gel. Extract the correct PCR product with the Zymoclean™ Gel DNA Recovery Kit.
Amplify the bla-gfp cassette from pUC19G using bla-gfp F and bla-gfp R primers, and Q5® High-Fidelity DNA Polymerase. Remove protein from the PCR reaction by phenol-chloroform extraction, and precipitate DNA by ethanol precipitation with sodium acetate.
Resuspend the pelleted bla-gfp amplicon in 8 µl water, and add 1 µl Cutsmart buffer and 1 µl DpnI restriction enzyme. Incubate at 37°C for 5 hours. Add 3 µl DNA loading dye, and separate the sample in 0.8%-agarose gel. Extract the correct PCR product with the Zymoclean™ Gel DNA Recovery Kit.
Phosphorylate the gel-purified bla-gfp PCR product with T4 Polynucleotide Kinase according to manufacturer’s instructions.
Ligate the bla-gfp cassette and the divergent-PCR product from step 8 using the Quick Ligation™ Kit, and transform the reaction into NEB® 10-beta Competent E. coli according to manufacturer’s instructions. Spread the transformation on LB-agar plates with 50 µg/ml carbenicillin, and incubate overnight at 37°C.
Inoculate 3 ml LB with 50 µg/ml carbenicillin with positive colonies as determined by PCR screening and observable green fluorescence. Allow cultures to grow to saturation at 37°C while shaking at 250 rpm, and extract the plasmid DNA with the Zyppy™ Plasmid Miniprep Kit. Confirm the plasmid with the bla-gfp cassette instead of the target gene is the correct size by separating on a 0.8%-agarose gel.
Amplify the homologous-recombination sequence (the region spanning the 3-kb arms surrounding the bla-gfp cassette) from the newly-generated plasmid using primers HomRR@pSUmC F and HomRR@pSUmC R with Q5® High-Fidelity DNA Polymerase according to manufacturer’s instructions. Remove protein from PCR reactions by phenol-chloroform extraction, and precipitate DNA by ethanol precipitation with sodium acetate. Resuspend the DNA pellet and separate the sample in 0.8%-agarose gel. Extract the correct PCR product with the Zymoclean™ Gel DNA Recovery Kit.
-
Insert the homologous-recombination sequence into pSUmC by insertion/deletion PCR.
Set up a 50-µl Q5® High-Fidelity DNA Polymerase reaction with 480 ng of pSUmC, 3.4 µg of homologous recombination sequence, and no additional primers, with an extension time of 13 minutes and a melting temperature of 60°C. Remove protein from the insertion/deletion PCR reaction by phenol-chloroform extraction, and precipitate DNA by ethanol precipitation with sodium acetate. Resuspend the DNA pellet in 8 µl water, and add 1 µl of Cutsmart buffer and 1 µl of DpnI. Incubate at 37°C for 5 hours. Heat-inactivate DpnI by incubating the sample at 80°C for 20 minutes.
Transform the sample into NEB® 10-beta Competent E. coli according to manufacturer’s instructions. Spread the transformation on LB-agar plates with 50 µg/ml carbenicillin, and incubate overnight at 37°C.
Inoculate 3 ml LB with 50 µg/ml carbenicillin with positive colonies as determined by PCR screening and observable dual green-and-red fluorescence. Allow cultures to grow to saturation at 37°C while shaking at 250 rpm, and extract the plasmid DNA with the Zyppy™ Plasmid Miniprep Kit. Confirm the plasmid is the correct size by separating on a 0.8%-agarose gel.
Transform the complete deletion construct into dam−/dcm− Competent E. coli according to manufacturer’s instructions. Inoculate 150 ml LB with 50 µg/ml carbenicillin with transformed E. coli, and grow to saturation at 37°C with shaking at 250 rpm. Extract unmethylated plasmid DNA using the QIAfilter Plasmid Maxi Kit.
