Abstract
Astrocyte activation is a hallmark of HIV infection and aging in the CNS. In chronically infected HIV patients, prolonged activation of astrocytes has been linked to accelerated aging including but not limited to neurocognitive impairment and frailty. The current study addresses the role of HIV protein Tat in inducing a set of small noncoding microRNAs (miRNA) that play critical role in astrogliosis. In our efforts to link astrocyte activation as an indicator of aging, we assessed the brains of both wild type and HIV transgenic rats for the expression of glial fibrillary acidic protein (GFAP). As expected, in the WT animals we observed age-dependent increase in astrogliosis in the older animals compared to the younger group. Interestingly, compared to the young WT group, young HIV Tg rats exhibited higher levels of GFAP in this trend was also observed in the older HIV Tg rats compared to the older WT group. Based on the role of SIRT1 in aging and the regulation of SIRT1 by miRNAs-34a and -138, we next assessed the expression levels of these miRs in the brains of both the young an old WT and HIV Tg rats. While there were no significant differences in the young WT versus the HIV Tg rats, in the older HIV Tg rats there was a significant upregulation in the expression of miRs-34a & -138 in the brains. Furthermore, increased expression of miRs-34a & -138 in the older Tg rats, correlated with a concomitant decrease in their common anti-aging target protein SIRT1, in the brains of these animals. To delineate the mechanism of action we assessed the role of HIV-Tat (present in the Tg rats) in inducing miRs-34a & -138 in both the primary astrocytes and the astrocytoma cell line A172, thereby leading to posttranscriptional suppression of SIRT1 with a concomitant up regulation of NF-kB driven expression of GFAP.
Keywords: Aging, HIV, CNS, Astrocyte, miRNA, SIRT1
INTRODUCTION
Despite the advent of effective combinatorial antiretroviral therapy (cART) and its success in suppressing viremia and increasing the lifespan of those afflicted, conditions associated with normal aging such as cardiovascular disease, neurocognitive impairment, lipodystrophy and diabetes are often observed in HIV-infected individuals[1–3]. In fact premature aging is emerging as a co-morbidity in the era cART [4–7]. In the CNS, a spectrum of neurocognitive disorders termed as HIV-associated neurocognitive disorders (HAND) is present in almost 50% of the infected individuals. Instead of frank dementia, minor cognitive motor disorders are more prevalent in the present era. While the underlying molecular mechanisms leading to premature aging are not well characterized, it is speculated that persistent chronic inflammation could be a major factor underlying the progression and pathology of HAND and premature aging in general. Mechanisms involved in neuroinflammation are likely attributable to the presence of viral proteins such as HIV transactivation of transcription (Tat) & the virus envelope (gp120) or cellular factors such as the pro-inflammatory cytokines [8–12]. Interestingly, although virus replication can be controlled in the presence of cART, the limitation of some of these drugs to penetrate the CNS coupled with ability of some of the viral proteins such as Tat (that is present despite cART) to induce cytotoxicity, induces a state of chronic inflammation in the CNS. In the CNS glia make up the majority of cells in the brain, and can be activated by the presence of either viral and/or cellular factors, leading in turn, to a cascade of neurodegeneration resulting in synaptodendritic injury, a pathological correlate of HAND [13, 14].
It is now becoming widely recognized that normal aging is accompanied by astroglial activation [15–17]. Several studies support the premise that activation of this reactive glial population leads to an amplified and prolonged neuroinflammatory response [15, 18]. To understand the role of astrolgiosis in the context of aging and HIV infection, we chose to use the well-established model, HIV Tg rat, developed by Reid et al [19]. In this rat model efficient (but non-infectious, due to deletion of Gag and Pol) viral gene expression has been demonstrated to occur in the lymph nodes, spleen, thymus, blood and the CNS [19–21]. This model has been shown to exhibit abnormalities that mimic aspects of HIV-associated neurocognitive disorders in the era of cART, including glial activation [22], dendritic damage and reduced spine density in the striatal neurons [13, 23], mitochondrial abnormalities [24] and behavioral abnormalities [25, 26]. This model also replicates aspects of human HAND in the persistence of Tat in brain and other tissues, Aging HIV Tg rat is thus used in this study as a surrogate for aging observed in the HIV-infected individuals in the presence of cART
Astrocytes, the most abundant cell type within the brain, provide a crucial neurotropic support for the neurons during normal homeostasis. Activation of these cells by either HIV or certain cellular proteins disrupts the balance of tropic support, resulting in generation of a cytokine storm within the CNS, which then sets a stage for cellular senescence and aging. Activation of astrocytes in response to HIV/HIV proteins has been well documented [27–30]. In keeping with this information, understanding how the viral proteins accelerate the aging process is of paramount importance in the field.
MicroRNAs (miRs) are small (~20–22) nucleotide (nt) regulatory RNA molecules, that are encoded by the genome but are not translated into protein(s). Instead, they play key roles in regulating gene expression by associating with the 3′ untranslated region (UTR) of the nascent messenger RNA (mRNA) molecules, thereby repressing their expression [31]. Recent studies have demonstrated that miRNAs are highly expressed in the CNS including the brain and spinal cord. For example, we have recently reported that miR-9 regulates glial activation via targeting the monocyte chemoattractant protein inducible peptide (MCPIP)-1 [32]. The role of miRNAs in regulating the aging process has also been well studied. For example miRNA lin-4, regulates lifespan of C. elegans [33]. To date, numerous miRNAs have been shown to be significantly up- or down-regulated during aging; many of these miRNAs such as miR-71 in C. elegans and miR-17–92 in mammals have been identified as regulators of aging and cellular senescence [34, 35]. These developments in the field have provided an understanding of specific factors that alter age-related signaling pathways in various species from C. elegans to humans. Interestingly, several miRNAs, such as, miR-34a, miR-138, miR-142, have been reported to regulate the expression of SIRT1, an ortholog of yeast Sir2, that is implicated in the regulation of life span, stress resistance, and metabolism [36–39]. Since SIRT1 plays a crucial role in the aging process, we sought to examine the regulation of its expression by miRNAs. In this regard, we chose to assess the levels of brain enriched miRs-34a and -138, both of which have been shown to regulate SIRT1 [40, 41], and, also been shown to be regulated by HIV/HIV protein [41, 44–47]. We thus hypothesized that based on the fact that HIV proteins induce premature aging, it is likely that levels of these miRs that regulate the anti-aging gene SIRT1, will be upregulated earlier in the HIV Tg rats compared to the WT animals, and concomitantly that their target SIRT1 will be downregulated, leading to astrocyte activation in the brains of aging Tg (young/old) and WT (old) rodents. Further, using an in vitro approach we demonstrate that HIV Tat contributed to upregulation of miRs-34a & -138 resulting in negative regulation of SIRT1, ultimately leading to astrogliosis.
MATERIALS AND METHODS
Animals
Male, young (3–6 months;) and old (>15 months) HIV transgenic rats [HIV Tg rat, Harlan Sprague Dawley: HIV-1 (F344) Harlan and F344 rats (age and background-matched controls) were used in this study. These two ages represent human ages of ~18 and >60 years, respectively.
HIV Tg rats expresses a non-replicative provirus under its own viral promoter. As a result, the provirus encodes for only one (env) of the three genes (gag, pol, and env) needed to produce viral particles, plus all of the regulatory (tat, rev) and supplementary (vif, vpr, vpu, and nef) genes. Thus, these animals are noninfectious due to the functional deletion of Gag and Pol within the HIV-1 provirus (carrying only 7 of the 9 HIV genes) [19]. Rats were housed in pairs in microisolator cages within an Association for Assessment and Accreditation of Laboratory Animal Care International-approved rodent housing facility for the entirety of the study, which maintained a constant temperature (20–24°C) and humidity (30–60% relative humidity). Rats had access to chow and water ad libitum throughout the study period. All animal procedures were performed according to the protocols approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center. All animals were euthanized by exsanguination under anesthesia (5% isoflurane by mask). Brains (prefrontal cortex was used in this study) were dissected and immediately snap frozen, and stored at -80°C until used for various assays.
