Abstract
Pexophagy is a selective autophagy process that degrades damaged and/or superfluous peroxisomes in the yeast vacuole or in mammalian lysosomes. The molecular mechanisms of pexophagy are well studied in yeast. Peroxisomes can be rapidly induced by oleate in the budding yeast, Saccharomyces cerevisiae, and by oleate or methanol in the methylotrophic yeast, Pichia pastoris. A number of peroxisomal matrix enzymes, such as 3-ketoacyl CoA thiolase (thiolase) and alcohol oxidase (AOX), are upregulated correspondingly to meet metabolic demands of the cells. Removal of these peroxisome-inducing carbon sources creates conditions wherein peroxisomes are superfluous and results in pexophagy and the degradation of these peroxisomal matrix enzymes. In this chapter, we discuss different assays to monitor pexophagy in yeast. These assays rely on tracking the localization of the BFP–SKL protein (a peroxisomally targeted version of the blue fluorescent protein) by microscopy, biochemical analysis of the degradation of peroxisomal matrix proteins, thiolase and AOX, and/or measuring the reduction of AOX activity during pexophagy.
1. INTRODUCTION
Peroxisomes are single-membrane-bound organelles present in most eukaryotic cells. They are involved in essential cellular metabolism, most notably the β-oxidation of fatty acids, as well as the production and degradation of toxic hydrogen peroxide and other reactive oxygen species. The human diseases caused by peroxisome dysfunction underscore the vital importance of this organelle (Ma, Agrawal, & Subramani, 2011).
The number of intracellular peroxisomes can be rapidly adjusted and tightly controlled in response to the changing environment and/or physiological conditions. This can be achieved through the coordination of peroxisome biogenesis and degradation. In rodents, the abundance of peroxisomes increases when the animals are administered with hypolipidemic drugs that are known peroxisome proliferators (Fahimi et al., 1982) or some other chemicals, such as phthalate esters (Yokota, 1986). The number of peroxisomes decreases rapidly and almost returns to the preproliferation state within a week if the drugs are discontinued (Yokota, 1993). Similar dynamic patterns of peroxisome homeostasis (balance between biogenesis and degradation) are also observed in yeast. Peroxisomes are induced when yeast cells are grown in the presence of the specific carbon sources like oleic acid or methanol, whose metabolism in that yeast requires peroxisomal enzymes and metabolism. However, when the favored peroxisome proliferation stimulus is removed, the superfluous and redundant peroxisomes are no longer required and are subjected instead to pexophagy, the selective degradation of peroxisome through autophagy.
Autophagy is a self-eating, dynamic, cellular process involving the sequestration of cytoplasmic constituents into a double-membrane structure called the autophagosome, followed by autophagosome/lysosome fusion and degradation of the cargos in the lysosome or vacuole (Levine & Klionsky, 2004). It is dramatically induced by various extra- and intracellular stresses, such as nutrient or growth factor depletion or mTOR kinase inhibition. Autophagy was initially described as a nonselective bulk degradation process. However, autophagy can also target selective cargos (Farré & Subramani, 2016). Pexophagy involves the general autophagy machinery, as well as specific elements such as the pexophagy receptor, Atg30 in Pichia pastoris (Farre, Manjithaya, Mathewson, & Subramani, 2008), and Atg36 in Saccharomyces cerevisiae (Motley, Nuttall, & Hettema, 2012). These receptors tag peroxisomes for degradation: Atg30 and Atg36 are associated with the peroxisomal membrane proteins (PMPs) and can recruit the core autophagy machinery to peroxisomes by binding the scaffold proteins (Atg11 and Atg17) and the ubiquitin-like protein Atg8 (Farre, Burkenroad, Burnett, & Subramani, 2013; Farre et al., 2008).
P. pastoris serves as an ideal cell model to study pexophagy due to (1) the large size and/or abundance of peroxisomes that can be induced by both methanol and oleate; (2) this process can be easily visualized by both biochemical and microscopy assays; and (3) convenient genetic manipulation of the cells is possible. In this chapter, we will discuss mainly the methods used to monitor pexophagy in P. pastoris. Similar methods can be also applied to S. cerevisiae.