-
BASIC PROTOCOL 2
Transformation of C. trachomatis
Chlamydiae, like some other bacteria, can be induced to take up foreign DNA upon treatment with a CaCl2 buffer. However, the transformation of C. trachomatis L2 does not require heat shocking the bacteria, and the entire protocol is performed at room temperature (Wang et al., 2011). Moreover, transformation of crude preparation of C. trachomatis elementary bodies (EBs) appears to be more efficient than transforming density gradient purified chlamydiae (Mueller et al., 2016)(Wang et al., 2011). Following incubation in CaCl2 buffer and infection of McCoy cell cultures, it is essential to let chlamydiae recover in media without penicillin G and anhydrotetracycline (ATc). Unlike commercially-available, competent E. coli that typically require 1 h of recovery post-transformation, the most optimal time for recovery of C. trachomatis appears to be ~7 hrs. Although growth and propagation of chlamydiae is routinely performed in HeLa cells, generation of chlamydial mutants is carried out in McCoy cells. Confluent McCoy cell cultures do not undergo the substantial level of infection-independent cell death at ≥48 h post infection that is typically observed with HeLa cells. After post-transformation recovery, adding penicillin G into the culture media selects for transformed bacteria. Successful C. trachomatis transformants will produce inclusions with red and green fluorescence and containing chlamydiae that no longer manifest the penicillin-induced persistent morphology. Although the protocol outlined below is presented for transformation of pSUmC-based plasmids, steps 1–9 can be performed, with appropriate selection (and no ATc), to transform Chlamydia with any stably maintained plasmid.
Materials
Crude stock of C. trachomatis elementary bodies
McCoy cells (CRL-1696, ATCC)
RPMI 1640 (11875119, Gibco) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) (16000044, Gibco)
Hanks’ Balanced Salt Solution (HBSS) (24020117, Gibco)
Cycloheximide (C7698, Sigma)
Anhydrotetracycline hydrochloride (13803-65-1, Alfa Aesar)
Penicillin G sodium salt (P3032, Sigma)
6-Well Cell Culture Plates (3516, Corning)
Cell Scrapers (353085, Corning)
2-ml Safe-Lock Microcentrifuge Tubes (022363352, Eppendorf)
CaCl2 buffer (10 mM Tris pH 7.4 and 50 mM CaCl2)
Chlamydia storage buffer (sucrose-phosphate-glutamate buffer; SPG)
-
Unmethylated deletion construct at 1.0–1.5 µg/µl
Seed 6-well culture plates with freshly trypsinized McCoy cells at a density to achieve 100% confluent monolayers within 24 hrs. This is accomplished by introducing at least 8 ×105 cells in 2 mls RPMI + 10% FBS into each well.
-
Centrifuge 2.6 × 106 infection forming units from a crude stock of C. trachomatis at 20,000 × g for 30 minutes at 4°C. Resuspend the EB pellet in 50 µl CaCl2 buffer and add 2 µl of 1 µg/µl unmethylated deletion construct produced from BASIC PROTOCOL 1. Incubate at room temperature for 30 minutes while flicking every 10 minutes to mix the transformation reaction.
Crude (non density-gradient purified) EB stocks are prepared by scraping C. trachomatis-infected McCoy cultures into 2 mls HBSS at 40–44 hours post infection. Transfer the entire sample to a 2-ml Safe-Lock Microcentrifuge Tube, and centrifuge at 20,000 × g for 30 minutes at 4°C. Resuspend the pellet in 1 ml ice-cold Chlamydia storage buffer (sucrose-phosphate-glutamate buffer; SPG), and then centrifuge at 200 × g for 5 minutes at 4°C. EB-containing supernatants are aliquoted and stored frozen at −80°C. EBs are titred for infectious content by establishing infection forming units using indirect immunofluorescence (Support Protocol 2, Unit 11A.1).In order to increase the chances of successful transformation, multiple simultaneous replicates are recommended. Add 2 ml HBSS to the transformation reaction. Remove the medium from a confluent McCoy cell monolayer in one well of a 6-well cell culture plate, and add the HBSS/transformation mixture.
Infect the McCoy cell monolayer by centrifugation at 900 × g for 60 minutes at 20°C.
Replace the HBSS/transformation mixture with 2 ml RPMI supplemented with 10% FBS and 1 µg/ml cycloheximide.