Cell culture and cell lines
The human astrocytic cell line A172 (no. CRL-1620; American Type Culture Collection (ATCC) was cultured as described previously [48] and maintained in Dulbecco’s modified Eagle’s medium (DMEM) high glucose medium containing 10% heated-inactivated fetal bovine serum, 2 mM glutamine, penicillin (100 units/ml), streptomycin (100 μg/ml), essential amino acids and vitamins. In this study, A172 cells were used within 30 passages. Human primary astrocytes were obtained from ScienCell Research Laboratories (Carlsbad, CA, USA) and were cultured in DMEM/F12 medium (Invitrogen Life Technologies, Carlsbad, CA, USA) containing 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, sodium bicarbonate, gentamicin, non-essential amino acids and vitamins. Rat primary astrocytes were prepared from whole brains of postnatal (1- to 2-day-old) Sprague-Dawley rats and plated on poly-D-lysine pre-coated cell culture flasks containing DMEM (10% fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin). The cells were grown in a humidified atmosphere of 5% CO2/95% air at 37 °C. When the astrocytes reached confluence, they were passaged by trypsinization and plated at a density of 106 cells/well on 24-well culture plates in a final volume of 1 ml of DMEM and grown in a humidified atmosphere of 5% CO2/95%air at 37 °C. Two days later, the astrocytes were used for experimentation. Immunocytochemical analyses demonstrated that the cultures comprised of >95% GFAP-positive astrocytes.
Western blotting
Treated cells or tissue were lysed using the Mammalian Cell Lysis kit (Sigma, St. Louis, MO, USA) and the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce, Rockford, IL, USA) and quantified using the micro BCA Protein Assay kit (Pierce, Rockford, IL, USA). Equal amounts of the corresponding proteins were electrophoresed in a sodium dodecyl sulfate-polyacrylamide gel (10–12%) under reducing conditions followed by transfer to PVDF membranes. The blots were blocked with 5% non-fat dry milk in phosphate buffered saline. Western blots were then probed with antibodies recognizing the HIV tat antibody (Abcam), GFAP, SIRT1 (1:1000; Santa Cruz, Dallas, TX, USA) and β-actin (1:4000; Sigma, St. Louis, MO, USA). The secondary antibodies were alkaline phosphatase conjugated to goat anti mouse/rabbit IgG (1:5000). Signals were detected by chemiluminescence and imaged on the FLA-5100 (Fujifilm, Valhalla, NY, USA) digital image scanner; densitometry was performed utilizing Image J software (NIH) [49].
Real-time PCR
For quantitative analysis of mRNA expression, comparative real-time PCR was performed with the use of the SYBR Green PCR Master Mix (Applied Biosystems). The sequences for the amplification of SIRT1 were: 5′-ACAGTGAGAAAATGCTGGCCTA -3′ (forward) and 5′-GCCATTGTAGAATTCTTCAATTTCAC -3′ (reverse); the sequences for the amplification of GFAP were: 5′-ATGGAGCTCAATGACCGCTTT-3′ (forward) and 5′-CGCCTTGTTTTGCTGTTCCA -3′ (reverse); the primer sequences for the amplification of GAPDH were as follows: 5′-TGCACCACCAACTGCTTAGC-3′ (forward); 5′-ATGCCAGTGAGCTTCCCGTT-3′ (reverse). For the analysis of miR-34a and miR-138, total RNA was isolated from cells as described above. Comparative real-time PCR was performed using the Taqman Universal PCR Master Mix (Applied Biosystems). Specific primers and probes for mature miR-34a, miR-138 and snRNA RNU6B were obtained from Applied Biosystems. All reactions were run in triplicate. The amount of miRNA was obtained by normalizing to snRNA RNU6B and relative to control as previously reported [50].
Immunofluorescence labeling and image analyses
For GFAP immunostaining, sections encompassing the entire midbrain were sectioned at 12 μm on a cryostat. Slides were fixed with 4% paraformaldehyde for 45 min at room temperature followed by permeabilization with 0.3% Triton X-100 in PBS followed by incubation with H2O2 for 10 min. Sections were incubated with a blocking buffer containing 10% NGS in PBS for 1 h at room temperature followed by addition of rabbit anti-GFAP (1:50; Santa Cruz Biotechnologies) antibody and incubated overnight at 4° C. Primary Abs were labeled with secondary anti-mouse and anti-rabbit Abs conjugated to the fluorescent probes Alexa Fluor 488, and nuclei were labeled with DAPI. Slides were covered with a coverslip with ProLong Gold antifade reagent (Invitrogen, Carlsbad, CA) and allowed to dry for 24 h at room temperature. Images were captured with a 20X objective.
MiRNA in situ hybridization
In situ hybridization for miRNA was performed as described previously [51]. Following deparaffinization and antigen retrieval with citrate, sections were pre-hybridized in hybridization buffer (50% formamide, 10 mM Tris-HCl, pH 7.4, 200 μg/ml yeast tRNA, 1 × Denhardt’s solution, 600 mM NaCl, 0.25% SDS, 1 mM EDTA, 100 μg/ml salmon sperm DNA) for 1 h at 52 °C in a humidified chamber. LNA-modified miRs-34a & -138, labeled at both the 5′- and 3′-ends with digoxigenin (Exiqon), were diluted to a final concentration of 4 pM in hybridization buffer, heated to 65 °C for 5 min and separately hybridized to the sections at 52 °C overnight. The slides were then washed three times in 2 × SSC and twice in 0.2 × SSC at 42 °C. They were then blocked with 1% BSA and 3% normal goat serum in 1 × PBS for 1 h at room temperature, and incubated with anti-digoxigenin conjugated with horseradish peroxidase (1: 100, Roche Diagnostics GmbH, Mannheim, Germany) and anti-GFAP (1: 500, abcam) antibody overnight at 4 °C. The slides were washed 3 times with TBS and incubated with Alexa Fluor 488 goat anti-mouse IgG (1: 500, Invitrogen) and Alexa Fluor 594 goat anti-Rabit IgG (1: 200, Invitrogen) antibody for 1 h at room temperature. This was followed by 3 times TBS washes and signal amplification (for the in situ, now labeled with horseradish peroxidae) using TSA Cy5 kit (Perkin Elmer, Waltham, MA, USA) according to the manufacturer’s protocol. The slides were mounted in Prolong gold anti-fade reagent with DAPI (Invitrogen).
Luciferase reporter constructs and luciferase assay
Transfection and luciferase assay were performed as described previously [50, 52]. R-luc-SIRT1-3′UTR (miR-138 binding site) [43] was kindly provided by Dr. Feng-Quan Zhou (Johns Hopkins University School of Medicine) and pmirGLO-SIRT1-3′UTR (miR-34a binding site) [42] from Dr. Mohan Mahesh (Tulane National Primate Research Center). Briefly, human astrocytoma A172 cells were transfected with the respective reporter constructs, as well as miR mimic or inhibitor, followed by assessment of luciferase activity 24 h after transfection. For functional study, A172 cells were transiently co-transfected with the pNF-κB reporter construct and the control or mimic/inhibitor of miRNA or pSIRT1 (Addgene) for 24 h. Transfected cells were then treated with the SIRT1 agonist, resveratrol (5 mM, Sigma–Aldrich), followed by assessment of luciferase activity. Luciferase activities were measured and normalized to the control renilla luciferase activity. The luciferase activity of each group was compared with that of the control group. All transfections were performed in triplicate and repeated at least twice.