2. ASSAYS TO MONITOR PEXOPHAGY
2.1 BFP–SKL Fluorescence Assay
2.1.1 Background
In peroxisome proliferation conditions, such as methanol or oleate medium, a large number of peroxisomal proteins are induced and imported into the peroxisomes utilizing one or more peroxisome-targeting signals (PTSs) present in the amino acid sequences of the proteins. The import of most peroxisomal matrix cargos depends on the PTS1 signal composed of a noncleavable, C-terminal tripeptide, Ser–Lys–Leu (SKL), or its conserved variants (Gould, Keller, Hosken, Wilkinson, & Subramani, 1989). Hence, peroxisomes can be labeled by fusing the BFP (blue fluorescent protein) tag to the SKL sequence (BFP–SKL). Alternatively, PMPs, which are targeted to peroxisomes via an mPTS (membrane PTS) sequence, or peroxisomal matrix proteins may also be fused to GFP or BFP to follow peroxisomes. The fluorescent dye FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide] stains the vacuole membrane through endocytosis and can be used as a marker for the vacuole (Vida & Emr, 1995). BFP–SKL and FM4-64 are used together to follow the localization of peroxisomes relative to the vacuole during pexophagy by fluorescence microscopy.
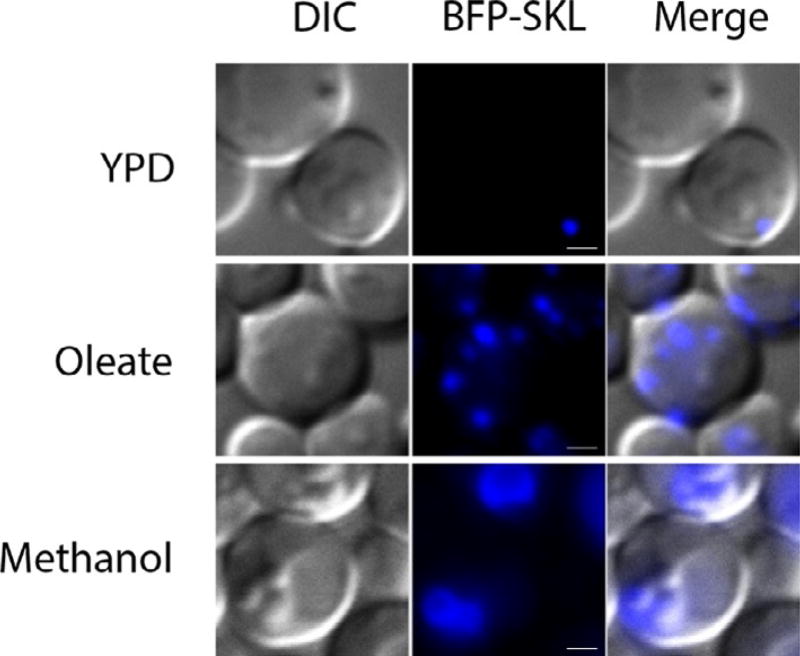
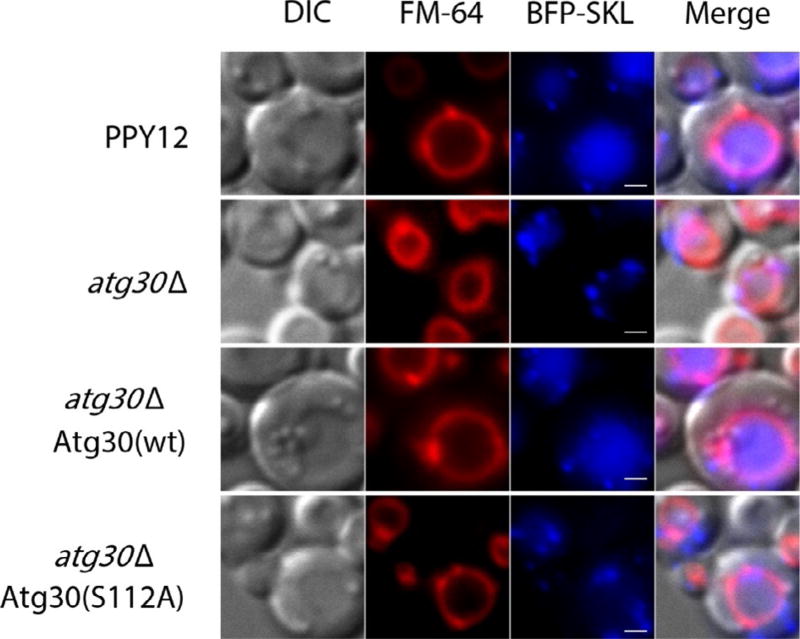
Peroxisomes are induced in oleate or methanol media, as evidenced by the increased number and size of punctuate BFP–SKL-positive dots compared to that in YPD medium (Fig. 1). Shifting cells from oleate to synthetic dextrose without nitrogen (SD-N) medium results in delivery of the BFP–SKL protein into the vacuole (Fig. 2). Atg30 acts as a selective receptor for pexophagy and its activity depends on the phosphorylation sites S71 and S112 (Farre et al., 2013, 2008). The atg30 deletion mutant strain (atg30Δ) is not able to deliver BFP–SKL to the vacuole and serves as a good negative control; reexpression of Atg30 (wt), but not of Atg30 (S112A), rescues the defect of atg30Δ strain (Fig. 2).