Incubate at 37°C in an atmosphere of 5% CO2 and 95% humidified air for 7 hours. Replace the medium with 2 ml RPMI supplemented with 10% FBS and 1 µg/ml cycloheximide, 0.6 µg/ml penicillin G sodium salt, and 50 ng/ml anhydrotetracycline, and return to the incubator.
48 hours after infection, harvest the infected McCoy cell monolayer into the original 2-ml medium with a cell scraper. Transfer the entire sample to a 2-ml Safe-Lock Microcentrifuge Tube, and centrifuge at 20,000 × g for 30 minutes at 4°C. Resuspend the pellet in 1 ml HBSS, and then centrifuge at 200 × g for 5 minutes at 4°C.
Remove the medium from a new McCoy cell monolayer in one well of a 6-well cell culture plate, and add the 1-ml supernatant from the previous step in addition to 1 ml fresh HBSS.
Infect the McCoy cell monolayer by centrifugation at 900 × g for 60 minutes at 20°C.
Replace the HBSS with 2 ml RPMI supplemented with 10% FBS and 1 ug/ml cycloheximide, 0.6 µg/ml penicillin G sodium salt, and 50 ng/ml anhydrotetracycline, and incubate at 37°C in an atmosphere of 5% CO2 and 95% humidified air.
-
Repeat steps 6 through 9 until inclusions develop which are both red- and green-fluorescence, as observed by microscopy (see Figure 2, below).
If after 10 passages, no fluorescent inclusions develop, restart the transformation protocol. -
Once dual-fluorescent inclusions develop, continue repassaging as described in steps 6 through 9 until the presence of inclusions which are only green-fluorescent are observed.
Throughout this process, maintain a multiplicity of infection at or below 0.5. If necessary, only use a portion of the harvested transformants for infecting new McCoy cell monolayers during repassaging. If after multiple passages (>5), no exclusively-green-fluorescent inclusions are observed, anhydrotetracycline may be removed from the medium to promote loss of the suicide plasmid. Once inclusions expressing only green fluorescence are identified among the dual-fluorescent populations, prepare a crude stock of the C. trachomatis transformants suspended in SPG, aliquot, and store at −80°C.
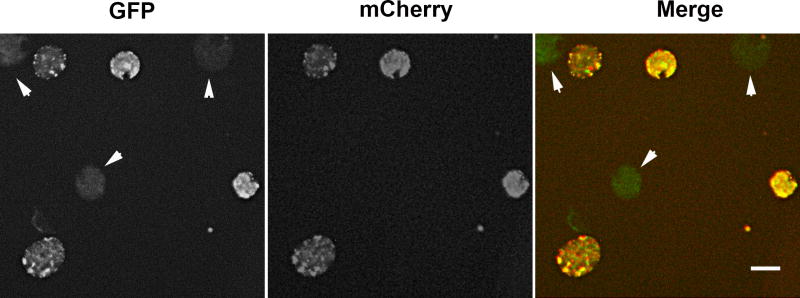
Figure 2.
Visualization of C. trachomatis transformants during screening for homologous recombination. Transformed chlamydiae, cultivated in medium lacking ATc, were examined for GFP and mCherry-mediated fluorescence. Transformants expressing both bright GFP and mCherry have not undergone homologous recombination. Mutants that have undergone homologous recombination and eliminated the pSUmC backbone lack mCherry signal and are comparatively dim for GFP fluorescence (indicated by arrows). Bar = 5 µm.
BASIC PROTOCOL 3
Isolation of deletion mutant by limiting dilution
Once chlamydial transformants are detected, it is essential to separate them from non-transformed chlamydiae that persist in the cultures despite the continual treatment with penicillin G. This goal is achieved by following the limiting dilutions protocol described below. Importantly, during separation of the transformed bacteria, ATc—the inducer of pgp6 expression—is no longer added into the culture media. This results in the continued generation of transformants that are unable to maintain the pSUmC plasmid (Mueller et al., 2016). The presence of penicillin G selects for chlamydiae that have undergone spontaneous homologous recombination, integrating the bla-gfp cassette into the chromosome in exchange for the chlamydial target gene. The fluorescence aspect of FRAEM represents a convenient marker to differentiate transformants from mutant chlamydiae. Initial transformants carry multiple copies of the suicide-vector construct and therefore appear bright with red and green fluorescence. Subsequent to homologous recombination, mutants will have lost the plasmid-encoded mCherry and only retain a single copy of the bla-gfp cassette in the genome. They will therefore exhibit no red fluorescence and comparably dim green fluorescence (Fig 2). An isogenic population of mutant chlamydiae is obtained by a second round of the limiting dilutions method. An alternative approach to the isolation of isogenic population would be the more routinely employed plaque purification. However, we have observed that some chlamydial mutants grow at a significantly slower rate than wild-type C. trachomatis. During plaque purification, non-mutant chlamydiae form robust plaques within 10–12 days post infection that interfere with the often slower growing mutant bacteria. Moreover, we found that some of the mutants do not form clearly distinguishable plaques at all. Therefore, plaque purification of Chlamydia transformants and mutants is not included in this protocol.