Statistical analysis
Statistical analysis was performed using Student’s t-test or one-way analysis of variance followed by Holm–Sidak test (SigmaPlot 11.0). The appropriate test was clarified in the figure legends. Results were judged statistically significant if P<0.05 by analysis of variance.
RESULTS
GFAP is significantly upregulated in the brains of young HIV Tg versus WT rats
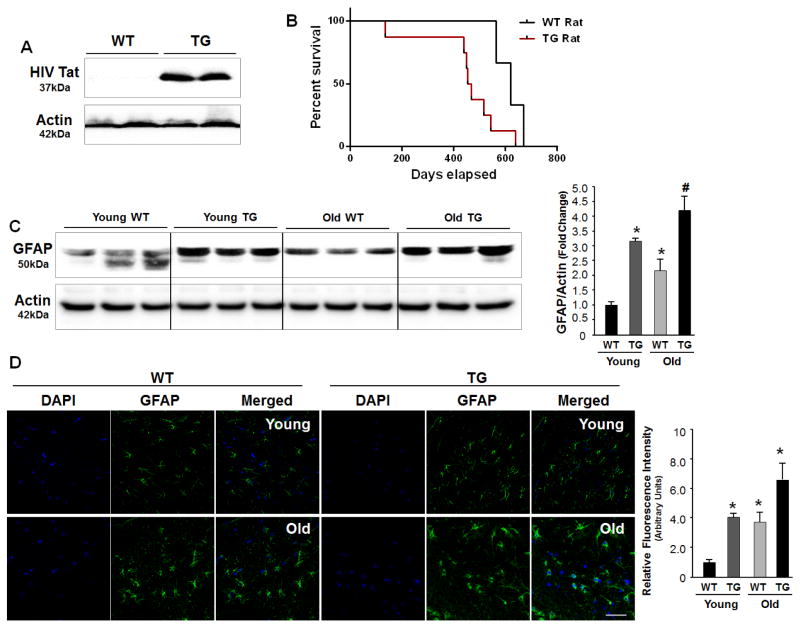
In this study we first sought to examine the expression of viral protein Tat in the brains of older HIV-1 transgenic rat. HIV Tg rat model expresses all of the HIV proteins except gag & pol [19, 53] and has been extensively characterized to study the effect of HIV Tat in the CNS by many labs [20, 54, 55]. This study was primarily to show the accumulation of viral proteins over time. As shown in Figure 1A, HIV-1 Tat was easily detectable in the brains of Tg rat. As expected there was no Tat present in the brains of WT rats. Furthermore, in agreement with previous reports [56], our findings also demonstrated a lower survival time in the Tg rats compared with the WT rats (mean ± SD: 671.4 ± 81.0 days vs 455.9 ± 145.6 days, p = 0.06) (Fig. 1B). Since astrocyte activation is a histological marker associated with HIV infection and aging [5, 57, 58], we next investigated the expression of GFAP in the brain homogenates isolated from young (3–6 months) vs old (> 12 months) WT and Tg rats. As shown in Figure 1C, expression levels of GFAP were significantly increased with aging in the both the WT and Tg rats. However, what was interesting was that in the Tg rats, there was higher expression of GFAP in the younger rats compared to the younger WT animals and this trend for GFAP increase continued even in the older rats; with older Tg rats expressing higher levels of GFAP compared with the older WT rats. GFAP protein expression studies were further validated by immunostaining brain sections from the WT and Tg rats of varying ages. As shown in Figure 1D, and in keeping with the western blot data, there was increased expression of GFAP in the brains of young Tg rats that was comparable with the GFAP levels in the older WT rats.
Figure 1. GFAP expression in the brains of WT and HIV Tg rats of varying ages.
(A) Western blot analyses of HIV Tat expression in the brains of HIV Tg rats. (B) Kaplan-Meier survival plot of WT and HIV Tg rats. (C) Western blot analyses of GFAP expression in the brains of young (3–6 months) and aged (>12 months) WT and age-matched HIV Tg rats. * p < 0.05 ANOVA vs young WT; # p < 0.05 ANOVA vs young TG. (D) Real-time PCR analysis of GFAP mRNA expression in the brains of young and aged WT and age-matched HIV Tg rats. (E) Immunostaining showing astrocyte activation in the brains of aged WT and young/aged Tg rats. Scale bar=20 μm. Relative mean fluorescence intensity calculated using Image J for GFAP. * p < 0.05 ANOVA vs young WT.
Downregulation of SIRT1 and upregulation of miRs-34a &-138 in the brains of WT and HIV transgenic rodents of varying ages
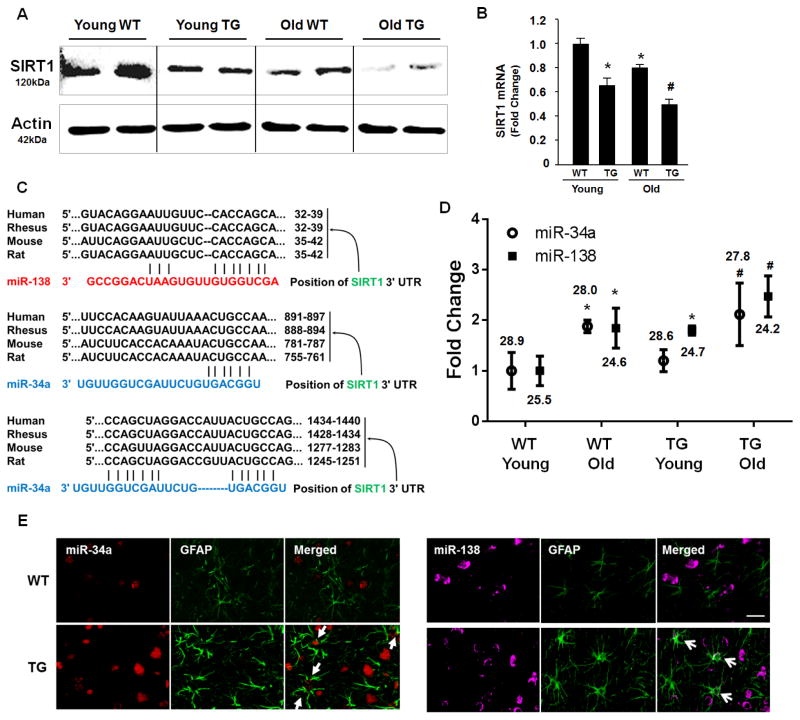
Next, we sought to investigate the expression of SIRT1, that plays an important role in the regulation of life span, stress resistance, and metabolism [59–61], in the brains of WT and Tg rats of varying ages. As shown in Figures 2A & B, SIRT1 mRNA and protein levels were significantly downregulated in the aging brains of both the WT and HIV Tg rats. Interestingly, the levels of SIRT1 was lower in the Tg rats at both the ages compared with the WT rats.
Figure 2. SIRT1 expression in the brains of WT and HIV Tg rats.
(A) Western blot analyses of SIRT1 expression in the brains of young and aged WT and age-matched HIV Tg rats. (B) Real-time PCR analyses of SIRT1 mRNA expression in the brains of young/aged WT and HIV Tg rats. Bars represent the mean±S.D. from three independent experiments. * p < 0.05 ANOVA vs young WT; # p < 0.05 ANOVA vs young TG. (C) The algorithms Targetscan program revealed complementary SIRT1 3-UTR potential binding sites in miRs-34a & -138. (D) The expression of miRs-34a & -138 in the brains of young/aged WT and HIV Tg rats. (E) In situ hybridization demonstrating increased miRs-34a & -138 expression in astrocytes (arrows) and neurons in the brain of HIV Tg rats. Scale bar=20 μm. Average Ct value is indicated. * p < 0.05 ANOVA vs young WT; # p < 0.05 ANOVA vs young TG.
