Abstract
Zinc cluster proteins (or binuclear cluster proteins) possess zinc fingers of the Zn(II)2Cys6-type involved in DNA recognition as exemplified by the well-characterized protein Gal4p. These fungal proteins are transcriptional regulators of genes involved in a wide variety of cellular processes including metabolism of compounds such as amino acids and sugars, as well as control of meiosis, multi-drug resistance etc. The yeast (Saccharomyces cerevisiae) sequencing project has allowed the identification of additional zinc cluster proteins for a total of 54. However, the role of many of these putative zinc cluster proteins is unknown. We have performed phenotypic analysis of 33 genes encoding (putative) zinc cluster proteins. Only two members of the GAL4 family are essential genes. Our results show that deletion of eight different zinc cluster genes impairs growth on non-fermentable carbon sources. The same strains are also hypersensitive to the antifungal calcofluor white suggesting a role for these genes in cell wall integrity. In addition, one of these strains (ΔYFL052W) is also heat sensitive on rich (but not minimal) plates. Thus, deletion of YFL052W results in sensitivity to a combination of low osmolarity and high temperature. In addition, six strains are hypersensitive to caffeine, an inhibitor of the MAP kinase pathway and phosphodiesterase of the cAMP pathway. In conclusion, our analysis assigns phenotypes to a number of genes and provides a basis to better understand the role of these transcriptional regulators.
INTRODUCTION
In yeast (Saccharomyces cerevisiae), a major class of transcriptional regulators is composed of a sub-family of zinc finger proteins called zinc cluster proteins or binuclear cluster proteins. These proteins contain a DNA binding domain which possesses the well-conserved motif CysX2CysX6CysX5–16-CysX2CysX6–8Cys with cysteines binding to two zinc atoms which coordinate folding of the domain (1).
This type of transcriptional regulator has only been identified in fungi and these proteins have been shown to be involved in a wide variety of cellular processes (reviewed in 2; also see Table 3). For example, Gal4p is involved in activation of genes that encode enzymes for galactose metabolism (3) while Hap1p activates genes involved in cellular respiration (4,5). In addition, other zinc cluster proteins increase expression of genes required for gluconeogenesis or metabolism of leucine, lysine, arginine, pyrimidine, thiamine, etc. (2). Other roles of zinc cluster proteins include the control of expression of genes required for use of γ-aminobutyric acid (GABA), serine, threonine or proline as a nitrogen source (2). In addition, some members of the family, such as Pdr1p and Pdr3p, are responsible for controlling expression of multi-drug resistance genes (6).
Table 3. Summary of the phenotypes and functions of zinc cluster genes.
| Systematic name |
Gene |
Function |
Phenotype |
Ref. |
| YAL051W | OAF1 (YAF1) | Activator of peroxisome proliferation along with Oaf2p | Impaired in growth of oleate as a carbon source, induction of β-oxidation enzymes is abolished | 19 |
| YBL005W | PDR3 | Activator related to Pdr1p | Pleiotropic drug resistance | 6,46 |
| YBL066C | SEF1 | Suppressor of essential function | Defective sporulation; high copy number suppressor of RPM2 | 47 |
| YBR033W | – | Unknown | Unknown | – |
| YBR150C | – | Unknown | Unknown | – |
| YBR239C | – | Unknown | Unknown | – |
| YBR240C | THI2 (PHO6) | Activator of thiamin biosynthetic genes | Thiamin auxotrophy, reduced expression of thiamin biosynthetic genes | 41 |
| YBR297W | MAL3R (MAL33) | Part of complex locus MAL3; MAL activator protein | Defective maltose fermentation | 48,49 |
| YCR106W | – | Unknown | Unknown | – |
| YDL170W | UGA3 | Activator necessary for GABA-dependent function of GABA genes | Exhibits defects in activation of UGA1 and UGA4 | 50,51 |
| YDR034C | LYS14 | Transcriptional activator of lysine pathway genes | Lysine requiring | 52,53 |
| YDR207C | UME6 | Regulator of both repression and induction of early meiotic genes | Exhibits defects in IME1-dependent activation and repression | 54 |
| YDR213W | UPC2 | Involved in sterol uptake | upc2-1 allele shows altered sterol uptake and increased sensitivity to NaCl and LiCl | 55,56 |
| YDR303C | – | Unknown | Null mutant is inviable | This study, 32,33 |
| YDR421W | – | Unknown | Moderately sensitive to SDS and benomyl | 45 |
| YDR520C | – | Unknown | Slight caffeine sensitivity | This study |
| YER184C | – | Unknown | Unable to grow on non-fermentable carbon sources; sensitive to calcofluor white | This study |
| YFL052W | – | Unknown | Unable to grow on non-fermentable carbon sources; sensitive to calcofluor white; heat sensitive on rich but not minimal medium | This study |
| YGL013C | PDR1 (CYH3) | General positive regulator of permeability genes | Pleiotropic drug resistance | 6,57 |
| YGR288W | MAL13 | Part of complex locus MAL1; MAL activator protein (non-functional in S228C and derivatives) | Defective maltose fermentation | 48,49 |
| YHR178W | STB5 | Binds Sin3p in two-hybrid assay | Cold and caffeine sensitive | This study, 40 |
| YIL130W | – | Unknown | Unable to grow on non-fermentable carbon sources; sensitive to calcofluor white, cyclheximide, benomyl, and MMS; slightly sensitive to hydoxyurea | This study, 45 |
| YIR023W | DAL81 (UGA35) | Positive regulator of multiple nitrogen catabolic genes such as allantoin and GABA catabolic genes | Unable to degrade allantoin | 51,58,59 |
| YJL089W | SIP4 | Involved in Snf1p regulated transcriptional activation | Required for maximal expression of carbon source responsive genes; moderate sensitivity to SDS | 60,61,45 |
| YJL103C | – | Unknown | Unknown | – |
| YJL206C | – | Unknown | Unknown | – |
| YKL015W | PUT3 | Positive regulator of PUT (proline utilization) genes | Unable to use proline as sole nitrogen source | 62,63 |