Materials
Crude stock of C. trachomatis transformed with the deletion construct
McCoy cells (CRL-1696, ATCC)
RPMI 1640 (11875119, Gibco) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) (16000044, Gibco)
Hanks’ Balanced Salt Solution (HBSS) (24020117, Gibco)
Cycloheximide (C7698, Sigma)
Penicillin G sodium salt (P3032, Sigma)
384-Well Tissue Culture Plates (781091, Greiner)
-
2-ml Safe-Lock Microcentrifuge Tubes (022363352, Eppendorf)
-
Titer (Support Protocol 2, Unit 11A.1) the crude stock of transformed C. trachomatis collected in BASIC PROTOCOL 2, and dilute a sample in HBSS to approximately 1 EB for every 500 µl HBSS. Infect McCoy cell monolayers in a 384-Well Tissue Culture Plate with the diluted crude stock.
Remove the medium from the 384-Well Tissue Culture Plate and apply 50 µl of dilute C. trachomatis transformant in HBSS to each well. This should result in approximately 1 EB for every 10 wells. Centrifuge the 384-Well Tissue Culture Plate at 900 × g for 60 minutes at 20°C. Replace the HBSS from the McCoy cell monolayers with 50 µl/well RPMI supplemented with 10% FBS, 1 µg/ml cycloheximide, and 0.6 µg/ml penicillin G sodium salt. Incubate at 37°C in an atmosphere of 5% CO2 and 95% humidified air.
7 to 14 days after infection (depending on the growth rate of the mutant), individual wells should contain McCoy cell monolayers which have become saturated with chlamydial inclusions. Identify wells containing green-fluorescent C. trachomatis by fluorescent microscopy. Harvest these samples by scraping the wells with pipette tips and transferring the contents to 2-ml Safe-Lock Microcentrifuge Tubes containing 2 ml HBSS. Mix samples by pipetting, and apply each to fresh McCoy cell monolayers in 6-well cell culture plates.
Infect the fresh McCoy cell monolayers by centrifuging at 900 × g for 60 minutes at 20°C. Replace the HBSS with 2 ml RPMI supplemented with 10% FBS, 1 µg/ml cycloheximide, and 0.6 µg/ml penicillin G sodium salt. Incubate at 37°C in an atmosphere of 5% CO2 and 95% humidified air.
-
36 hours post infection, take note of the percentage of exclusively green-fluorescent inclusions in each well, harvest each separately, and freeze at −80°C. Titer the sample which contained the highest percentage of green-fluorescent inclusions, dilute it to approximately 1 EB for every 5 ml HBSS, and infect fresh McCoy cell monolayers in a 384-Well Tissue Culture Plate.
Remove the medium from the 384-Well Tissue Culture Plate and apply 50 µl of dilute green-fluorescent C. trachomatis to each well. This should result in approximately 1 EB for every 100 wells. Centrifuge the 384-Well Tissue Culture Plate at 900 × g for 60 minutes at 20°C. Replace the HBSS from the McCoy cell monolayers with 50 µl/well RPMI supplemented with 10% FBS and 1 µg/ml cycloheximide, only. Incubate at 37°C in an atmosphere of 5% CO2 and 95% humidified air.
-
Repeat steps 3 and 4 without penicillin G sodium salt.