MiRs are widely being recognized as regulators of gene expression. It has been reported that several miRNAs, including miR-34a and miR-138, can regulate the expression of SIRT1 [36–39]. Figure 2C depicts the conserved miRs-34a & -138 binding sites within SIRT1 3′UTR in most species. Having determined the levels of SIRT1, we thus next sought to assess the expression of its regulatory miRs-34a & -138 in the brains of young versus old WT and Tg rats. As shown in Figure 2D, real time PCR analyses demonstrated increased expression of both miRs-34a & -138 in the brains of aged WT rats compared with younger WT animals. Similar to the findings with GFAP and SIRT1, both the miRs-34a & -138 were expressed at a higher level in the young and old Tg ts compared to their corresponding age-matched WT rats. These findings were further validated by in situ hybridization, demonstrating increased expression of miRs-34a & -138 both in the astrocytes (Fig. 2E) and neurons (Data not shown) in the brains of HIV Tg rats compared with WT rats.
HIV Tat decreases expression of SIRT1 but induces GFAP in human astrocytes
Since HIV proteins accumulate with respect to aging in the HIV Tg rats, and since astrocytic activation in the Tg rats correlated with the aging process, we hypothesized that exposure of both human astrocytic cell line A172 and human primary astrocytes to HIV protein Tat (that has been shown to result in increased astrocyte activation) [62–65], could also lead to downregulation of SIRT1. The rationale for keeping this study focused on HIV Tat is based on the fact that despite ART-mediated near complete virus replication suppression, HAND still exists in patients [66] with sufficient data validating the presence of Tat and other viral proteins in both lymph node & CNS compartments [67]. Furthermore, exogenous Tat injections in the rodent brain have been shown to result in a multitude of neuropathological [68] and behavioral abnormalities mimicking aspects of HAND [69, 70].
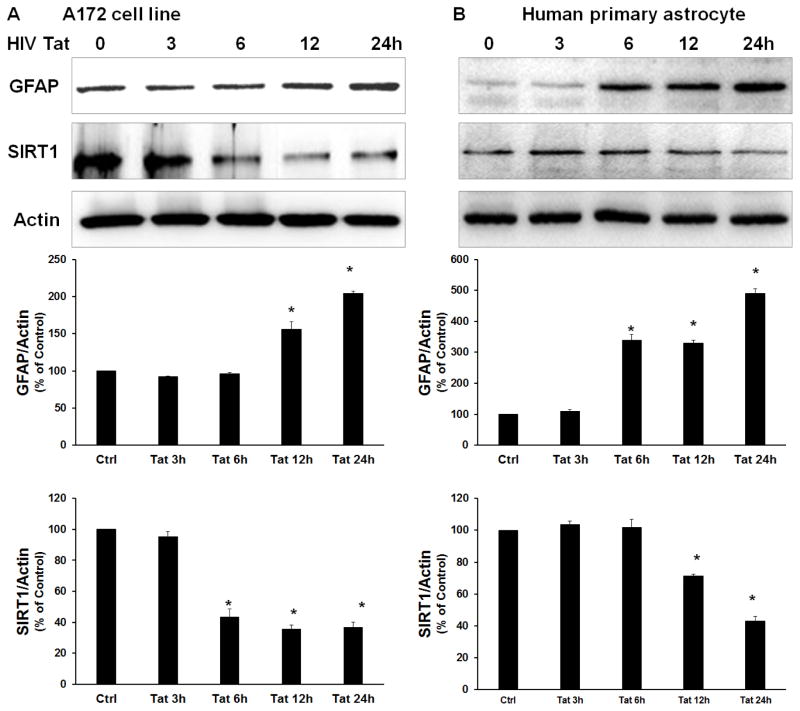
Human A172 astrocytes and primary astrocytes were exposed to HIV Tat (100 ng/ml) for varying times and analyzed for the expression levels of SIRT1 and GFAP proteins. As shown in Figure 3A & B, a significant decrease in SIRT1 protein content was detected in both A172 and human primary astrocytes following stimulation with HIV Tat. As expected, a concomitant and significant increase in GFAP was detected in the cells exposed to Tat (Fig. 3A & B).
Figure 3. HIV Tat downregulates the expression of SIRT1 and induces GFAP expression in A172 astrocytes.
Western blot analyses of SIRT1 and GFAP expression in A172 cells (A) and human primary astrocytes (B) treated with HIV Tat for various times. Representative immunoblots (upper panel) and the densitometric analyses (lower panel) of SIRT1 or GFAP/Actin from four separate experiments. All the data are indicated as mean±s.d. of four independent experiments. *p<0.05 versus control group using one-way analysis of variance followed by Holm–Sidak test.
HIV Tat induces miRs-34a & -138 expression in both human and rat astrocytes
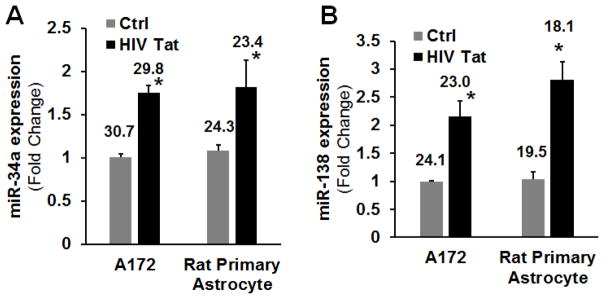
Since miRs-34a and -138 regulate levels of SIRT1, we next assessed the expression levels of these miRs in both A172 and rat primary astrocytes exposed to Tat, using the mature miRNA-specific quantitative PCR. As shown in Figures 4A & B, and in keeping with the in vivo data, there was increased expression of miRs-34a & -138 in the both cell line and primary astrocytes treated with HIV Tat compared with untreated control cells.
Figure 4. miRs-34a & -138 expression in human A172 cells following stimulation with HIV Tat.
Expression of miRs-34a (A) & -138 (B) by qPCR in the A172 and rat primary astrocytes following stimulation with HIV Tat. Bars represent mean ± SD from 3 independent experiments. Average Ct value is indicated. *P<0.05 vs ctrl.
