Zhang et al.1 state that they were unable to repeat findings presented in our 2012 publication in Nature Medicine regarding the characterization of oogonial stem cells (OSCs) in mouse and human ovaries2, using methods further detailed a year later3. Separately, Hernandez et al.4 question the specificity of antibodies that target the C terminus of DDX4 (DEAD box polypeptide 4) to viably sort OSCs from adult mouse, monkey and human ovaries, as we reported2,3. Although these two correspondences focus on our work from 2012, DDX4-specific antibody–based sorting of OSCs was first published in 2009 by another laboratory5. A year before this publication, Richards et al.6 reported isolation of viable germ cells from cultures of human embryonic stem cells using fluorescence-activated cell sorting (FACS) coupled with DDX4-specific antibodies. Our 2012 study therefore represents independent methodological verification of these two earlier reports.
We also note that Hernandez et al.4 corroborated the gene expression profile, in vitro growth characteristics and oocyte-forming properties of OSCs reported by others and us2,3,5,7–16. This latter point is relevant to weighing the merits of the claim by Zhang et al.1 that OSCs do not exist because they were unable to show new oocyte formation in transplantation-based assays using ovarian cell preparations prepared by DDX4 antibody-based sorting. Because, however, the ovarian cells sorted and used by Zhang et al.1 differ from the OSCs others and we have isolated and described, it is not surprising that their downstream endpoint analyses would not reproduce what has been already reported using purified OSCs as starting material.
With that said, apparent differences between our findings and those of Zhang et al.1 and Hernandez et al.4 regarding the ability of DDX4-specific antibodies to isolate OSCs highlight a fundamental issue raised by both sets of authors. Namely, is DDX4 entirely cytoplasmic in all germ cells at all stages of differentiation or do OSCs differ from other germ cells in their membrane localization of DDX4, thus making the protein available to be targeted in purification schemes involving magnetic-assisted cell sorting or FACS? If the latter case is true, why have some groups been able to repeat the DDX4-specific antibody–based approach to isolate OSCs while others have failed?
In our published study2, we provided extensive FACS-based validation data to show that DDX4-positive cells isolated from mouse ovaries through C-terminal DDX4 antibody-binding to viable (non-permeabilized) cell fractions are, in turn, recognized by an entirely different (N-terminal) DDX4-specific antibody only after the purified cells, which are free of oocyte contamination, are permeabilized. Given that both DDX4-specific antibodies specifically label oocytes in fixed ovarian tissue sections2, it is reasonable to conclude that the inability of C-terminal DDX4-specific antibodies to recognize immature oocytes during FACS, when OSCs are recognized and sorted in parallel, is due to an absence of the C-terminal antigenic sequence on non-permeabilized oocytes rather than an inability of the antibody to target DDX4 in oocytes.
Even so, Hernandez et al.4 used several approaches to conclude that DDX4 is not expressed by OSCs. In evaluating their FACS-based experiments to characterize C-terminal DDX4-specific antibody–positive (‘Ab+ve’) ovarian cells, we immediately noticed a large range in the relative frequency of Ab+ve cells obtained from human (4.5–24% positive) and rhesus macaque (2.5–50.6% positive) ovaries. The wide variability in yield of Ab+ve cells, along with a remarkably high number of cells deemed Ab+ve in some of their experiments, point to issues with antibody blocking, excess secondary antibody, and/or scoring of damaged or dead cells or debris as Ab+ve cells. By comparison, we reported that OSCs sorted from dispersed mouse and human ovarian tissue represent only 1.5% (±0.2%; n = 15) and 1.7% (±0.6%; n = 6), respectively, of the total viable cells sorted2.