Due to the high dilution factor used in step 5, there should only be 3 or 4 wells containing inclusions. If there are more, the sample used in step 5 must be re-titered, and the infection repeated. -
36 hours post infection, harvest each sample separately, suspend in SPG, and freeze at −80°C.
Mutant populations lacking the target gene may now be assessed for purity by whichever means are convenient, such as quantitative real-time PCR detection of the presence of the gene sequence or antibody-detection of the gene product by Western blot. If trace levels of gene or gene product are detected, repeat steps 5 through 8 as needed. Once purity is confirmed, analyze mutants by whole genome sequencing. Expand and prepare stocks of the mutants as needed. No selective pressures are required during chlamydial growth as the deletion mutations are stable. Readers are referred to detailed protocols (Scidmore, 2005; Unit 11A.1) for propagation and maintenance of Chlamydia strains.
-
REAGENTS AND SOLUTIONS
Use cell-culture rated, endotoxin-free water for in all recipes for solutions to be used during tissue culture.
Sucrose-phosphate-glutamate (SPG) buffer
75 g sucrose
87 ml 0.2 M Na2HPO4 (dibasic sodium phosphate)
13 ml 0.2 M NaH2PO4 (monobasic sodium phosphate)
0.72 g L-glutamic acid
H2O to 1 liter
Adjust pH to 7.4 with 2 N NaOH if necessary
Filter sterilize
Store up to 1 year at 4°C
CaCl2 Buffer
0.55 g CaCl2
10 mls 100 mM Tris pH 7.4
H2O to 100 ml
Filter sterilize
Store up to 1 year at 4°C.
Cycloheximide (1000X stock), 1 mg/ml
Dissolve 10 mg cycloheximide into 10 ml 95% ethanol
Dispense into 100 µl aliquots and store up to 6 months at −20°C
Penicillin G (10,000X stock), 6 mg/ml
Dissolve 6 mg Penicillin G into 1 ml H2O
Dispense into 20 µl aliquots and store up to 6 months at −20°C
Anhydrotetracycline (10,000X stock), 0.5 mg/ml
Dissolve 1 mg anhydrotetracycline in 2 mls tissue-culture rated DMSO
Dispense into 25 µl aliquots and store up to 6 months at −20°C
COMMENTARY
Background Information
Deletion of specific genes from the genome of any pathogenic bacterium has the potential to provide valuable information concerning the strategies the microorganism employs during host recognition, tissue tropism, and colonization, as well as the onset and progression of disease. The lack of a tractable genetic system in Chlamydia has hampered progress in studying and understanding the mechanisms underlying the pathogenicity of these bacteria. Numerous research groups have attempted to develop tools for successful manipulation of the chlamydial genome for decades. Until recently it was essentially impossible to transform and stably maintain exogenous DNA in chlamydiae. The first successful transformation of Chlamydia was reported by Tam et. al. (Tam, Davis, & Wyrick, 1994), where chimeric plasmid, pPBW100 containing chloramphenicol acetyltransferase (cat), was electroporated into C. trachomatis elementary bodies (Tam et al., 1994). Although the authors were able to detect chloramphenicol resistant chlamydiae, the bacteria lost the transformed plasmid within four passages. Electroporation was also used to introduce DNA into Chlamydia psittaci, 6BC. This chlamydial species was transformed with a derivative of the pUC plasmid carrying a copy of C. psittaci 16S rRNA gene modified with unique single-nucleotide changes which, upon exchange with the genomic 16S rRNA, conferred the transformed chlamydiae resistance to spectinomycin and kasugamacin (Binet & Maurelli, 2009). This study clearly showed that allelic exchange via homologous recombination does occur amongst chlamydiae. Delivery of the cryptic 7.5 kb chlamydial plasmid into a plasmid-free C. trachomatis strain L2 (25667R) and Chlamydia pneumoniae was reported employing polyamidoamine dendrimers (Kannan et al., 2013) (Gerard et al., 2013). However, it was the establishment of robust and reproducible transformation of C. trachomatis, L2 using calcium-chloride treatment (Wang et al., 2011) and the generation of shuttle vectors that led to a major breakthrough in the development of molecular tools for chlamydial genetics. Initially, the pBR325::L2 shuttle vector was generated by ligating pBR325 vector