Fig. 1.
Induction of peroxisomes in P. pastoris. The PPY12 strain overexpressing BFP–SKL was cultured in YPD medium to mid-log phase and shifted to oleate or methanol medium to induce peroxisomes (15 h). Scale bars, 1 µm.
Fig. 2.
BFP–SKL localization during pexophagy. Strain PPY12 (wt) and the mutants were cultured in oleate medium to induce peroxisome and then shifted to nitrogen starvation medium (SD-N) to induce pexophagy. The pictures are taken after 2 h of starvation. The vacuoles are labeled red with FM4-64 and peroxisomes blue with BFP–SKL. Scale bars, 1 µm.
2.1.2 Methods
2.1.2.1 Growth Medium Components
YPD medium (2% (w/v) glucose, 2% (w/v) Bactopeptone, 1% (w/v) yeast extract).
SD (defined synthetic dextrose medium) (0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 1 × complete supplement mixture (CSM) of amino acids (#1001-010, Sunrise Science), 2% (w/v) glucose).
SD-N (nitrogen starvation medium) (0.17% (w/v) yeast nitrogen base without ammonium sulfate and amino acids, 2% (w/v) glucose).
Ethanol medium: (0.17% (w/v) yeast nitrogen base without ammonium sulfate and amino acids, 0.5% (w/v) ammonium sulfate, 1 × CSM, 0.5% (v/v) ethanol).
Methanol medium (0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 1 × CSM, 0.5% (v/v) methanol, 0.05% (w/v) yeast extract).
Oleate medium for P. pastoris (0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 1 × CSM, 0.2% (v/v) oleate, 0.02% (v/v) Tween 40, 0.05% (w/v) yeast extract).
Oleate medium for S. cerevisiae (0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 1 × CSM, 0.1% (v/v) oleate, 0.5% (v/v) Tween 40, 0.1% (w/v) yeast extract).
2.1.2.2 Strains
| Strains Name |
Description | Genotype | Source |
|---|---|---|---|
| PPY12 | wt P. pastoris strain | his4 arg4 | Gould, McCollum, Spong, Heyman, and Subramani (1992) |
| BFP–SKL | Overexpression of BFP–SKL in PPY12 | PPY12 PrGAPDH-BFP–SKL::ARG4 his4 arg4 | Farre et al. (2008) |
| atg30Δ | atg30 deletion in BFP–SKL-expressing strain | PPY12 atg30Δ::KanMX, PrGAPDH-BFP–SKL::ARG4 his4 arg4 | Farre et al. (2008) |
| atg30Δ+Atg30 (wt) | Reexpress Atg30 (wt) in atg30Δ strain | PPY12 atg30Δ::KanMX, PrATG30-Atg30(wt)-HA::HIS4, PrGAPDH-BFP–SKL::ARG4 his4 arg4 | Farre et al. (2008) |
| atg30Δ+Atg30 (S112A) | Reexpress Atg30 (S112A) mutant in atg30Δ strain | PPY12 atg30Δ::KanMX, PrATG30-Atg30(S112A)-HA::HIS4, PrGAPDH-BFP–SKL::ARG4 his4 arg4 | Farre et al. (2013) |
2.1.2.3 Protocol
-
Inoculate the P. pastoris strains in a 50 mL YPD culture overnight at 30°C.