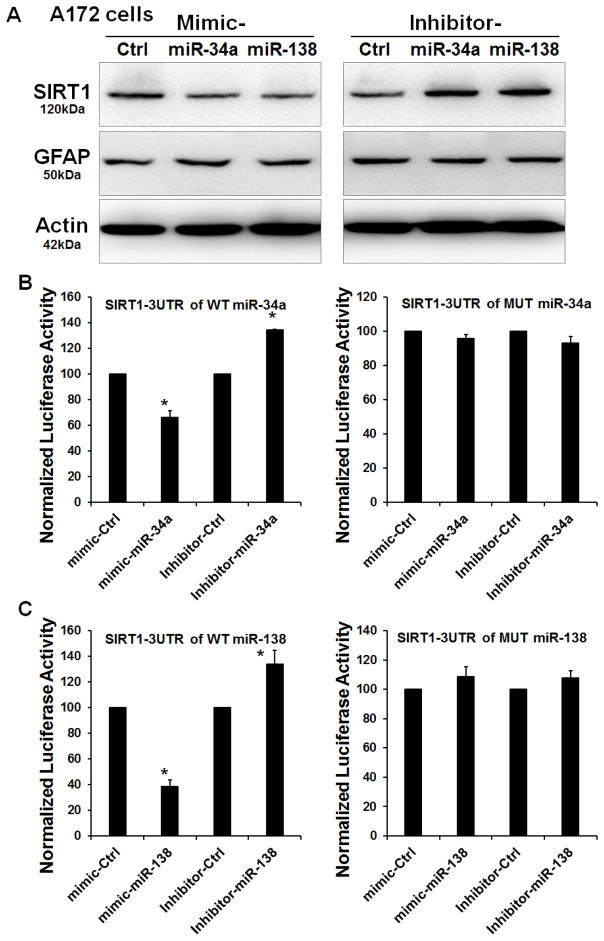
SIRT1 is a target of miR-34a and miR-138 in astrocytes
To establish a link between miRs-34a and -138, SIRT1 and GFAP, we first transfected human A172 cells with either the mimic or inhibitor for miR-34a or miR-138 for 72 h, followed by assessment of SIRT1 protein expression by western blot. Transfection of A172 cells with the mimic specific for either miR-34a or -138 resulted in decreased expression of SIRT1 protein with a concomitant upregulation of GFAP (Fig. 5A). In contrast, an increase in SIRT1 expression and a concomitant decrease in GFAP protein was observed in A172 cells treated with the inhibitor of either miR-34a or -138 (Fig. 5B). To further validate the binding of miRs-34a and -138 to the 3untranslated region (3′-UTR) of SIRT1 mRNA, human A172 cells were transfected with the SIRT1 Luc reporter constructs encoding for either SIRT1 3′-UTR binding sequences for either miRs-34a & or -138 (Fig. 5C). Cotransfection of A172 cells with both the mimic miR-34a or miR-138 and the respective SIRT1-Luc construct resulted in a significant decrease in Luc activity, suggesting thereby the preferential binding of miR-34a and miR-138 with the 3′-UTR of SIRT1. As a negative control, astrocytes were also cotransfected with a construct containing mutations in the miR-34a or miR-138 -binding region of the 3′-UTR–SIRT1. As expected, transfection of cells with the SIRT1 3′-UTR Mut failed to downregulate Luc activity (Fig. 5D). Reciprocally, these findings were also validated by knocking down the expression of endogenous miRs-34a & -138 by cotransfecting the cells with the respective anti-miRs. As expected, cotransfection with anti-miRs-34a or -138 resulted in significant enhancement of Luc activity (Fig. 5B). These data thus suggested that SIRT1 is a target for both miRs-34a & -138 in human astrocytes.
Figure 5. SIRT1 is a target of miR-34a and miR-138.
(A) Western blot analyses of SIRT1 and GFAP in lysates of A172 transfected with either miR-34a/miR-138 mimics or inhibitor. (B) Relative luciferase activity of WT/3′UTR mutant constructs of SIRT1 co-transfected with either the mimic of inhibitor of miR-34a/miR-138 in A172 cells.
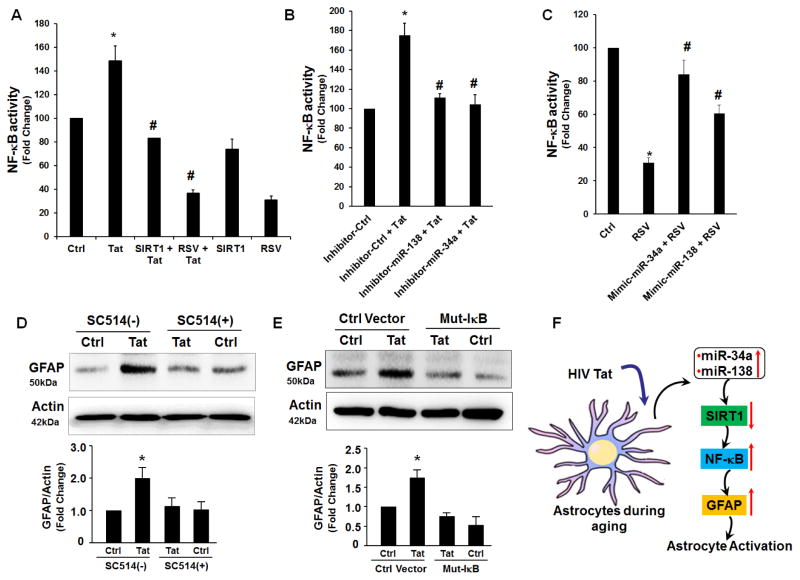
Inhibition of SIRT1 restores HIV Tat -induced NF-kB activation
Since activation of the NF-κB signaling pathway is known to contribute to activation of GFAP we next sought to examine whether Tat-mediated downregulation of SIRT1 affected NF-κB activity (target of miRs-34a & -138). SIRT1 is a known repressor of NF-κB and an anti-aging protein. To test whether overexpression of SIRT1 via resveratrol (RSV), a SIRT1 agonist, could inhibit HIV Tat-induced NF-κB activity, A172 cells were transfected with a NF-κB-driven luciferase reporter construct and either also transfected with a SIRT1 overexpressing plasmid or treated with RSV followed by assessment of the luciferase activity. Intriguingly, we found that activation of SIRT1 either by overexpressing SIRT1 construct or RSV treatment inhibited HIV Tat-induced NF-κB-dependent luciferase reporter activity (Fig. 6A). To next validate the role of miRs-34a and -138 we cotransfected A172 cells with either of miR-34a or miR-138 inhibitor (as a means to upregulate SIRT1) and a NF-κB-driven luciferase reporter plasmid followed by assessment of luciferase activity. Consistently, we found that increased SIRT1 expression via transfection of cells with miRNA inhibitors also resulted in inhibition of HIV Tat-induced NF-κB activity (Fig. 6B). Finally, to confirm that miR-34a and miR-138 regulated NF-κB activity via targeting SIRT1, A172 cells were cotransfected with the NF-κB reporter construct and either miR-34a or miR-138 mimic in the absence or presence of the SIRT1 agonist, RSV. Interestingly, exposure to RSV alone resulted in decreased activity of NF-κB that was partially restored with miR-34a or miR-138 mimics (Fig. 6C). We next sought to validate the role of NF-κB in HIV Tat-mediated GFAP expression. As shown in Fig. 6D, pretreatment of A172 cells with Ikk-2 inhibitor SC514 (5 μM) significantly decreased HIV Tat-mediated induction of GFAP. This was further confirmed by the fact that transfection of A172 cells with mutant IκB resulted in amelioration of HIV Tat-mediated induction of GFAP (Fig. 6E). Our data thus demonstrate that HIV Tat-induced expression of miRs-34a and -138, regulates NF-κB activity via targeting SIRT1 (Fig. 6F).
Figure 6. Inhibition of SIRT1 restores HIV Tat -induced NF-kB activation.
(A) Activation of SIRT1 inhibits HIV Tat-induced NF-kB activation. Cells were co-transfected with pNF-kB reporter construct and the pSIRT1 plasmid. Luciferase activity reflecting NF-kB activation was monitored. *p < 0.05 ANOVA vs the control cells; #p < 0.05 ANOVA vs Tat treated cells. (B) Suppression of miR-34a or miR-138 inhibited Tat-induced NF-kB activation. Cells were co-transfected with pNF-kB reporter construct and the miR-34a or miR-138 inhibitor for 24 h followed by exposure to Tat for 4 h and assessment of luciferase activity. (C) Cells were co-transfected with pNF-kB reporter construct and the miR-34a or miR-138 mimic in the presence or absence of RSV for 24 h, followed by assessment of luciferase activity. Overexpression of miR-34a or miR-138 in A172 cells restored reseveratrol-mediated downregulation of NF-kB activity. (D, E) A172 cells were pre-treated with Ikk-2 inhibitor-SC514 (5 μM) for 1h (D) or transfected with mutant IkB plasmid for 12h (E) followed by exposure to Tat for 24 h and assessment of GFAP expression by western blot. (F) Schematic demonstrating Tat-mediated activation of astrocytes involving upregualtion of miRs-34a & -138 with subsequent downregulation of their target SIRT1, leadin in turn, to activation of NFkB. MiRs-34a & -138 are positively regulated by HIV Tat and function to repress antiaging factor SIRT1. SIRT1 negatively regulates NFkB activation, ultimately leading to astrogliosis. (↑) Represents upregulation; (↓) represents downregulation. *p < 0.05 ANOVA vs the control cells; #p < 0.05 ANOVA vs RSV treated cells.