with the native pL2 plasmid. Plasmid-free C. trachomatis strain (25667R) was stably transformed, conferring β-lactam resistance to chlamydiae. This large transforming vector appeared to have an effect on the chlamydial developmental cycle, yielding a lower number of infectious elementary bodies. Therefore a smaller shuttle vector, pGFP::SW2, was engineered that contained sequences encoding β-lactamase as well as red-shifted green fluorescent protein fused to chloramphenicol acetyl transferase under control of a neisserial promoter (Wang et al., 2011). Interestingly, pGFP::SW2 also contained an E.coli origin of replication in addition to the native origin, indicating that E.coli cannot utilize the chlamydial origin of replication and vice versa. pGFP::SW2 has been used as a backbone in the generation of numerous shuttle plasmids employed in studying overexpression of various molecules in C. trachomatis (Agaisse & Derre, 2013) (Agaisse & Derre, 2014) (Bauler & Hackstadt, 2014) (Mueller & Fields, 2015). Importantly, it has been also determined that transformation of C. trachomatis L2 with a chlamydial shuttle vector does not require the use of a plasmid-free strain, as the transformed bacteria lose the original pL2 plasmid after multiple passages (Bauler & Hackstadt, 2014) (Mueller et al., 2016). Although the exogenous genes of interest were constitutively expressed either under a Neisseria meningitidis or chlamydial promoter, the development of conditional expression vectors was essential in order to investigate the effects of potentially toxic molecules that may play a role during chlamydial infection. Thus, shuttle vectors employing the tetracycline-inducible system were engineered (Wickstrum, Sammons, Restivo, & Hefty, 2013) (Bauler & Hackstadt, 2014). C. trachomatis transformed with the pBOMB4-Tet-mCherry plasmid were capable of producing visually detectible levels of mCherry fluorescent protein 2 h after induction with as little as 10 ng/ml ATc, demonstrating the extreme sensitivity of the tetracycline-inducible promoter in chlamydia (Bauler & Hackstadt, 2014). Although overexpression of a gene product may prove informative, the precise function of a specific molecule and fulfillment of the molecular Koch’s postulate confirming causation can only be achieved by deletion of the entire gene encoding that molecule from the bacterial chromosome. The approach for generating a bacterial null mutant by allelic exchange using homologous recombination and subsequent complementation has provided valuable information concerning the pathogenesis of numerous bacteria for decades. In Chlamydia, modifications of the native plasmid such as placing pgp6, a key gene for maintenance of the chlamydial plasmid, under the tet-inducible system led to the production of the suicide vector, pSUmC. This vector is controllably maintained by the bacterium and can thus be used to introduce sequences homologous to the chlamydial genome, permitting allelic exchange and the successful generation of specific chlamydial deletion mutants via FRAEM (Mueller et al., 2016). Other approaches have previously been employed to disrupt genes in the chlamydial genome such as chemical mutagenesis with ethyl methanesulfonate (EMS). However, this method disrupts expression through the introduction of random point mutations making chlamydiae with mutations restricted to only a single gene of interest extremely rare to produce and difficult to isolate (Kari et al., 2011) (Nguyen & Valdivia, 2012) (Kokes et al., 2015). Moreover, the TargeTron system, which disrupts gene expression by introducing a group II intron into the target ORF, requires the use of proprietary algorithms and limits integration to sites evaluated to be efficient (Johnson & Fisher, 2013; Lowden, Yeruva, Johnson, Bowlin, & Fisher, 2015). In mutants generated by EMS or TargeTron, the region of the ORF upstream of the mutation site remains intact, free to express the truncated portion of the gene product. For these reasons mutagenesis by deletion of an entire gene via allelic exchange using homologous recombination is so far the most reliable and specific approach in generating chlamydial mutants.