Note: (i) The flask for the culture should be large enough to provide good aeration. (ii) Use an autophagy- (e.g. atg1Δ) or pexophagy-defective mutant strain (e.g. atg30Δ) as a negative control.
-
Measure the cell density (OD600) and make sure the cells are in log phase.
Note: It is important to keep the cells in log phase (OD600 of 0.4–0.8); autophagy would occur if the cells enter starvation phase due to nutrient depletion in medium. If cells are overgrown next day, dilute them to 0.1–0.2 OD600/mL and culture them till they reach the log phase.
Collect cell pellets and wash them two times with autoclaved H2O by centrifugation at 3000 rpm, 3 min. The purpose is to wash away the carbon source that may be associated with the cells.
-
Resuspend the pellets in the methanol or oleate medium at starting OD600 of 0.1/mL in the presence of FM4-64 (Molecular Probes, 2 µg/mL). Grow cells overnight to 1–2 OD/mL at 30°C with shaking (250 rpm, ~15 h).
Note: The medium would turn purple with addition of FM4-64. Adjust the pH to 6.0–6.8 with dilute KOH if it turns yellow.
-
Harvest 10 OD600 cells and wash twice with autoclaved H2O, pelleting cells after each wash using centrifugation. Resuspend the cell pellets in 10 mL of SD-N medium and incubate in the shaker at 30°C to induce pexophagy.
Note: As is the case with general autophagy, there are two modes of pexophagy, i.e. macro- and micropexophagy which depend on the availability of the carbon source in the medium (Tuttle & Dunn, 1995). Moving cells from methanol to glucose medium or alternatively to ethanol medium induces micropexophagy and macropexophagy, respectively, in P. pastoris.
Take 1 mL of cells at 0, 0.5, 2, 4, and 6 h, respectively. Centrifuge at 3000 rpm for 3 min and wash once with autoclaved H2O.
Discard the supernatant, and resuspend the cell pellets in 100 µL SD-N medium.
Mount the cells by spotting 5 µL of the cells on a dust-free slide and then drop the cover glass and move it horizontal until stop to have a single layer.
-
Observe the cells under a fluorescence microscope, using the Blue GFP (Ex: 402 nm; Em: 457 nm) and Rhodamine filter (Ex: 515 nm; Em: 640 nm) sets to view the BFP–SKL and FM4-64 signal, respectively.
Notes: (i) Two negative controls can be used to avoid the “bleed through” of signals in the channels: BFP–SKL cells with no FM4-64 staining; cells (no BFP–SKL expression) stained with FM4-64. (ii) Pictures taken at different time points reflect the different stages of pexophagy, which can be observed by colocalization of BFP–SKL with FM4-64, as well as of other autophagy markers, such as the isolation membrane resident, Atg8 fused with GFP (GFP-Atg8). (iii) Peroxisomes can be induced by oleate or methanol in P. pastoris as shown earlier. However, peroxisomes can be induced only by oleate in S. cerevisiae. The components of the oleate medium also differ as shown in Section 2.1.2.1. In the case of oleate-induced peroxisomes, methods to monitor pexophagy in P. pastoris (including the BFP–SKL localization and the following thiolase degradation and thiolase-GFP processing assays) can be also applied to S. cerevisiae.