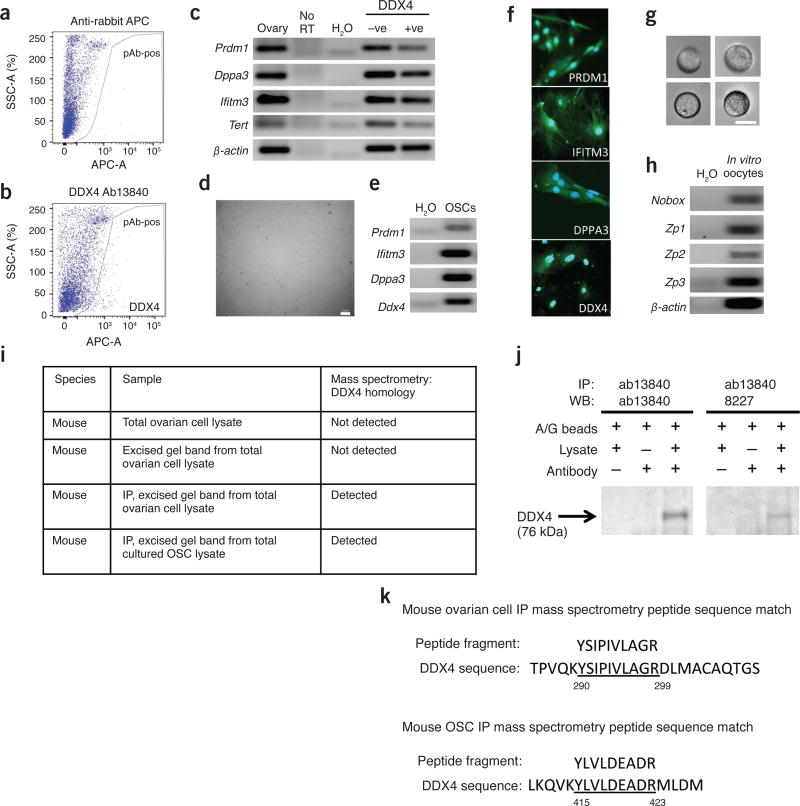
In new experiments using ovarian cortical tissue collected from female baboons (Papio anubis) during peak reproductive life (12–15 years of age; cryopreserved tissue provided by the University of Oklahoma Health Sciences Center), a comparable yield of C-terminal DDX4-positive cells was also obtained (Fig. 1a,b; 3.8% ± 1.4% of total viable cells sorted; mean ± s.e.m., n = 4 animals) that possessed the hallmark features of OSCs reported for other species (Fig. 1c–h). Although it is not possible for us to retrospectively troubleshoot the FACS protocol used by Hernandez et al.4, it is clear that their adaptation of our OSC isolation protocol2,3 produces cell yield outcomes that are inconsistent across their own experimental replicates, and which also diverge widely from what we have observed using mouse, baboon and human ovarian tissue2 (Fig. 1a,b). As a consequence, their use of cell preparations that contain up to 50% of the total ovarian cells sorted for downstream analysis of protein trafficking or gene expression unfortunately offers little, if any, clarity on the properties of purified OSCs or the comparative relationship of their findings to those we reported2.
Figure 1.
Characterization of viable DDX4-positive ovarian cells isolated by FACS. (a,b) Identification by FACS of a rare population of viable cells in dispersed adult baboon ovarian cell fractions that are recognized by antibodies against the C terminus of DDX4 (a, secondary antibody–only control; b, population shift of extracellular DDX4-positive cells; representative of results from four independent experimental replicates, using ovarian tissue from different animals for each replicate). (c) Primitive germ cell gene expression profile analysis of freshly isolated DDX4-positive cells from baboon ovaries (see ref. 2 for methodological details, and Supplementary Table 1 for PCR primer information; representative of results from three independent experimental replicates, using ovarian tissue from different animals for each replicate). (d) Bright-field image of baboon OSCs established in culture (scale bar, 100 mm; representative of results from analysis of four independent OSC lines, using ovarian tissue from different animals to establish each line). (e) Primitive germ cell gene expression analysis of cultured DDX4-positive cells (OSCs) from baboon ovaries (representative of results from analysis of three independent OSC lines). (f) Immunofluorescence-based detection of germ cell marker proteins (green, indicated protein in white text; blue, DAPI nuclear stain) in cultured baboon OSCs (see ref. 2 for methodological details; representative of results from independent analysis of 2 different OSC lines). (g) Appearance of in vitro–derived oocytes generated by baboon cultured OSCs (scale bar, 25 µm; images shown are representative of results from analysis of four independent OSC lines). (h) Analysis of oocyte marker gene expression (Nobox, newborn ovary homeobox; Zp1–3, zona pellucida proteins 1–3) in oocytes derived in vitro from cultured baboon OSCs (see ref. 2 for methodological details, and Supplementary Table 1 for PCR primer information; representative of results from independent analysis of 2 different OSC lines). (i) Mass spectrometric analysis of peptide sequences matched to mouse DDX4 in protein samples prepared from adult female mice as total ovarian cell lysates (dispersed unsorted cell fraction), the excised 76-kDa band area after denaturing gel electrophoresis of total ovarian cell lysates, the excised 76-kDa band area after denaturing gel electrophoresis of C-terminal DDX4 antibody-immunoprecipitated (IP) total ovarian cells lysates and the excised 76-kDa band area following denaturing gel electrophoresis of C-terminal DDX4 antibody-immunoprecipitated OSC lysates (representative of results from three independent experimental replicates using different mice for each replicate). (j) Western blot (WB) analysis of mouse total ovarian cell lysates after immunoprecipitation using protein A/G-conjugated beads with a rabbit polyclonal antibody targeting the C terminus of DDX4 (ab13840) and detection using the same antibody (left) or a different polyclonal antibody recognizing an internal epitope (8227; right); both approaches resulted in detection of a single 76-kDa band, corresponding to DDX4 (representative of results from four independent experimental replicates using different mice for each replicate). (k) Examples of mass spectrometry–generated peptide sequences (see i) from C-terminal DDX4-specific antibody–immunoprecipitated mouse total ovarian cell lysates (top) and mouse OSCs (bottom) matched with 100% homology to the mouse DDX4 protein sequence (underlined).