| YKL038W | RGT1 | Transcriptional repressor and activator of genes involved in glucose metabolism | Constitutive expression of glucose-induced HXT genes; sensitive to calcofluor white; sensitive to SDS, unable to grow on non–fermentable carbon source; slight sensitivity to MMS | 11,45 |
| YKL222C | – | Unknown | Moderately sensitive to caffeine | This study |
| YKR064W | – | Unknown | Unknown | – |
| YLL054C | – | Unknown | Unable to grow on non–fermentable carbon sources; sensitive to calcofluor white | This study |
| YLR014C | PPR1 | Activator of URA1 and URA3 | Deficient in pyrimidine biosynthetic pathway | 64,65 |
| YLR098C | CHA4 (SIL2) | Activator of CHA1 | Unable to grow with serine or threonine as sole nitrogen source | 66 |
| YLR228C | ECM22 | Unknown | Moderately sensitive to caffeine | This study, 42 |
| YLR256W | HAP1 (CYP1) | Activator of respiration genes | Essential for anaerobic or heme deficient growth; sensitive to SDS and EGTA; moderately sensitive to hygromycin | 4,5,45 |
| YLR266C | – | Unknown | Unable to grow on non-fermentable carbon sources; sensitive to calcofluor white | This study |
| YLR278C | – | Unknown | Moderately sensitive to caffeine | This study |
| YLR451W | LEU3 | Regulates expression of genes involved in branched chain amino acid biosynthesis and ammonia assimilation | Leaky leucine auxotroph | 67,68 |
| YML076C | – | Unknown | Unknown | – |
| YML099C | ARG81 (ARGR2) | Positive and negative regulator of many arginine-responsive genes | 69 | |
| YMR019W | STB4 | Binds Sin3p in two-hybrid assay | Sensitive to caffeine | This study, 40,45 |
| YMR168C | CEP3 | Cbf3 kinetochore complex binds CDE III centromere element | Essential gene | 35,36 |
| YMR280C | CAT8 | Involved in activation of gluconeogenic genes | Unable to grow on non-fermentable carbon sources and ethanol | 70 |
| YNR063W | – | Unknown | Unknown | – |
| YOL089C | HAL9 | Involved in salt tolerance | Exhibits decreased salt tolerance and ENA1 expression | 71 |
| YOR162C | YRR1 | Activator of multi-drug resistance genes | Hypersensitive to 4-nitroquinoline oxide (4-NQO) and benomyl; unable to grow on non-fermentable carbon sources; sensitive to calcofluor white | This study, 39,45 |
| YOR172W | – | Unknown | Unknown | – |
| YOR337W | TEA1 | Activator of Ty1 elements | Diminished Ty1 expression; sensitive to SDS and moderately sensitive to MMS | 72,45 |
| YOR363C | OAF2 (PIP2) | Activator of peroxisome proliferation along with Oaf1p | Impaired in growth of oleate as a carbon source, induction of β-oxidation enzymes is abolished | 19,73 |
| YOR380W | – | Unknown | Unable to grow on non-fermentable carbon sources; sensitive to calcofluor white | This study |
| YPL133C | – | Unknown | Unable to grow on non-fermentable carbon sources; sensitive to calcofluor white | This study |
| YPL248C | GAL4 | Activator of GAL genes | Cannot utilize galactose as sole carbon source | 3 |
| YPR094W | – | Unknown | Unknown | – |
| YPR196W | MAL63 | Activator of maltose genes | Unable to ferment maltose | 48,49 |
Quite often, the DNA binding domain (comprising the cysteine-rich region) of zinc cluster proteins is located at the N-terminus while an acidic activating domain is located at the C-terminus. A region of low homology of ∼80 amino acids, termed the middle homology region, is found among many zinc cluster proteins and is located between the DNA binding and activation domains and may be involved in controlling the transcriptional activity of zinc cluster proteins (7). In many cases, deletion of the region that bridges the DNA binding domain to the activation domain results in constitutive activity of the transcriptional activator. For example, deletion of the middle region of Hap1p renders the protein active even in the absence of the inducer heme (4). Similar results were obtained with Leu3p, a protein implicated in leucine biosynthesis (8,9). Many of the characterized zinc cluster proteins are transcriptional activators. Well-known exceptions are Ume6p and Rgt1p which are both repressors and activators of early meiotic and glucose transport genes, respectively (10,11).
Many zinc cluster proteins bind to DNA as homodimers through a coiled-coil dimerization domain located at the C-terminus of the zinc finger. Three types of DNA binding sites have been identified: inverted, direct and everted repeats (reviewed in 12). For example, Gal4p binds as a homodimer to inverted repeat DNA sequences (13 and references therein) while Hap1p binds to a direct repeat (14–16). Analysis of the binding sites of Leu3p showed that it recognizes repeats oriented in opposite directions, an everted repeat (17). These observations imply that the two zinc fingers of Leu3p must be oriented in opposite directions unlike those of Gal4p where they have been shown to face each other (13).
Alternate modes of DNA binding by zinc cluster proteins have also been described. For example, AlcR, a transcriptional activator of ethanol oxidation genes in Aspergillus, binds to DNA as a monomer (18). In addition, the two zinc cluster proteins Oaf1p and Oaf2p (Pip2p) bind as heterodimers to target sequences of genes for peroxisome proliferation (19,20). Heteromeric formation is also observed between members of different families of transcription factors. For example, ArgRII, a member of the family of zinc cluster proteins, heterodimerizes with members of the MADS family, ArgRI and Mcm1p, to activate genes for arginine metabolism (21). In summary, zinc cluster proteins perform a wide variety of functions through transcriptional activation or repression by binding to target genes as homodimers, heterodimers or monomers.
The S.cerevisiae sequencing project has allowed the identification of additional zinc cluster proteins for a total of 54 (2). However, the function of many of these putative zinc cluster proteins is unknown. In an effort to understand the roles of the uncharacterized zinc cluster proteins, we examined the phenotypes of 33 strains carrying deletions of genes encoding zinc cluster proteins under various growth conditions.