2.2 Thiolase and AOX Degradation Assay
2.2.1 Background
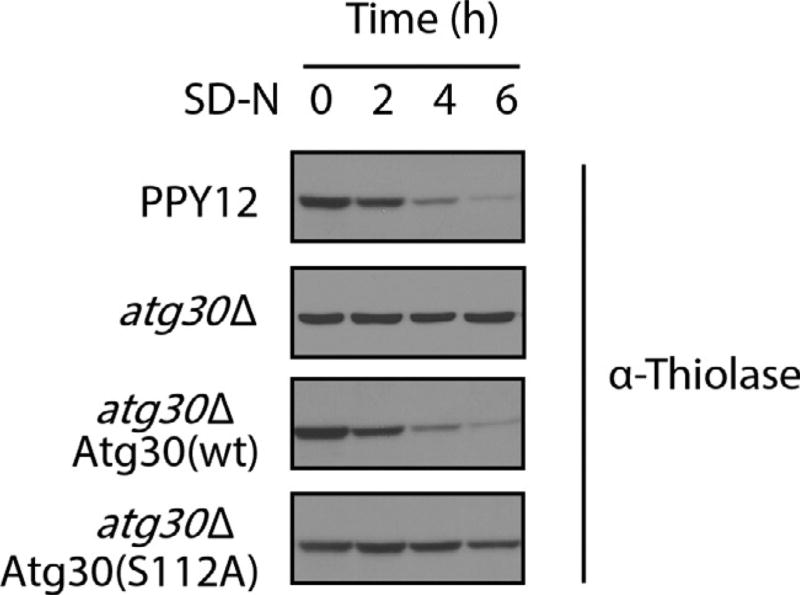
Peroxisomes are induced rapidly by shifting methylotrophic yeast from glucose to methanol or oleate medium. Correspondingly, a number of enzymes are dramatically upregulated to allow cells to metabolize these carbon sources. Examples are the alcohol oxidase (AOX) for methanol utilization and 3-ketoacyl CoA thiolase (thiolase) for β-oxidation of oleate. Replacing these carbon sources by glucose or ethanol results in the induction of pexophagy and degradation of these enzymes. Hence the protein levels of AOX or thiolase can be used to monitor pexophagy. Below is an example of thiolase detection during pexophagy. In the wt strain, thiolase protein is degraded upon moving cells from oleate to SD-N medium. Consistent with the results based on BFP–SKL localization (Fig. 2), thiolase degradation was blocked in atg30Δ cells and rescued by reexpressing Atg30 (wt) but not the mutant, Atg30 (S112A) (Fig. 3).
Fig. 3.
Thiolase degradation assay. The P. pastoris strains were grown in oleate medium to induce peroxisomes and shifted to nitrogen starvation medium (SD-N) for the indicated times. Proteins were extracted by TCA precipitation, resolved by SDS-PAGE, and detected with antithiolase antibody.
2.2.2 Method
2.2.2.1 Growth Media and Strains
The growth media and strains are the same as in Section 2.1.2.
2.2.2.2 Protocol
Induce peroxisomes in methanol or oleate media as in Section 2.1.2.
Harvest 10 OD600 cells from either methanol or oleate medium and wash twice with autoclaved H2O, pelleting cells after each wash by centrifugation.
Resuspend the cell pellets in 10 mL of SD-N starvation medium and incubate in the shaker at 30°C to induce pexophagy.
For detection of AOX, take 1 mL of the methanol-induced culture at 0, 2, 4, and 6 h (starvation time), respectively, to a new tube containing 150 µL 100% trichloroacetic acid (TCA). For detection of thiolase, take 1 mL of oleate-induced cell culture at 0, 2, 4, and 6 h (starvation time), respectively, to a new tube containing 150 µL 100% TCA.
Mix and store at −80°C for at least 1 h before TCA precipitation.
Thaw the frozen TCA cell mixture at room temperature and pellet the protein lysates by centrifugation at 12,000 rpm, 10 min.
-
Aspirate off the supernatants and add 500 µL 80% ice cold acetone. The pellets are fully suspended using a vortex mixer or water bath sonication. Centrifuge contents at 12,000 rpm, 3 min. Discard the supernatants and wash the pellets again. Air-dry the pellets in a hood (~1 h).
Note: Water bath sonication offers better disruption of the pellets. Remove as much of the supernatants as possible before air-drying the pellets. Be careful not to touch or disturb the pellets.
Resuspend the pellets in 100 µL of the following buffer (1% sodium dodecyl sulfate or SDS, 0.1 N NaOH) followed by water bath sonication.
Add 20 µL 6 × SDS loading buffer (10% SDS, 50% Glycerol, 0.5 M DTT; 0.25% bromophenol blue) and boil the samples for 5 min.
-
Centrifuge the samples at 12,000 rpm, 5 min. Load 10 µL of the samples in the 10% SDS-PAGE gel and detect AOX or thiolase proteins by standard Western blot assays.
Note: We use nitrocellulose membranes and wet transfer (100 V, 1 h). The antibodies for AOX and thiolase detection are rabbit polyclonal antiserum made in our lab and incubated with the membrane overnight. An enhanced chemiluminescent (ECL) kit (RPN2106, GE Healthcare) is used to detect the signal.