Critical Parameters and troubleshooting
Basic Protocol 1: The suicide vector pSUmC is 11.2 kb is size while completed constructs containing flanking arms exceed 16 kb. Therefore, great care in carrying out molecular biology techniques is essential. In the event that insertion/deletion PCR fails to yield desired constructs, we have also had success using to Gibson Assembly (NEB E5510S; Gibson Assemble® Kit) to generate constructs. In order to successfully amplify the required > 6kb regions of chlamydial DNA, genomic DNA should be high quality. Avoiding manipulations potentially causing sheer and repeated freeze-thaw cycles are especially important. The use of NEB® 10-beta E. coli as a cloning host has been the most productive in our hands. All final constructs should be checked for size to ensure that deletions have not occurred within the pSUmC backbone.
Basic Protocols 2 and 3: As with all Chlamydia-based work, aseptic technique is essential. These methods have been established for C. trachomatis serovar L2 (LGV 434) and have not been verified for other chlamydial species. As noted above, crude preparations of EBs yield better results than density-gradient-pure preparations. It is important to utilize the fluorescence reported feature of FRAEM to discriminate between transformants and instances where single and double crossover events have occurred. Transformants will be bright red and green, whereas inclusion containing meridiploids will appear noticeably dimmer. Second crossover events accompanied by loss of the pSUmC backbone with appear as faint green (Fig. 2). In many instances 40–48 hrs of infection are required to reliably discern faint fluorescence. In instances where singly green fluorescence is not achieved, even after cultivation of transformants in the absence of ATc, it is likely that deletion of the targeted gene is lethal for the bacteria. Potential issues along with suggested troubleshooting approaches are listed below in Table 1.
Table 1.
| Problem | Possible cause | Solution |
|---|---|---|
| No transformants | Low transformation rate | Repeat protocol with as many replicates as manageable. |
| Degraded anhydrotetracycline | Prepare fresh stocks of anhydrotetracycline. Avoid freeze-thaw cycles. | |
| Poor quality of deletion construct plasmid DNA | Confirm plasmid size and quality by separation through agarose gel. Extract plasmid from larger volumes of liquid culture of E. coli for cleaner, more-concentrated plasmid DNA. | |
| Deletion construct contains errors in plasmid genes essential for chlamydial maintenance | Sequence the deletion construct. Re-clone if necessary. | |
| Over-passaged McCoy cells hindering rate of chlamydial infection | Use low-passage number McCoy cells. | |
| Recovery time insufficient for propagation of the particular deletion construct | Extend the recovery time of BASIC PROTOCOL 2, step 5 from 7 hours to under 12 hours. | |
| Deletion construct is not depleted from the chlamydial population upon removal of anhydrotetracycline from medium | Deletion construct contains sequence errors interfering with expression of tetR or repression of pgp6 | As a technical positive control, transform with empty pSUmC, and ensure regulation of plasmid stability is functioning properly. Sequence the deletion construct for errors. Re-clone if necessary. |
| No exclusively green-fluorescent inclusions develop after multiple rounds of re-infection of transformants in the absence of anhydrotetracycline | Homologous recombination arms of deletion construct do not match chlamydial genomic sequence | Amplify and sequence arms from chlamydial genome and deletion construct, and ensure sequences match exactly. Re-clone if necessary. |
| Target gene may be essential or deletion may cause severe growth defect | Clone a deletion construct targeting a non-coding, intergenic region of the chlamydial genome as a technical positive control. If the positive control functions properly in C. trachomatis, propagate the transformants carrying the desired deletion construct under varying conditions, ie, different host cells, concentrations of selective pressures, and times between rounds of harvesting progeny and re-infecting new monolayers, in order to provide the required growth conditions for propagation of the chlamydial mutant. |
Anticipated Results
We estimate that, in our hands, average transformation efficiency with pSUmC-based constructs is on the order of between 1 in 106 and 107. Since efficiency can vary, we typically infect a total of two 6-well plates during chlamydial transformations. Depending on the impact of the deletion of chlamydial development several attempts may be required. Unless an essential gene is being targeted, this protocol is expected to yield deletion mutants in the gene of interest that are viable and stable.