We feel it is important, however, to comment further on one of the most critical approaches used by these authors—namely, the mass spectrometry-based analysis of protein fractions prepared from Ab+ve ovarian cells of mouse, monkey and human origin. Total cellular proteins were prepared from each source, as well as from dispersed unsorted cells from macaque ovary and monkey testis included as positive controls, and resolved by one-dimensional (1D) gel electrophoresis, after which undefined “gel bands were cut into 1-mm cubes and pieces” for mass spectrometry. Evidence of peptide fragments with matched sequence homology to DDX4 appeared only in the testis sample. Peptides matched to DDX4 were not detected in any of the Ab+ve ovarian cell samples or in any of the dispersed unsorted ovarian cell fractions. Despite the fact that a key positive control—namely dispersed unsorted ovarian cell fractions containing oocytes, failed to show the presence of DDX4 protein, Hernandez et al.4 still concluded that Ab+ve ovarian cells lack DDX4, and as a consequence protocols that sort OSCs on the basis of cell surface DDX4 expression are erroneous.
We undertook experiments with 2-month-old C57BL/6 female mice (Charles River Laboratories), using protocols reviewed and approved by the Northeastern University Institutional Animal Care and Use Committee, to discern why these authors failed to detect DDX4 in dispersed ovaries, and what could be done to more rigorously test whether C-terminal DDX4-positive ovarian cells (OSCs) contain detectable levels of DDX4 protein. In agreement with Hernandez et al.4, we failed to detect peptide fragments homologous to DDX4 in total proteins prepared from dispersed unsorted ovarian cell fractions (Fig. 1i) and analyzed by the Taplin Mass Spectrometry Facility of Harvard Medical School (http://taplin.med.harvard.edu) and the Proteomics and Mass Spectrometry Facility of the University of Massachusetts Medical School (http://www.umassmed.edu/proteomics/). We next subjected our protein fractions to denaturing 1D gel electrophoresis, and excised gel bands closely surrounding the expected 76-kDa size of the DDX4 protein for analysis. These samples also failed to show the presence of peptide sequences homologous to DDX4 (Fig. 1i), despite the fact that these ovarian cell samples must contain the DDX4 protein contributed by oocytes.
We then subjected protein samples prepared from dispersed unsorted ovarian cell fractions to immunoprecipitation with the C-terminal DDX4-specific antibody used for FACS-based isolation of OSCs (Abcam 13840), and analyzed the immunoprecipitates by western blotting using either the same antibody or a different DDX4-specific antibody directed against an internal sequence of the protein (Cell Signaling 8227). Both antibodies detected a single immunoreactive band of approximately 76-kDa (Fig. 1j), confirming the presence of DDX4 in unsorted ovarian cell fractions. Based on this, we used C-terminal DDX4-specific antibodies to prepare immunoprecipitates from dispersed unsorted ovarian cells and from ex vivo–expanded OSCs. After denaturing electrophoresis, silver-stained bands corresponding to the predicted size of DDX4 (76 kDa; Fig. 1j) were excised and submitted for mass spectrometry. Under these conditions, both samples yielded peptide fragments with 100% sequence homology to DDX4 (Fig. 1k). These results not only clarify why Hernandez et al.4 failed to detect peptides corresponding to DDX4 in any of their ovarian cell samples, but also highlight that extensive preparative work (immunoprecipitation, denaturing gel electrophoresis, directed band excision) is needed to enable identification of peptide fragments homologous to DDX4 in a sample that, by virtue of the presence of oocytes, must contain DDX4 protein.