MATERIALS AND METHODS
Strains
Wild-type S.cerevisiae strains used to generate the gene deletions were: FY73 (22), MATα his3-Δ200 ura3-52; YPH499 (23), MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1; YPH500 (23), MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1; YPH501 (23), a cross between YPH499 and YPH500; BY4742 (24), MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0. Deletions were obtained by the PCR method of Baudin et al. (25) using HIS3 as a marker for selection (template for PCR was pMHIS3; 26). Oligos had the sequence N43CAGGGTTTTCCCAGTCA and N43GCGGATAACAATTTCAC with N corresponding to sequences of the target genes. Some open reading frames (ORFs) were entirely deleted and, for others, the deletion spanned the zinc finger (cysteine-rich region) located at the N-terminus of the putative ORFs, as indicated in Table 1. Deletions in FY73 were obtained by direct transformation of the haploid strain. Deletions in the YPH background were obtained by transforming the diploid strain YPH501 followed by sporulation and selection of HIS+ spores. For sporulation, diploid strains were plated on sporulation plates (1% potassium acetate, 0.1% yeast extract, 0.05% glucose, 2% agar) for 1 week followed by random spore analysis (27). Diploid strains carrying deletions of the ORF YPR094W or YDR303C were obtained from Research Genetics (Huntsville, AL). Haploid strains were obtained by sporulation.
Table 1. Strains used in this study.
| Syst. name |
Strain name |
Background |
Deletion |
Marker |
Source |
| YBL066C | BE | BY4742 | ORF | KAN | Research Genetics |
| YBR033W | FI | FY73 | aa 27–200 | HIS3 | This study |
| YBR150C | FA | FY73 | aa 24–167 | HIS3 | This study |
| YBR239C | YD | YPH499 | ORF | HIS3 | This study |
| YBR240C | FB | FY73 | aa 25–166 | HIS3 | This study |
| YCR106W | FC | FY73 | aa 23–206 | HIS3 | This study |
| YDR213W | BZY | BY4742 | ORF | KAN | Research Genetics |
| YDR303C | BZX | BY4743 | ORF | KAN | Research Genetics |
| YDR421W | BZD | BY4742 | ORF | KAN | Research Genetics |
| YDR520C | YZN | YPH499 | ORF | HIS3 | This study |
| YER184C | FZT | FY73 | aa 26–225 | HIS3 | This study |
| YFL052W | YZS | FY73 | aa 24–173 | HIS3 | This study |
| YHR178W | BT | BY4742 | ORF | KAN | Research Genetics |
| YIL130W | FZG | FY73 | aa 24–246 | HIS3 | This study |
| YJL089W | BR | BY4742 | ORF | KAN | Research Genetics |
| YJL103C | YZL | YPH499 | ORF | HIS3 | This study |
| YJL206C | FZQ | FY73 | aa 28–149 | HIS3 | This study |
| YKL222C | YH | YPH499 | ORF | HIS3 | This study |
| YKR064W | YK | YPH499 | ORF | HIS3 | This study |
| YLL054C | FZI | FY73 | aa 25–220 | HIS3 | This study |
| YLR228C | YN | YPH499 | ORF | HIS3 | This study |
| YLR266C | FS | FY73 | aa 25–246 | HIS3 | This study |
| YLR278C | YO | YPH499 | ORF | HIS3 | This study |
| YML076C | BZM | BY4742 | aa 41–912 | KAN | Research Genetics |
| YMR019W | YZE | YPH499 | ORF | HIS3 | This study |
| YNR063W | FZO | FY73 | aa 25–166 | HIS3 | This study |
| YOL089C | FZJ | FY73 | aa 115–319 | HIS3 | This study |
| YOR162C | FZU | FY73 | aa 28–332 | HIS3 | This study |
| YOR172W | YZV | YPH499 | ORF | HIS3 | This study |
| YOR380W | FZP | FY73 | aa25–196 | HIS3 | This study |
| YPL133C | FZH | FY73 | aa 25–266 | HIS3 | This study |
| YPR094W | BZZA | BY4742 | ORF | KAN | Res. Genetics |
| YPR196W | YZ | YPH499 | ORF | HIS3 | This study |
Strains used in this study are listed. The names of the targeted ORFs are given (Syst. name) as well as the wild-type strains used to perform the deletions (Background). The entire ORFs or, alternatively, the sequences encompassing the cysteine-rich region (putative DNA binding domain) were deleted as indicated in the Deletion column. Markers used to select for recombination events are also listed. For more details, see Materials and Methods.
Proper recombination events were verified by PCR using two pairs of primers. One set consisted of the primer GCCTCGTTCAGAATGACACG (located in the 3′-end of the HIS3 marker) and a primer in the promoter region of the target gene and the second set of a primer located downstream of the ORF and the primer TTACTCTTGGCCTCCTCTAG (located in the 5′-end of the HIS3 marker). Single integration events were verified by Southern blot analysis for all strains using the HIS3 or the kanamycin markers as probes. In all cases, the sizes of the detected bands were in agreement with homologous recombination at the targeted gene. Genomic DNA was isolated according to Philippsen et al. (28). Southern blots were done according to standard procedures (27). Hybridizations were performed at 42°C in 50% formamide, 1 M NaCl, 2.8× Denhardt’s solution, 0.5% SDS and 10% dextran sulphate.
Media
Media were prepared according to Adams et al. (29). YPD contained 1% yeast extract, 2% peptone, 2% glucose. SD contained 2% glucose, 0.67% yeast nitrogen base without amino acids. Adenine, histidine, leucine, lysine, tryptophan and uracil were added to the media at a final concentration of 0.004%. Nitrogen source requirements were tested on minimal media as described above, except that yeast nitrogen base lacked ammonium sulphate. A source for nitrogen was supplied by adding GABA at a final concentration of 2% or 1 mg/ml of either urea, serine, threonine, proline as specified in Table 2. YEP contained 1% yeast extract and 2% peptone with either 2% glycerol or 2% lactic acid. Sensitivity to drugs was assayed on YPD plates supplemented with 0.15% caffeine (Sigma) or 70 µg/ml calcofluor white (Sigma).