DISCUSSION
Our findings herein demonstrate that while neuroinflammation is a part of the aging process in both the WT as well as HIV Tg rats, there was an early onset of markers of aging such as astrocyte activation and downregulation of SIRT1 in the HIV Tg rats compared with the WT controls. Since one of the key features of HIV Tg rats is a progressive accumulation of viral proteins with the aging process, we recapitulated the in vivo findings in vitro using cell culture models such as the human astrocyte cell line A172 and primary human astrocytes exposed to viral protein Tat (known to accumulate with time in the HIV Tg rats). Our cell culture findings demonstrated that similar to in vivo findings, in both the astrocyte cells exposed to HIV protein Tat there was increased expression of GFAP, a marker of aging.
Besides GFAP, many other factors have been identified to be differentially expressed in the aging and/or HIV-infected brain [71, 72]. Among these, SIRT1, the anti-aging protein, is of particular interest. SIRT1 is a multifunctional protein which participates in the development of cancer, metabolic disorders, HIV infection and aging [38, 73, 74]. It has in fact been proposed as a therapeutic target for the treatment of various disorders [75, 76]. Under normal homeostasis, expression of SIRT1 is tightly controlled by many factors, a reflection perhaps of its many functions in cell biology that, when deregulated, can influence the development of a number of pathologies. Recently, post-transcriptional regulatory mechanisms of SIRT1 by miRNAs have been postulated to play critical roles in the aging process. Currently, more than twenty miRNAs have been identified in regulating SIRT1 translation in various biological processes [37, 38, 77]. The rationale for focusing our studies on brain enriched miRs-34a & -138 is based on the fact that: a) these age-related miRs are significantly elevated in the brains of SIV-infected macaques, b) have also been implicated in regulating neurogenesis, and c) they target SIRT1 3′-UTR, thereby regulating SIRT1 expression at the post-transcriptional level [42, 50, 78–81]. Our data clearly demonstrated that exposure of astrocytes to HIV Tat resulted in decreased expression of SIRT1 with a concomitant increase in astrocyte marker, GFAP. Furthermore, overexpression of miR-34a or miR-138 resulted in decreased expression of SIRT1 protein. Reciprocally, increased expression of SIRT1 protein was observed in astrocytes transfected with inhibitors of miR-34a or miR-138. In keeping with these data, our in vivo study also demonstrated increased expression of both miRs-34a as well as miR-138 in the aging brain of WT rats. Interestingly, younger HIV Tg rats demonstrated an even more increase in both the miRs compared with the corresponding age-matched WT rats. This was also reflected in the downregulation of the miR-target, SIRT1, that also was concomitantly and significantly decreased with aging in both the WT and HIV Tg rats, although the downregulation of SIRT1 in HIV Tg rats much more pronounced than in the age-matched WT rats.
Another novel finding of this study was the role of miR-34a and miR-138 mediated downregulation of SIRT1 as a contributor of astrocyte activation. A large body of literature suggests that chronic inflammation plays an important role in age-related immune dysregulation in older adults in the general population [82, 83]. Moreover, excessive production and/or accumulation of proinflammatory mediators such as TNF-α, IL-1β, and IL-6, as a consequence of HIV infection and the aging process, suggests that immune activation likely plays a role in accelerated aging associated with HIV disease. Astrocytes, the abundant cell type within the brain, provide an important reservoir for the generation of inflammatory mediators in response to HIV/HIV proteins [27, 28, 65, 84, 85]. As part of brain parenchyma, astrocytes are continuously exposed to viral (Tat, gp120) & cellular toxins (cytokines & chemokines), secreted from HIV-1-infected macrophages/microglia. Once activated, following insult/injury, astrocytes undergo astrogliosis characterized by proliferation and the release of toxic mediators. Although HIV-1 does not productively infect astrocytes, early viral proteins such as Tat, Rev, and Nef are expressed by the astrocytes in brain tissue of HAND patients [29]. In our earlier studies we have demonstrated that astrocytes exposed to viral and cellular factors can activate several signaling pathways leading to dysregulation of chemokine release by these cells [29, 30]. Herein, we further demonstrated that activation of SIRT1 using either pharmacological or genetic approaches significantly inhibited HIV Tat induced activation of NF-kB and GFAP expression in astrocytes. In parallel, inhibition of either miR-34a or miR-138 blocked Tat induced activation of NF-kB activation in astrocytes. Taken together, our findings suggest that accumulation of HIV Tat protein in the CNS could be a contributing factor leading to premature aging in the Tg rats. Furthermore, since Tat is known to induce astrogliosis our findings also suggest that exposure of astrocytes to Tat could induce expression of miRs-34a & -138, leading in turn, to downregulation of SIRT1 with a concomitant upregulation of GFAP. These data are consistent with previous studies which indicate that both miRs-34a & -138 play important roles in CNS diseases and the immune system [41, 79, 86–90].
In summary, the current findings demonstrate that both miRs-34a & -138 play a crucial role in astrocyte activation by targeting the 3′-UTR of SIRT1, subsequently affecting the aging process in HIV-infected individuals. Therapeutic approaches using inhibitors miRs-34a & -138, that could significantly dampen HIV Tat-induced astrocyte activation, could thus serve as potential targets to abrogate premature aging during HIV infection.
Acknowledgments
This work was supported by the pilot grant (GH) from CHAIN Center grant (P30 MH062261) (HF) and a grant MH-068212 (SB) from the National Institutes of Health and the NRI grant 15-17: 31-3205-0913 (SB & GP) from UNMC. Support by the Nebraska Center for Substance abuse research is also acknowledged.
Footnotes
Conflict of Interest
The authors declare no competing financial interests in relation to the work described.