Although we are pleased that Hernandez et al.4 have independently verified past observations from others and us regarding the existence of OSCs in mouse, monkey and human ovarian tissue2,3,5–17, the overall conclusion offered by these authors that the C-terminal DDX4 antibody cross-reacts with some unknown protein, rather than DDX4, to enable sorting of OSCs is questionable. Not only has our OSC isolation protocol been replicated with a monoclonal antibody targeting the C terminus of human DDX4 protein17, but computational analysis indicates that only one other protein in the National Center for Biotechnology Information (NCBI) database—termed ATP-binding cassette subfamily C member 12 (ABCC12), shares any significant sequence homology with the C-terminal region of DDX4 that was recognized by the antibody used for OSC sorting3. This sequence in ABCC12 is located in a well-documented intracellular region of the protein, and it would not be recognized on viable cells by C-terminal DDX4–specific antibodies3. Likewise, recent studies using Ddx4-Cre;Rosa26tdTm/+ mice have demonstrated that C-terminal DDX4–positive ovarian cells isolated from ovaries by FACS exhibit Cre-mediated activation of the Ddx4 promoter–driven reporter, indicative of DDX4 gene expression in these cells15. We therefore believe the preponderance of evidence continues to weigh in favor of the Ddx4 gene being expressed in OSCs, and of externalized expression of the C terminus of the DDX4 protein by OSCs.
Acknowledgments
We thank K. Chandrasekhar and D. Navaroli for generating the new data presented in Figure 1, and the we thank the US National Institutes of Health (NIH) for funding support (R37-AG012279, R21-HD072280).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper (doi:10.1038/nm.3964).
References
- 1.Zhang H, et al. Nat. Med. 2015;21:1116–1118. doi: 10.1038/nm.3775. [DOI] [PubMed] [Google Scholar]
- 2.White YAR, et al. Nat. Med. 2012;18:413–421. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woods DC, Tilly JL. Nat. Protoc. 2013;8:966–988. doi: 10.1038/nprot.2013.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez SF, et al. Nat. Med. 2015;21:1114–1116. doi: 10.1038/nm.3966. [DOI] [PubMed] [Google Scholar]
- 5.Zou K, et al. Nat. Cell Biol. 2009;11:631–636. doi: 10.1038/ncb1869. [DOI] [PubMed] [Google Scholar]
- 6.Richards M, Fong C-Y, Bongso A. Fertil. Steril. 2008;93:986–994. doi: 10.1016/j.fertnstert.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 7.Pacchiarotti J, et al. Differentiation. 2010;79:159–170. doi: 10.1016/j.diff.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. J. Mol. Cell Biol. 2011;3:132–141. doi: 10.1093/jmcb/mjq043. [DOI] [PubMed] [Google Scholar]
- 9.Zou K, Hou L, Sun K, Xie W, Wu J. Stem Cells Dev. 2011;20:2197–2204. doi: 10.1089/scd.2011.0091. [DOI] [PubMed] [Google Scholar]
- 10.Imudia AN, et al. Fertil. Steril. 2013;100:1451–1458. doi: 10.1016/j.fertnstert.2013.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L, et al. Mol. Hum. Reprod. 2014;20:271–281. doi: 10.1093/molehr/gat081. [DOI] [PubMed] [Google Scholar]
- 12.Grieve KM, McLaughlin M, Dunlop CE, Telfer EE, Anderson RA. [26 July 2015];Maturitas. doi: 10.1016/j.maturitas.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Silvestris E, D’Oronzo S, Cafforio P, D’Amato G, Loverno G. J. Ovarian Res. 2015;8:55. doi: 10.1186/s13048-015-0184-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie W, Wang H, Wu J. Sci. Reports. 2014;4:5580. doi: 10.1038/srep05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park ES, Tilly JL. Mol. Hum. Reprod. 2015;21:58–65. doi: 10.1093/molehr/gau071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khosravi-Farsani S, Amidi F, Roudkenar MH, Sobhani A. Cell J. 2015;16:406–415. doi: 10.22074/cellj.2015.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fakih MH, et al. JFIV Reprod. Med. Genet. 2015;3:154. [Google Scholar]