Table 2. Phenotypes resulting from deletions of genes encoding putative zinc cluster proteins.
| Strain | Syst. Name | Gene | Carbon Source | Nitrogen source | Minimal (Halvorson) | Sensitivity to Temperature | Calcofluor | Caffeine | Anaerobic | ||||||||||||
| Conditions | |||||||||||||||||||||
| |
|
|
Glycerol |
Lactate |
Urea |
GABA |
Serine |
Threonine |
Proline |
‘All’ |
No biotin |
No Pantho. |
No folate |
37°C
YPD |
37°C
SD |
20°C
YPD |
20°C
SD |
|
|
YPD |
SD |
| BE | YBL066C | SEF1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FI | YBR033W | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FA | YBR150C | TBS1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| YD | YBR239C | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FB | YBR240C | THI2 (PHO6) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FC | YCR106W | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| BZY | YDR213W | UPC2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| BZD | YDR421W | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| YZN | YDR520C | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | X | + | + |
| FZT | YER184C | – | XXX | XXX | + | + | + | + | + | + | + | + | + | + | + | + | + | XXX | + | + | + |
| YZS | YFL052W | – | XXX | XXX | + | + | + | + | + | + | + | + | + | XXX | + | + | + | XX | + | + | + |
| BT | YHR178W | STB5 | + | + | X | X | X | X | X | X | X | X | X | + | + | XXX | XXX | + | XXX | + | + |
| FZG | YIL130W | – | XXX | XXX | + | + | + | + | + | + | + | + | + | + | + | + | + | XXX | + | + | + |
| BR | YJL089W | SIP4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| YZL | YJL103C | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FZQ | YJL206C | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| YH | YKL222C | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | XX | + | + |
| YK | YKR064W | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FZI | YLL054C | – | XXX | XXX | + | + | + | + | + | + | + | + | + | + | + | + | + | XXX | + | + | + |
| YN | YLR228C | ECM22 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | XX | + | + |
| FS | YLR266C | – | XXX | XXX | + | + | + | + | + | + | + | + | + | + | + | + | + | XXX | + | + | + |
| YO | YLR278C | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | XX | + | + |
| BZM | YML076C | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| YZE | YMR019W | STB4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | XXX | + | + |
| FZO | YNR063W | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FZJ | YOL089C | HAL9 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FZU | YOR162C | YRR1 | XXX | XXX | + | + | + | + | + | + | + | + | + | + | + | + | + | XXX | + | + | + |
| YZV | YOR172W | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FZP | YOR380W | – | XXX | XXX | + | + | + | + | + | + | + | + | + | + | + | + | + | XXX | + | + | + |
| FZH | YPL133C | – | XXX | XXX | + | + | + | + | + | + | + | + | + | + | + | + | + | XXX | + | + | + |
| BZZA | YPR094W | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| YZ | YPR196W | MAL63 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
Growth of deletion strains was assayed under various conditions as described in Materials and Methods. In the column ‘Halvorson’, No Pantho. refers to the lack of panthoneic acid in the Halvorson medium and ‘All’ corresponds to Halvorson medium containing biotin, panthoneic acid and folate. In the column ‘Calcofluor’, sensitivity of the deletion strains to the compound calcofluor white is listed.
XXX, no growth (or severely impaired growth); XX, moderate growth; X, slightly inhibited growth; +, normal growth.
Growth assays
Wild-type and deletion strains were grown overnight in liquid (YPD), spun and resuspended in water. Cells were then serially diluted (approximately 5 × 104, 5 × 103, 5 × 102 and 5 × 101 cells) and spotted on appropriate plates. For assay of growth under anaerobic conditions, an anaerobic chamber was used along with a BBL GasPak (Becton Dickinson) and a palladium catalyst activated by heating at 160°C for 2 h. An oxygen indicator (BBL Becton Dickinson) showed that cells were actually grown under anaerobic conditions.
Requirement for specific compounds
Requirement for specific compounds were tested in a minimal medium (Halvorson; 29) containing the following components: 2% glucose, 2% ammonium sulphate, 0.25 M K2HPO4, 0.25 M succinic acid, 0.002% sodium carbonate, 14 mM CaCl2, 21 mM MgSO4, 3.8 µM FeCl3, 8.3 µM MnCl2 7.7 µM ZnSO4, 7.8 µM CuSO4, 1 × 10–7% d-biotin, 3 × 10–5% calcium pantotheanate, 3 × 10–5% folic acid, 3.3 × 10–4% myo-inositol, 3.3 × 10–4% pyridoxine–HCl, 3.3 × 10–4% nicotinic acid, 3.3 × 10–4% p-amino benzoic acid, 3.3 × 10–4% thiamine–HCl, 0.004% adenine, 0.004% histidine, 0.004% leucine, 0.004% lysine, 0.004% methionine, 0.004% tryptophan and 0.004% uracil. Specific components were omitted as specified in Table 2.
RESULTS AND DISCUSSION
Many yeast genes encode putative zinc cluster proteins of totally unknown function while others have not been well characterized. Thus, we performed a phenotypic analysis of yeast strains carrying deletions of genes encoding members of the Gal4p family of zinc cluster proteins. The yeast genome contains 54 ORFs that potentially encode zinc cluster proteins containing the consensus sequence CysX2CysX6CysX5–16-CysX2CysX6–8Cys. Two ‘zinc cluster-like proteins’ were excluded from our analysis since they do not conform to the consensus amino acid sequence given above. YPR009W and YGL162W (SUT1) have 63 and 68 amino acids between the third and fourth cysteines of the zinc cluster, respectively, as opposed to 5–16 for other zinc cluster proteins found in S.cerevisiae, as well as other fungi (2). YGL162W was shown to be involved in sterol uptake (30). In addition, the cysteine-rich regions of YGL162W and YPR009W share homology to the glucose transporters encoded by the SUT1, SUT2 and SUT3 genes of yeast Pichia stipitis (31 and data not shown). Thus, the YPR009W and YGL162W genes may not encode DNA binding proteins. In addition, well-characterized zinc cluster proteins such as Gal4p, Hap1p, Leu3p, Uga3p etc. were also excluded from our analysis. Thus, we phenotypically analyzed 33 genes encoding (putative) zinc cluster proteins under various conditions including alternate carbon sources, growth temperature, presence of caffeine etc.
We deleted ORFs of genes encoding putative zinc cluster proteins by the PCR method of Baudin et al. (25) using HIS3 as selectable marker and haploid strains (see Materials and Methods). Deletions were verified by using two pairs of primers for PCR analysis (data not shown). Moreover, all deletion strains were also verified by Southern blot analysis (data not shown). Most zinc cluster genes could be deleted in a haploid background. However, no colonies were obtained using a PCR product aimed at deleting ORF YDR303C in the haploid strain FY73. Random spore analysis using a heterozygote strain (BY4743) carrying a deletion of ORF YDR303C revealed that all spores (∼50) were sensitive to G418 showing that YDR303C is an essential gene. Similar results were obtained with transposon insertion in the YDR303C gene (32) or deletion of the ORF (33). In addition, the zinc cluster protein Cep3p, a component of the Cbf3 kinetochore complex that binds centromeric elements (34) was shown to be encoded by an essential gene (35,36). Deletion of the gene YHR178W encoding a putative zinc cluster protein is not lethal in the BY4742 background (Table 2); however, this gene was scored as essential in the YM4587 background (37). This discrepancy may be explained by the use of the different strain. For example, strain YM4587, unlike strain BY4742, carries a mutant allele of the TYR1 gene involved in tyrosine synthesis. Thus, according to our data, only two members of the Gal4p zinc cluster family are encoded by essential genes.