References
- 1.Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–55. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzinger JJ, et al. Monocytes as regulators of inflammation and HIV-related comorbidities during cART. J Immunol Res. 2014;2014:569819. doi: 10.1155/2014/569819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandkovsky U, et al. Pilot study of younger and older HIV-infected adults using traditional and novel functional assessments. HIV Clin Trials. 2013;14(4):165–74. doi: 10.1310/hct1404-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross AM, et al. Methylome-wide Analysis of Chronic HIV Infection Reveals Five-Year Increase in Biological Age and Epigenetic Targeting of HLA. Mol Cell. 2016;62(2):157–68. doi: 10.1016/j.molcel.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen RA, Seider TR, Navia B. HIV effects on age-associated neurocognitive dysfunction: premature cognitive aging or neurodegenerative disease? Alzheimers Res Ther. 2015;7(1):37. doi: 10.1186/s13195-015-0123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuya-Kanamori L, Kelly MD, McKenzie SJ. Co-morbidity, ageing and predicted mortality in antiretroviral treated Australian men: a quantitative analysis. PLoS One. 2013;8(10):e78403. doi: 10.1371/journal.pone.0078403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caron-Debarle M, et al. HIV-associated lipodystrophy: from fat injury to premature aging. Trends Mol Med. 2010;16(5):218–29. doi: 10.1016/j.molmed.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Berman JW, et al. HIV-tat alters Connexin43 expression and trafficking in human astrocytes: role in NeuroAIDS. J Neuroinflammation. 2016;13(1):54. doi: 10.1186/s12974-016-0510-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh KA, et al. Antioxidant protection from HIV-1 gp120-induced neuroglial toxicity. J Neuroinflammation. 2004;1(1):8. doi: 10.1186/1742-2094-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louboutin JP, Strayer D. Role of Oxidative Stress in HIV-1-Associated Neurocognitive Disorder and Protection by Gene Delivery of Antioxidant Enzymes. Antioxidants (Basel) 2014;3(4):770–97. doi: 10.3390/antiox3040770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields JA, et al. Neuroprotective effects of the immunomodulatory drug FK506 in a model of HIV1-gp120 neurotoxicity. J Neuroinflammation. 2016;13(1):120. doi: 10.1186/s12974-016-0585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutta R, Roy S. Chronic morphine and HIV-1 Tat promote differential central nervous system trafficking of CD3+ and Ly6C+ immune cells in a murine Streptococcus pneumoniae infection model. J Neuroinflammation. 2015;12:120. doi: 10.1186/s12974-015-0341-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Festa L, et al. Induction of Interleukin-1beta by Human Immunodeficiency Virus-1 Viral Proteins Leads to Increased Levels of Neuronal Ferritin Heavy Chain, Synaptic Injury, and Deficits in Flexible Attention. J Neurosci. 2015;35(29):10550–61. doi: 10.1523/JNEUROSCI.4403-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorrell ME, Hauser KF. Ligand-gated purinergic receptors regulate HIV-1 Tat and morphine related neurotoxicity in primary mouse striatal neuron-glia co-cultures. J Neuroimmune Pharmacol. 2014;9(2):233–44. doi: 10.1007/s11481-013-9507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wynne AM, Henry CJ, Godbout JP. Immune and behavioral consequences of microglial reactivity in the aged brain. Integr Comp Biol. 2009;49(3):254–66. doi: 10.1093/icb/icp009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robillard KN, et al. Glial cell morphological and density changes through the lifespan of rhesus macaques. Brain Behav Immun. 2016 doi: 10.1016/j.bbi.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham LC, et al. Chronic consumption of a western diet induces robust glial activation in aging mice and in a mouse model of Alzheimer’s disease. Sci Rep. 2016;6:21568. doi: 10.1038/srep21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry CJ, et al. Minocycline attenuates lipopolysaccharide (LPS)-induced neuroinflammation, sickness behavior, and anhedonia. J Neuroinflammation. 2008;5:15. doi: 10.1186/1742-2094-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid W, et al. An HIV-1 transgenic rat that develops HIV-related pathology and immunologic dysfunction. Proc Natl Acad Sci U S A. 2001;98(16):9271–6. doi: 10.1073/pnas.161290298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar S, et al. Age- and ethanol concentration-dependent effects of acute binge drinking in the HIV-1 transgenic rat. Alcohol Clin Exp Res. 2013;37(Suppl 1):E70–8. doi: 10.1111/j.1530-0277.2012.01860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vigorito M, Connaghan KP, Chang SL. The HIV-1 transgenic rat model of neuroHIV. Brain Behav Immun. 2015;48:336–49. doi: 10.1016/j.bbi.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Royal W, 3rd, et al. Immune activation, viral gene product expression and neurotoxicity in the HIV-1 transgenic rat. J Neuroimmunol. 2012;247(1–2):16–24. doi: 10.1016/j.jneuroim.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao JS, et al. Increased neuroinflammatory and arachidonic acid cascade markers, and reduced synaptic proteins, in brain of HIV-1 transgenic rats. J Neuroinflammation. 2011;8:101. doi: 10.1186/1742-2094-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Villeneuve LM, et al. HIV-1 transgenic rats display mitochondrial abnormalities consistent with abnormal energy generation and distribution. J Neurovirol. 2016 doi: 10.1007/s13365-016-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid WC, et al. Neurobehavioral Abnormalities in the HIV-1 Transgenic Rat Do Not Correspond to Neuronal Hypometabolism on 18F-FDG-PET. PLoS One. 2016;11(3):e0152265. doi: 10.1371/journal.pone.0152265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.June HL, et al. Vitamin A deficiency and behavioral and motor deficits in the human immunodeficiency virus type 1 transgenic rat. J Neurovirol. 2009;15(5–6):380–9. doi: 10.3109/13550280903350200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong Y, Benveniste EN. Immune function of astrocytes. Glia. 2001;36(2):180–90. doi: 10.1002/glia.1107. [DOI] [PubMed] [Google Scholar]
- 28.Minagar A, et al. The role of macrophage/microglia and astrocytes in the pathogenesis of three neurologic disorders: HIV-associated dementia, Alzheimer disease, and multiple sclerosis. J Neurol Sci. 2002;202(1–2):13–23. doi: 10.1016/s0022-510x(02)00207-1. [DOI] [PubMed] [Google Scholar]
- 29.Dou H, et al. Neuropathologic and neuroinflammatory activities of HIV-1-infected human astrocytes in murine brain. Glia. 2006;54(2):81–93. doi: 10.1002/glia.20358. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Effects of human immunodeficiency virus type 1 on astrocyte gene expression and function: potential role in neuropathogenesis. J Neurovirol. 2004;10(Suppl 1):25–32. doi: 10.1080/753312749. [DOI] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Yao H, et al. MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun. 2014;5:4386. doi: 10.1038/ncomms5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310(5756):1954–7. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 34.de Lencastre A, et al. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20(24):2159–68. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grillari J, Grillari-Voglauer R. Novel modulators of senescence, aging, and longevity: Small non-coding RNAs enter the stage. Exp Gerontol. 2010;45(4):302–11. doi: 10.1016/j.exger.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy BK, Smith ED, Kaeberlein M. The enigmatic role of Sir2 in aging. Cell. 2005;123(4):548–50. doi: 10.1016/j.cell.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Yamakuchi M. MicroRNA Regulation of SIRT1. Front Physiol. 2012;3:68. doi: 10.3389/fphys.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhuri AD, Yelamanchili SV, Fox HS. MicroRNA-142 reduces monoamine oxidase A expression and activity in neuronal cells by downregulating SIRT1. PLoS One. 2013;8(11):e79579. doi: 10.1371/journal.pone.0079579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudhuri AD, et al. Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1. FASEB J. 2013;27(9):3720–9. doi: 10.1096/fj.13-232678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe M, Bonini NM. MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol. 2013;23(1):30–6. doi: 10.1016/j.tcb.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, et al. MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 2015;589(6):726–9. doi: 10.1016/j.febslet.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Mohan M, et al. Dysregulated miR-34a-SIRT1-acetyl p65 axis is a potential mediator of immune activation in the colon during chronic simian immunodeficiency virus infection of rhesus macaques. J Immunol. 2015;194(1):291–306. doi: 10.4049/jimmunol.1401447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu CM, et al. MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev. 2013;27(13):1473–83. doi: 10.1101/gad.209619.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dombkowski AA, et al. Cortical Tubers: Windows into Dysregulation of Epilepsy Risk and Synaptic Signaling Genes by MicroRNAs. Cereb Cortex. 