2.3 Thiolase–GFP Processing Assay
2.3.1 Background
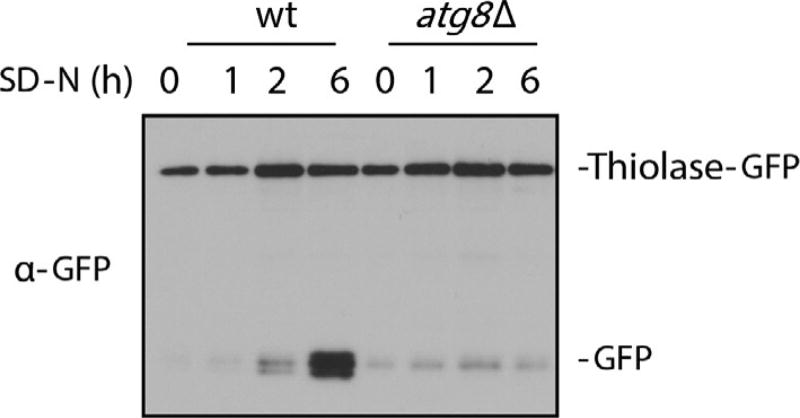
Another way to view thiolase degradation is to detect the cleavage of a thiolase-GFP fusion protein in the vacuole. The GFP tag is fused to the C-terminus of thiolase and its endogenous promoter drives the expression of the fusion protein. During pexophagy, thiolase-GFP protein is delivered to the vacuole, where the GFP moiety is cleaved off by vacuolar hydrolases. GFP is relatively stable in vacuoles compared to the GFP–fusion protein. This allows pexophagy to be monitored through the appearance of free GFP, which would appear and accumulate after shifting the strains from oleate to starvation medium (Fig. 4). The atg8 deletion mutant has a defect in the processing of thiolase-GFP due to blockage of the general autophagy. Hence, the abundance of free GFP reflects the flux of pexophagy and alternatively, the ratio of full-length thiolase-GFP/GFP can give a semiquantitative measure of pexophagy. Our lab used this assay to test the essential phosphorylation sites of Atg30/Atg36 for pexophagy (Farre et al., 2013). In this section, we discuss the methods used to establish stable thiolase-GFP overexpressing strains, as well as detection of processing of thiolase-GFP during pexophagy.
Fig. 4.
Thiolase–GFP processing assay. The P. pastoris strains, PPY12 (wt or atg8Δ overexpressing thiolase–GFP), were grown in oleate medium to induce peroxisomes and shifted to starvation medium (SD-N) for the indicated times. Proteins were extracted by TCA precipitation, resolved by SDS-PAGE, and detected with anti-GFP antibody. The figure was kindly provided by Dr. Jean-Claude Farré.
2.3.2 Methods
2.3.2.1 Strains
| Strains Name |
Description | Genotype | Source |
|---|---|---|---|
| wt | PPY12 strain overexpressing thiolase–GFP | PPY12 PrTHIOLASE-thiolase–GFP::ARG4 his4 arg4 | Farre et al. (2013) |
| atg8Δ | atg8 deletion in thiolase–GFP expressing strain | PPY12 atg8Δ::KanMX, PrTHIOLASE-thiolase–GFP::ARG4 his4 arg4 | Farre et al. (2013) |
2.3.2.2 Preparation of Competent Cells
Inoculate PPY12 in a 50 mL YPD culture in a flask large enough to provide good aeration overnight.
Measure the OD600 of cells (the ideal OD should be in the range of log phase). Harvest 30 OD cells by centrifugation at 3000 rpm, 3 min.
Remove the supernatant, resuspend, and vortex in 5 mL fresh YPD medium.
Add 100 µL Hepes buffer (1 M, pH 8.0) and 125 µL DTT (1 M), vortex the cells.
Incubate the cells with shaking at 30°C for 15 min.
Wash with ice cold sterile H2O (~45 mL) and centrifuge at 3000 rpm, 3 min (repeat the above wash step).
Discard the supernatant, vortex in 5 mL sorbitol (1 M), and centrifuge cells at 3000 rpm, 3 min.
-
Remove the supernatant, resupend the cells in 500 µL sorbitol (1 M), and incubate them on ice for 1 h. The cells are ready for transformation.
Note: For highest efficiency, transform the cells directly without freezing. The cells can be also stored in small aliquots at −80°C freezer.