Growth on non-fermentable carbon sources
Respiratory-deficient mutants are unable to grow on non-fermentable carbon sources such as glycerol and lactate (38). Thus, we tested the ability of the deletion strains to grow on non-fermentable carbon sources. Cells were serially diluted and spotted on YEP–glycerol and YEP–lactate plates and grown at 30°C for 3 days. Deletion of ORFs YER184C, YFL052W, YIL130W, YLL054C, YLR266C, YOR162C (YRR1), YOR380W and YPL133C impaired growth on both glycerol and lactate plates (Table 2) while normal growth was observed on plates containing glucose as the carbon source (Fig. 1 and data not shown). An example of data is provided in Figure 1 with strains carrying deletions of ORFs YER184C, YFL052W, YLL054C and YOR380W. Thus, deletion of ORFs YER184C, YFL052W, YIL130W, YLL054C, YLR266C, YOR162C (YRR1), YOR380W and YPL133C prevents growth in medium containing lactate or glycerol as the only carbon source. In addition, deletion of YOR162C (YRR1) leads to hypersensitivity to the mutagen 4-nitroquinoline N-oxide due to reduced expression of the ABC transporter SNQ2 (39).
Figure 1.
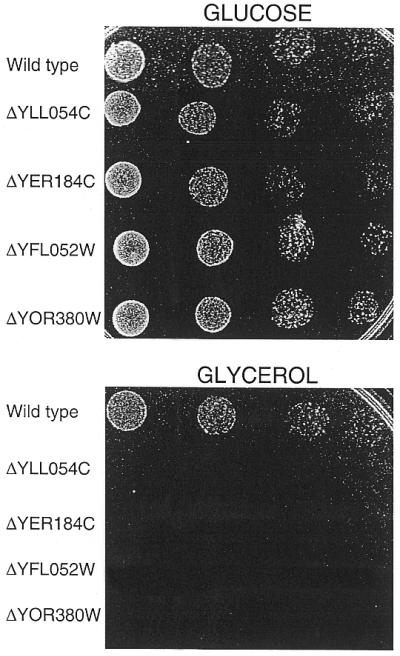
Growth on non-fermentable carbon source of selected strains. Wild-type and deletion strains were grown overnight in YEP–glucose, washed, serially diluted and spotted on YEP–glucose or YEP–glycerol plates, as indicated. Wild-type strain is FY73. For more details, see Materials and Methods.
Temperature sensitivity
Growth of the deletion strains was tested at high and at low temperatures (Table 2). The strains were spotted on YPD and minimal plates and grown at 37 or 20°C. One strain, YZS (YFL052W), did not grow in rich medium at 37°C but grew on minimal medium at 37°C. Since salt concentration is higher in minimal than in rich medium, deletion of the YFL052W gene may render the cells sensitive to a combination of both high temperature and low salt concentration. In addition, knockout of the YFL052W gene renders the cell hypersensitive to calcofluor white (see below and Fig. 3), a phenotype associated with cell wall mutants (38). Thus, YFL052W may be involved in maintenance of cell wall integrity. Deletion of YHR178W (STB5) resulted in a cold sensitive phenotype since the strain did not grow on either YPD or SD at 20°C. Both Stb5p and Stb4p (encoded by YMR019W) were shown to interact with Sin3p, a protein that recruits the histone deacetyltransferase Rpd3p involved in repression of transcription (40). Thus, Stb4p and Stb5p may encode transcriptional repressors.
Requirement for specific compounds
A number of zinc cluster proteins, such as Dal81p, Uga3p, Put3p and Cha4p have been shown to be involved in the use of alternate nitrogen sources (Table 3). We tested if other genes encoding zinc cluster proteins would play a similar role by growing deletion strains on plates containing urea, GABA, serine, threonine or proline as the sole nitrogen source. Only one strain carrying a deletion of the YHR178W (STB5) gene showed reduced growth (Table 2). We also tested growth of deletion strains on minimal Halvorson medium (Table 2). Again, only deletion of the gene YHR178W (STB5) resulted in reduced growth. Moreover, besides deletion strain YHR178W, omission of folic acid, panthoneic acid or biotin in the Halvorson medium did not alter growth of any deletion strain (data not shown). As expected (41), a strain carrying a deletion of the THI2 gene (YBR240C) was auxotrophic for thiamine but not sensitive to the omission of folic acid, panthoneic acid or biotin (data not shown). In conclusion, deletion of YHR178W results in slightly slower cell growth under minimal growth conditions such as ‘Halvorson’ medium or when nitrogen sources other than ammonium sulphate are used.
Sensitivity to caffeine
Caffeine, a purine analog, has a toxic effect on cells through inhibition of the MAP kinase pathway and phosphodiesterase of the cAMP pathway (38). Deletion strains were tested for hypersensitivity to caffeine. Although wild-type strains are all derived from SC228, they showed different sensitivities to caffeine: YPH499 was more sensitive than FY73 and BY4742. All the FY73 and BY4742-based strains grew well on caffeine plates with the exception of ΔYHR178W which was highly sensitive to caffeine (Table 2). An example of results is provided in Figure 2. Normal growth of the strain deleted of YHR178W is seen on YPD plates but greatly reduced growth is observed if caffeine is added (Fig. 2). A strain carrying a deletion of YDR520C was slightly more sensitive to caffeine than wild-type YPH499 while deletions of YKL222C, YLR228C or YLR278C resulted in moderate sensitivity (Table 2). Severe sensitivity to caffeine was observed with a deletion of ORF YMR019W as compared to wild-type YPH499. In addition, deletion of the latter ORF resulted in cells clumping when grown in YPD liquid media (data not shown).
Figure 2.