Time considerations
Although now genetically malleable, it is important to remember that Chlamydia is not E. coli with regard to ease of manipulation. The time considered for completing the final pSUmC construct containing the bla-gfp cassette surrounded by the region required for allelic exchange by homologous recombination depends entirely of the chlamydial sequences being targeted. Some C. trachomatis genes and/or 3 kb sequences flanking the gene of interest may impair the growth of E.coli and optimal propagation of the cloning vector pUC18A. Thus, the time required for successful completing of Basic Protocol 1 may vary from several weeks to several months. Chlamydial transformation and production of mutants can generally be completed in several weeks. Although not always the case, evidence of transformants can be detected as early as the second passage under selective pressure (6 days). So long as the targeted gene is not essential, evidence of double recombination (green only inclusions) can typically be detected by passage 5. In our experience, these are minimum time requirements. Time-frames can obviously be extended based on other parameters. For example, more time will be required if the null mutation has a significant impact on the chlamydial developmental cycle.
SIGNIFICANCE.
Chlamydia trachomatis is a prevalent and medically significant human pathogen. Definitively establishing how gene products contribute to virulence was historically challenging due to an inability to genetically manipulate chlamydiae. The ability to reproducibly transform chlamydiae with exogenous DNA has recently propelled Chlamydia into the family of genetically tractable obligate intracellular bacteria. The ability to specifically delete complete gene sequences represents an essential capability to study respective protein function. These protocols describe techniques to transform Chlamydia trachomatis serovar L2 and employ allelic exchange mutagenesis to engineer null mutations in targeted genes. It is anticipated that these techniques will prove efficacious in promoting rapid and reliable progress in understanding an important infectious disease.
Acknowledgments
This work was supported by Public Health Service grants from the National Institutes of Health, NIAID (AI065530 and AI124649), to K.A. Fields.
LITERATURE CITED
- Agaisse H, Derre I. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One. 2013;8(2):e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse H, Derre I. Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infect Immun. 2014;82(5):2037–2047. doi: 10.1128/IAI.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauler LD, Hackstadt T. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol. 2014;196(7):1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Maurelli AT. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc Natl Acad Sci U S A. 2009;106(1):292–297. doi: 10.1073/pnas.0806768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser M, Cebe R, Drewello D, Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31(1):88–90. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Hudson AP. Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine. 2013;9(7):996–1008. doi: 10.1016/j.nano.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Gong S, Yang Z, Lei L, Shen L, Zhong G. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol. 2013;195(17):3819–3826. doi: 10.1128/JB.00511-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Fisher DJ. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One. 2013;8(12):e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan RM, Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Hudson AP. Dendrimer-enabled transformation of Chlamydia trachomatis. Microb Pathog. 2013;65:29–35. doi: 10.1016/j.micpath.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Caldwell HD. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A. 2011;108(17):7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokes M, Dunn JD, Granek JA, Nguyen BD, Barker JR, Valdivia RH, Bastidas RJ. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe. 2015;17(5):716–725. doi: 10.1016/j.chom.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden NM, Yeruva L, Johnson CM, Bowlin AK, Fisher DJ. Use of aminoglycoside 3’ adenyltransferase as a selection marker for Chlamydia trachomatis intron-mutagenesis and in vivo intron stability. BMC Res Notes. 2015;8:570. doi: 10.1186/s13104-015-1542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KE, Fields KA. Application of beta-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS One. 2015;10(8):e0135295. doi: 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KE, Wolf K, Fields KA. Gene Deletion by Fluorescence-Reported Allelic Exchange Mutagenesis in Chlamydia trachomatis. MBio. 2016;7(1):e01817–01815. doi: 10.1128/mBio.01817-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BD, Valdivia RH. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci U S A. 2012;109(4):1263–1268. doi: 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore M. Cultivation and laboratory maintenance of Chlamydia trachomatis. Curr Protoc Microbiol. 2005 doi: 10.1002/9780471729259. Chapter 11:Unit 11A.1. [DOI] [PubMed] [Google Scholar]
- Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Caldwell HD. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun. 2013;81(3):636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam JE, Davis CH, Wyrick PB. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can J Microbiol. 1994;40(7):583–591. doi: 10.1139/m94-093. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7(9):e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstrum J, Sammons LR, Restivo KN, Hefty PS. Conditional gene expression in Chlamydia trachomatis using the tet system. PLoS One. 2013;8(10):e76743. doi: 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G. Chlamydial Plasmid-Dependent Pathogenicity. Trends Microbiol. 2016 doi: 10.1016/j.tim.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]