2016;26(3):1059–71. doi: 10.1093/cercor/bhu276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilakka-Kanthikeel S, Nair MP. Interaction of drugs of abuse and microRNA with HIV: a brief review. Front Microbiol. 2015;6:967. doi: 10.3389/fmicb.2015.00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCubbrey AL, et al. MicroRNA-34a Negatively Regulates Efferocytosis by Tissue Macrophages in Part via SIRT1. J Immunol. 2016;196(3):1366–75. doi: 10.4049/jimmunol.1401838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivetti di Val Cervo P, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A. 2012;109(4):1133–8. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bethel-Brown C, et al. Platelet-derived growth factor (PDGF)-BB-mediated induction of monocyte chemoattractant protein 1 in human astrocytes: implications for HIV-associated neuroinflammation. J Neuroinflammation. 2012;9:262. doi: 10.1186/1742-2094-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu G, et al. Exosome-mediated shuttling of microRNA-29 regulates HIV Tat and morphine-mediated neuronal dysfunction. Cell Death Dis. 2012;3:e381. doi: 10.1038/cddis.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaudhuri AD, Yelamanchili SV, Fox HS. Combined fluorescent in situ hybridization for detection of microRNAs and immunofluorescent labeling for cell-type markers. Front Cell Neurosci. 2013;7:160. doi: 10.3389/fncel.2013.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu G, et al. Release of luminal exosomes contributes to TLR4-mediated epithelial antimicrobial defense. PLoS Pathog. 2013;9(4):e1003261. doi: 10.1371/journal.ppat.1003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vigorito M, LaShomb AL, Chang SL. Spatial learning and memory in HIV-1 transgenic rats. J Neuroimmune Pharmacol. 2007;2(4):319–28. doi: 10.1007/s11481-007-9078-y. [DOI] [PubMed] [Google Scholar]
- 54.Midde NM, et al. Genetically expressed HIV-1 viral proteins attenuate nicotine-induced behavioral sensitization and alter mesocorticolimbic ERK and CREB signaling in rats. Pharmacol Biochem Behav. 2011;98(4):587–97. doi: 10.1016/j.pbb.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu J, et al. HIV-1 transgenic rats display an increase in [H]dopamine uptake in the prefrontal cortex and striatum. J Neurovirol. 2015 doi: 10.1007/s13365-015-0391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng J, et al. The HIV-1 transgenic rat as a model for HIV-1 infected individuals on HAART. J Neuroimmunol. 2010;218(1–2):94–101. doi: 10.1016/j.jneuroim.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 57.Cassol E, et al. Cerebrospinal fluid metabolomics reveals altered waste clearance and accelerated aging in HIV patients with neurocognitive impairment. AIDS. 2014;28(11):1579–91. doi: 10.1097/QAD.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lynch AM, et al. The impact of glial activation in the aging brain. Aging Dis. 2010;1(3):262–78. [PMC free article] [PubMed] [Google Scholar]
- 59.Pagans S, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3(2):e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Satoh A, et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18(3):416–30. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soo Youn G, et al. HDAC6 mediates HIV-1 tat-induced proinflammatory responses by regulating MAPK-NF-kappaB/AP-1 pathways in astrocytes. Glia. 2015 doi: 10.1002/glia.22865. [DOI] [PubMed] [Google Scholar]
- 63.Darbinian N, et al. Functional interaction between cyclin T1/cdk9 and Puralpha determines the level of TNFalpha promoter activation by Tat in glial cells. J Neuroimmunol. 2001;121(1–2):3–11. doi: 10.1016/s0165-5728(01)00372-1. [DOI] [PubMed] [Google Scholar]
- 64.Fan Y, et al. Activation of Egr-1 expression in astrocytes by HIV-1 Tat: new insights into astrocyte-mediated Tat neurotoxicity. J Neuroimmune Pharmacol. 2011;6(1):121–9. doi: 10.1007/s11481-010-9217-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bethel-Brown C, et al. HIV-1 Tat-mediated induction of platelet-derived growth factor in astrocytes: role of early growth response gene 1. J Immunol. 2011;186(7):4119–29. doi: 10.4049/jimmunol.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simioni S, et al. Cognitive dysfunction in HIV patients despite long-standing suppression of viremia. AIDS. 2010;24(9):1243–50. doi: 10.1097/QAD.0b013e3283354a7b. [DOI] [PubMed] [Google Scholar]
- 67.Cowley D, et al. Genetic and functional heterogeneity of CNS-derived tat alleles from patients with HIV-associated dementia. J Neurovirol. 2011;17(1):70–81. doi: 10.1007/s13365-010-0002-5. [DOI] [PubMed] [Google Scholar]
- 68.Bansal AK, et al. Neurotoxicity of HIV-1 proteins gp120 and Tat in the rat striatum. Brain Res. 2000;879(1–2):42–9. doi: 10.1016/s0006-8993(00)02725-6. [DOI] [PubMed] [Google Scholar]
- 69.Rappaport J, et al. Molecular pathway involved in HIV-1-induced CNS pathology: role of viral regulatory protein, Tat. J Leukoc Biol. 1999;65(4):458–65. doi: 10.1002/jlb.65.4.458. [DOI] [PubMed] [Google Scholar]
- 70.Bruce-Keller AJ, et al. Synaptic transport of human immunodeficiency virus-Tat protein causes neurotoxicity and gliosis in rat brain. J Neurosci. 2003;23(23):8417–22. doi: 10.1523/JNEUROSCI.23-23-08417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stauch KL, et al. Proteomic analysis and functional characterization of mouse brain mitochondria during aging reveal alterations in energy metabolism. Proteomics. 2015;15(9):1574–86. doi: 10.1002/pmic.201400277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stauch KL, et al. Data for mitochondrial proteomic alterations in the aging mouse brain. Data Brief. 2015;4:127–9. doi: 10.1016/j.dib.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25(3):138–45. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9(2):123–8. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Villalba JM, Alcain FJ. Sirtuin activators and inhibitors. Biofactors. 2012;38(5):349–59. doi: 10.1002/biof.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mellini P, Valente S, Mai A. Sirtuin modulators: an updated patent review (2012 - 2014) Expert Opin Ther Pat. 2015;25(1):5–15. doi: 10.1517/13543776.2014.982532. [DOI] [PubMed] [Google Scholar]
- 77.Xie H, et al. Cryptosporidium parvum induces SIRT1 expression in host epithelial cells through downregulating let-7i. Hum Immunol. 2014;75(8):760–5. doi: 10.1016/j.humimm.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harries LW. MicroRNAs as Mediators of the Ageing Process. Genes (Basel) 2014;5(3):656–70. doi: 10.3390/genes5030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu K, et al. MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 2012;13:115. doi: 10.1186/1471-2202-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Bak M, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–44. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Witwer KW, et al. A plasma microRNA signature of acute lentiviral infection: biomarkers of central nervous system disease. AIDS. 2011;25(17):2057–67. doi: 10.1097/QAD.0b013e32834b95bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hearps AC, et al. Aging is associated with chronic innate immune activation and dysregulation of monocyte phenotype and function. Aging Cell. 2012;11(5):867–75. doi: 10.1111/j.1474-9726.2012.00851.x. [DOI] [PubMed] [Google Scholar]
- 83.Nasi M, et al. Ageing and inflammation in patients with HIV infection. Clin Exp Immunol. 2016 doi: 10.1111/cei.12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boska MD, et al. Associations between brain microstructures, metabolites, and cognitive deficits during chronic HIV-1 infection of humanized mice. Mol Neurodegener. 2014;9:58. doi: 10.1186/1750-1326-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li J, et al. Human immunodeficiency virus type 1 efficiently binds to human fetal astrocytes and induces neuroinflammatory responses independent of infection. BMC Neurosci. 2007;8:31. doi: 10.1186/1471-2202-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Christofidou-Solomidou M, et al. Dietary flaxseed modulates the miRNA profile in irradiated and non-irradiated murine lungs: a novel mechanism of tissue radioprotection by flaxseed. Cancer Biol Ther. 2014;15(7):930–7. doi: 10.4161/cbt.28905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kou X, et al. Ampelopsin attenuates brain aging of D-gal-induced rats through miR-34a-mediated SIRT1/mTOR signal pathway. Oncotarget. 2016 doi: 10.18632/oncotarget.12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chan XH, et al. Targeting glioma stem cells by functional inhibition of a prosurvival oncomiR-138 in malignant gliomas. Cell Rep. 2012;2(3):591–602. doi: 10.1016/j.celrep.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 89.Schroder J, et al. MicroRNA-138 is a potential regulator of memory performance in humans. Front Hum Neurosci. 2014;8:501. doi: 10.3389/fnhum.2014.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kisliouk T, Cramer T, Meiri N. Heat stress attenuates new cell generation in the hypothalamus: a role for miR-138. Neuroscience. 2014;277:624–36. doi: 10.1016/j.neuroscience.2014.07.047. [DOI] [PubMed] [Google Scholar]