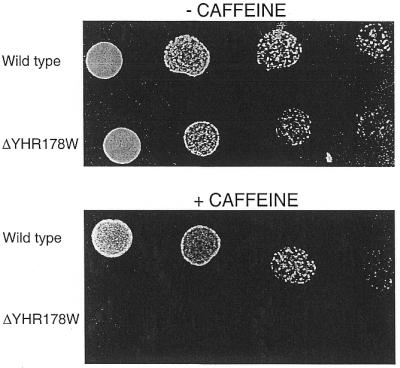
Caffeine sensitivity in YHR178W (STB5) deleted strain. Wild-type (BY4742) and deletion strain (BT, Table 1) were grown overnight in YPD, washed, serially diluted and spotted on YPD plates either with or without 0.15% caffeine.
Sensitivity to calcofluor white
Calcofluor white is a compound that has high affinity for the cell wall component chitin. Hypersensitivity to that compound has been associated with cell wall mutants (38,42). Deletion of eight ORFS [YER184C, YFL052W, YIL130W, YLL054C, YLR266C, YOR162C (YRR1), YOR380W and YPL133C] rendered the cells hypersensitive to calcofluor white (Table 2 and Fig. 3). A strain carrying a transposon insertion in the promoter of the YLR2228C has been shown to be hypersensitive to calcofluor (42). However, in our assay with a deletion strain, no effect of calcofluor was observed. In summary, eight deletion strains are hypersensitive to calcofluor white. Interestingly, the same deletion strains were unable to grow on non-fermentable carbon sources. We do not know the relationship (if any) between these two phenotypes.
Figure 3.
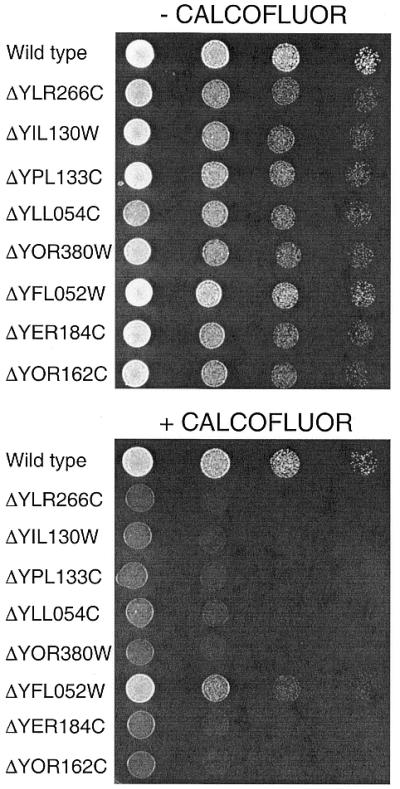
Calcofluor white sensitivity in deletion strains. Wild-type (FY73) and deletion strains were grown overnight in YPD, washed, serially diluted and spotted on YPD plates with or without 70 µg/ml calcofluor white. ORF deletions are indicated on left.
Anaerobic conditions
Since yeast is a facultative anaerobe, we tested the effect of growing deletion strains under anaerobic conditions to identify zinc cluster genes necessary for growth under these conditions. Growth was assayed on rich plates as well as minimal plates. All the tested knockout strains were able to grow in both media under these conditions (Table 2). Thus, Hap1p is the only zinc cluster protein which may be necessary for anaerobic growth (43).
CONCLUSIONS
The S.cerevisiae sequencing project has allowed the identification of many new (putative) members of the zinc cluster protein family. Only two members (YDR303C and YMR168C) of the GAL4 family of zinc cluster proteins are essential genes. Our deletion analysis has revealed phenotypes for a number of genes encoding zinc cluster proteins (Table 2). The most prevalent class of phenotypes is the inability to grow on non-fermentable carbon sources and sensitivity to calcofluor white. Other phenotypes observed include temperature and caffeine sensitivity. A number of deletions (e.g., YFL052W, YHR178W, YMR019W) resulted in multiple phenotypes. For example, deletion of ORF YFL052W results in an inability to grow on non-fermentable carbon sources, sensitivity to high temperature and calcofluor white (Table 2). Similarly, deletion of ORF YMR019W renders the cells clumpy and sensitive to caffeine. It is difficult to establish a relationship (if any) between these various phenotypes. Since our analysis focused on a class of transcriptional regulators, deletion of their genes may have a widespread effect on gene expression and some phenotypes observed may be due to greatly altered cell physiology. For example, whole-genome analysis with DNA microarrays revealed that expression of a large number of genes is affected by deletion of ORF YMR019W including most genes encoding ribosomal proteins (B.Akache and B.Turcotte, unpublished results). Thus, multiple phenotypes may be due to indirect effects.
Although many conditions were tested in our analysis, no phenotypes could be identified for a number of GAL4 family members. These proteins may perform highly specialized functions or, alternatively, there may be redundant genes encoding zinc cluster proteins. Analysis of double deletion mutants as well as whole-genome analysis of gene expression with microarrays should help to identify functions of zinc cluster proteins.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr J.Cook (Millennium Pharmaceuticals, Cambridge, MA) for his very generous gift of strains. We thank members of the H.Bussey laboratory (McGill University) for advice and material. We also thank Drs P.Belhumeur (Université de Montréal, Canada) and C.Taron (New England Biolabs, Beverly, MA) for comments and Dr S.Ali (McGill University) for critical review of the manuscript. This work was supported by grants from the Medical Research Council of Canada and the National Sciences and Engineering Research Council of Canada. B.T. is a scholar from the Fonds de la Recherche en Santé du Québec. B.A. and K.W. were supported by the Research Institute of the Royal Victoria Hospital.
References
- 1.Vallee B.L., Coleman,J.E. and Auld,D.S. (1991) Zinc fingers, zinc clusters, and zinc twists in DNA-binding protein domains. Proc. Natl Acad. Sci. USA, 88, 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd R.B. and Andrianopoulos,A. (1997) Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol ., 21, 388–405. [DOI] [PubMed] [Google Scholar]
- 3.Lohr D., Venkov,P. and Zlatanova,J. (1995) Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J., 9, 777–787. [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer K., Kim,K.S., Kogan,S. and Guarente,L. (1989) Functional dissection and sequence of yeast HAP1 activator. Cell, 56, 291–301. [DOI] [PubMed] [Google Scholar]
- 5.Creusot F., Verdiere,J., Gaisne,M. and Slonimski,P.P. (1988) CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J. Mol. Biol., 204, 263–276. [DOI] [PubMed] [Google Scholar]
- 6.Koloczkowska A. and Goffeau,A. (1999) Regulation of pleiotropic drug resistance in yeast. Drug Resist. Updates, 2, 403–414. [DOI] [PubMed] [Google Scholar]
- 7.Schjerling P. and Holmberg,S. (1996) Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res., 24, 4599–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou K.M. and Kohlhaw,G.B. (1990) Transcriptional activator LEU3 of yeast. Mapping of the transcriptional activation function and significance of activation domain tryptophans. J. Biol. Chem., 265, 17409–17412. [PubMed] [Google Scholar]
- 9.Friden P., Reynolds,C. and Schimmel,P. (1989) A large internal deletion converts yeast LEU3 to a constitutive transcriptional activator. Mol. Cell. Biol., 9, 4056–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson J.C. and Lopes,J.M. (1996) The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic Acids Res., 24, 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozcan S., Leong,T. and Johnston,M. (1996) Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol., 16, 6419–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwabe J.W. and Rhodes,D. (1997) Linkers made to measure. Nat. Struct. Biol., 4, 680–683. [DOI] [PubMed] [Google Scholar]
- 13.Marmorstein R., Carey,M., Ptashne,M. and Harrison,S.C. (1992) DNA recognition by GAL4: structure of a protein–DNA complex. Nature, 356, 408–414. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L. and Guarente,L. (1994) The yeast activator HAP1—a GAL4 family member-binds DNA in a directly repeated orientation. Genes Dev., 8, 2110–2119. [DOI] [PubMed] [Google Scholar]
- 15.Ha N., Hellauer,K. and Turcotte,B. (1996) Mutations in target DNA elements of yeast HAP1 modulate its transcriptional activity without affecting DNA binding. Nucleic Acids Res., 24, 1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King D.A., Zhang,L., Guarente,L. and Marmorstein,R. (1999) Structure of a HAP1–DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein. Nat. Struct. Biol., 6, 64–71. [DOI] [PubMed] [Google Scholar]
- 17.Hellauer K., Rochon,M.-H. and Turcotte,B. (1996) A novel DNA binding motif for yeast zinc cluster proteins: the Leu3p and Pdr3p transcriptional activators recognize everted repeats. Mol. Cell. Biol., 16, 6096–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolaev I., Lenouvel,F. and Felenbok,B. (1999) Unique DNA binding specificity of the binuclear zinc AlcR activator of the ethanol utilization pathway in Aspergillus nidulans. J. Biol. Chem., 274, 9795–9802. [DOI] [PubMed] [Google Scholar]
- 19.Karpichev I.V., Luo,Y., Marians,R.C. and Small,G.M. (1997) A complex containing two transcription factors regulates peroxisome proliferation and the coordinate induction of β-oxidation enzymes in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rottensteiner H., Kal,A.J., Hamilton,B., Ruis,H. and Tabak,H.F. (1997) A heterodimer of the Zn2Cys6 transcription factors Pip2p and Oaf1p controls induction of genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Eur. J. Biochem., 247, 776–783. [DOI] [PubMed] [Google Scholar]
- 21.Amar N., Messenguy,F., El Bakkoury,M. and Dubois,E. (2000) ArgRII, a component of the ArgR–Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol., 20, 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winston F., Dollard,C. and Ricupero-Hovasse,S.L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]
- 23.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brachmann C.B., Davies,A., Cost,G.J., Caputo,E., Li,J., Hieter,P. and Boeke,J.D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- 25.Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noël J. and Turcotte,B. (1998) Zinc cluster proteins Leu3p and Uga3p recognize highly related but distinct DNA targets. J. Biol. Chem., 273, 17463–17468. [DOI] [PubMed] [Google Scholar]
- 27.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York, NY.
- 28.Philippsen P., Stotz,A. and Scherf,C. (1991) DNA of Saccharomyces cerevisiae. Methods Enzymol., 194, 169–182. [DOI] [PubMed] [Google Scholar]
- 29.Adams A., Gottschling,D.E. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Bourot S. and Karst,F. (1995) Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene, 165, 97–102. [DOI] [PubMed] [Google Scholar]
- 31.Weierstall T., Hollenberg,C.P. and Boles,E. (1999) Cloning and characterization of three genes (SUT1–3) encoding glucose transporters of the yeast Pichia stipitis. Mol. Microbiol., 31, 871–883. [DOI] [PubMed] [Google Scholar]
- 32.Burns N., Grimwade,B., Ross-Macdonald,P.B., Choi,E.Y., Finberg,K., Roeder,G.S. and Snyder,M. (1994) Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev., 8, 1087–1105. [DOI] [PubMed] [Google Scholar]
- 33.Lucau-Danila A., Wysocki,R., Roganti,T. and Foury,F. (2000) Systematic disruption of 456 ORFs in the yeast Saccharomyces cerevisiae. Yeast, 16, 547–552. [DOI] [PubMed] [Google Scholar]
- 34.Clarke L. (1998) Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev., 8, 212–218. [DOI] [PubMed] [Google Scholar]
- 35.Lechner J. (1994) A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J., 13, 5203–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strunnikov A.V., Kingsbury,J. and Koshland,D. (1995) CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol., 128, 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niedenthal R., Riles,L., Guldener,U., Klein,S., Johnston,M. and Hegemann,J.H. (1999) Systematic analysis of S.cerevisiae chromosome VIII genes. Yeast, 15, 1775–1796. [DOI] [PubMed] [Google Scholar]
- 38.Hampsey M. (1997) A review of phenotypes in Saccharomyces cerevisiae. Yeast, 13, 1099–1133. [DOI] [PubMed] [Google Scholar]
- 39.Cui Z., Shiraki,T., Hirata,D. and Miyakawa,T. (1998) Yeast gene YRR1, which is required for resistance to 4-nitroquinoline N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol. Microbiol., 29, 1307–1315. [DOI] [PubMed] [Google Scholar]
- 40.Kasten M.M. and Stillman,D.J. (1997) Identification of the Saccharomyces cerevisiae genes STB1–STB5 encoding Sin3p binding proteins. Mol. Gen. Genet., 256, 376–386. [DOI] [PubMed] [Google Scholar]
- 41.Nishimura H., Kawasaki,Y., Kaneko,Y., Nosaka,K. and Iwashima,A. (1992) Cloning and characteristics of a positive regulatory gene, THI2 (PHO6), of thiamin biosynthesis in Saccharomyces cerevisiae. FEBS Lett., 297, 155–158. [DOI] [PubMed] [Google Scholar]
- 42.Lussier M., White,A.M., Sheraton,J., di Paolo,T., Treadwell,J., Southard,S.B., Horenstein,C.I., Chen-Weiner,J., Ram,A.F., Kapteyn,J.C., Roemer,T.W., Vo,D.H., Bondoc,D.C., Hall,J., Zhong,W.W., Sdicu,A.M., Davies,J., Klis,F.M., Robbins,P.W. and Bussey,H. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics, 147, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chantrel Y., Gaisne,M., Lions,C. and Verdiere,J. (1998) The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics, 148, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross-Macdonald P., Coelho,P.S., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.H., Sheehan,A., Symoniatis,D., Umansky,L., Heidtman,M., Nelson,F.K., Iwasaki,H., Hager,K., Gerstein,M., Miller,P., Roeder,G.S. and Snyder,M. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 45.Kumar A., Cheung,K.H., Ross-Macdonald,P., Coelho,P.S., Miller,P. and Snyder,M. (2000) TRIPLES: a database of gene function in Saccharomyces cerevisiae. Nucleic Acids Res., 28, 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delaveau T., Delahodde,A., Carvajal,E., Subik,J. and Jacq,C. (1994) PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol. Gen. Genet., 244, 501–511. [DOI] [PubMed] [Google Scholar]
- 47.Groom K.R., Heyman,H.C., Steffen,M.C., Hawkins,L. and Martin,N.C. (1998) Kluyveromyces lactis SEF1 and its Saccharomyces cerevisiae homologue bypass the unknown essential function, but not the mitochondrial RNase P function, of the S.cerevisiae RPM2 gene. Yeast, 14, 77–87. [DOI] [PubMed] [Google Scholar]
- 48.Charron M.J., Read,E., Haut,S.R. and Michels,C.A. (1989) Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics, 122, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Needleman R. (1991) Control of maltase synthesis in yeast. Mol. Microbiol., 5, 2079–2084. [DOI] [PubMed] [Google Scholar]
- 50.André B. (1990) The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol. Gen. Genet., 220, 269–276. [DOI] [PubMed] [Google Scholar]
- 51.Vissers S., Andre,B., Muyldermans,F. and Grenson,M. (1990) Induction of the 4-aminobutyrate and urea-catabolic pathways in Saccharomyces cerevisiae. Specific and common transcriptional regulators. Eur. J. Biochem., 187, 611–616. [DOI] [PubMed] [Google Scholar]
- 52.Feller A., Dubois,E., Ramos,F. and Pierard,A. (1994) Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol., 14, 6411–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feller A., Ramos,F., Pierard,A. and Dubois,E. (1999) In Saccharomyces cerevisae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur. J. Biochem., 261, 163–170. [DOI] [PubMed] [Google Scholar]
- 54.Strich R., Surosky,R.T., Steber,C., Dubois,E., Messenguy,F. and Esposito,R.E. (1994) UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev., 8, 796–810. [DOI] [PubMed] [Google Scholar]
- 55.Crowley J.H., Leak,F.W.,Jr, Shianna,K.V., Tove,S. and Parks,L.W. (1998) A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol ., 180, 4177–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leak F.W.,Jr, Tove,S. and Parks,L.W. (1999) In yeast, upc2-1 confers a decrease in tolerance to LiCl and NaCl, which can be suppressed by the P-type ATPase encoded by ENA2. DNA Cell Biol., 18, 133–139. [DOI] [PubMed] [Google Scholar]
- 57.Balzi E., Chen,W., Ulaszewski,S., Capieaux,E. and Goffeau,A. (1987) The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem., 262, 16871–16879. [PubMed] [Google Scholar]
- 58.Bricmont P.A., Daugherty,J.R. and Cooper,T.G. (1991) The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 1161–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coornaert D., Vissers,S. and Andre,B. (1991) The pleiotropic UGA35(DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene, 97, 163–171. [DOI] [PubMed] [Google Scholar]
- 60.Lesage P., Yang,X. and Carlson,M. (1996) Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol. Cell. Biol., 16, 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincent O. and Carlson,M. (1998) Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J., 17, 7002–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.des Etages S.A., Falvey,D.A., Reece,R.J. and Brandriss,M.C. (1996) Functional analysis of the PUT3 transcriptional activator of the proline utilization pathway in Saccharomyces cerevisiae. Genetics, 142, 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siddiqui A.H. and Brandriss,M.C. (1989) The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol. Cell. Biol., 9, 4706–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Losson R. and Lacroute,F. (1981) Cloning of a eukaryotic regulatory gene. Mol. Gen. Genet., 184, 394–399. [DOI] [PubMed] [Google Scholar]
- 65.Roy A., Exinger,F. and Losson,R. (1990) cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol. Cell. Biol., 10, 5257–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmberg S. and Schjerling,P. (1996) Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics, 144, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Friden P. and Schimmel,P. (1987) LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol. Cell. Biol., 7, 2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou K.M., Bai,Y.L. and Kohlhaw,G.B. (1990) Yeast regulatory protein LEU3: a structure–function analysis. Nucleic Acids Res., 18, 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qui H.F., Dubois,E. and Messenguy,F. (1991) Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol. Cell. Biol., 11, 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hedges D., Proft,M. and Entian,K.D. (1995) CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 1915–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mendizabal I., Rios,G., Mulet,J.M., Serrano,R. and de Larrinoa,I.F. (1998) Yeast putative transcription factors involved in salt tolerance. FEBS Lett., 425, 323–328. [DOI] [PubMed] [Google Scholar]
- 72.Gray W.M. and Fassler,J.S. (1996) Isolation and analysis of the yeast TEA1 gene, which encodes a zinc cluster Ty enhancer-binding protein. Mol. Cell. Biol., 16, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rottensteiner H., Kal,A.J., Filipits,M., Binder,M., Hamilton,B., Tabak,H.F. and Ruis,H. (1996) Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J., 15, 2924–2934. [PMC free article] [PubMed] [Google Scholar]