2.3.2.3 Electroporation and Screening the Positive Thiolase–GFP Colonies
-
Mix 50 µL of competent cells with 2 µg linearized thiolase-GFP plasmid on ice. Incubate electroporation cuvettes (2 mm) on ice for 10 min.
Note: Plasmids need to be linearized in order to integrate into the genome of P. pastoris. The thiolase-GFP construct contains the ARG4 selection marker and is integrated into the ARG4 genomic locus of PPY12. Hence, SD-Arg plates are used as the selection plate as described later.
Pulse cells at (1500 V, 200 Ω, 25 µF) using electroporation instrument (BTX Electro Cell Manipulator 630).
Add 1 mL ice cold sorbitol immediately and place the cuvettes in 30°C incubator overnight.
Transfer cells from the cuvettes to a 1.5 mL tube and centrifuge at 3000 rpm, 3 min.
-
Wash cells with autoclaved H2O and plate one-fourth of the cells on the SD-Arg selection plates. Place the plates upside down in 30°C incubator till visible colonies forms (~2 days).
Note: Components for SD-Arg plate: 0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 1 × CSM without Arg (#1031-010, Sunrise Science), 2% (w/v) glucose, 2% Agar.
Streak the colonies using sterile toothpicks to a new SD-Arg plate and grow them in a 30°C incubator. This provides another round of selection to the colonies and also makes sure that the colony is derived from a single one.
Inoculate a single colony to a 5 mL YPD media overnight and transfer cells to oleate medium to induce peroxisomes as in Section 2.1.2.
Extract proteins by TCA precipitation and detect thiolase-GFP expression by Western blot (anti-GFP antibody). They can be also confirmed by observing the thiolase-GFP signal under the fluorescence microscope.
Make glycerol stocks of the positive colony by generating a 1:1 mix of the cell culture with 50% autoclaved glycerol. Store them at −80°C freezer for future use.
2.3.2.4 Thiolase–GFP Processing Detection
Induce peroxisomes by oleate medium as in Section 2.1.2.
Harvest 10 OD600 cells from the oleate medium and wash cells twice with autoclaved H2O and collecting cells after each wash by centrifugation.
Resuspend the cell pellets in 10 mL of SD-N starvation (1 OD/mL) medium and incubate in the shaker at 30°C to induce pexophagy.
Collect 1 OD600 of cells at time 0, 1, 2, and 6 h, respectively, and extract the proteins by TCA precipitation (see more details in Section 2.2.2.2).
Detect the thiolase–GFP fusion and free GFP proteins by standard Western blot assay using anti-GFP antibody (#11814460001, Roche, dilution: 1:1000). The molecular weight of thiolase and GFP protein are 43 kDa and 26 kDa, respectively. Hence thiolase–GFP should be at 69 kDa.
2.4 AOX Plate Assay
2.4.1 Background
All methylotrophic yeasts possess a common methanol-utilizing pathway. Methanol is first oxidized by AOX in peroxisomes to form formaldehyde and hydrogen peroxide (H2O2), both of which are toxic to cells and are broken down by the enzymes located in the peroxisome (Yurimoto, Oku, & Sakai, 2011). Catalase is a peroxidase that catalyzes and decomposes H2O2 into H2O and oxygen. We can use a similar chemical reaction in vitro to measure the activity of AOX. ABTS (2,2′ -Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) can be used as a substrate of peroxidase: oxidized by peroxidase in the presence of the oxidizing agent, H2O2. The oxidized ABTS turns green, which is a measure of the activity of AOX and indirectly the abundance of peroxisomes in the AOX plate assay. When cells are shifted from methanol to glucose or ethanol, pexophagy occurs and results in the degradation of AOX protein and disappearance of the green color in the strain patches. Our lab used the assay to demonstrate that Atg37 is required for pexophagy as the atg37 mutant still turns green even under pexophagy-inducing conditions (Nazarko et al., 2014).
2.4.2 Methods
2.4.2.1 Growth Plates
YPD plates (2% (w/v) glucose, 2% (w/v) Bactopeptone, and 1% (w/v) yeast extract, 2% agar).
Methanol plates (0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 0.5% (v/v) methanol, supplemented with 1 × CSM, 2% agar).
SD plates (glucose plate) (0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 1 × CSM of amino acids (#1001-010, Sunrise Science), 2% (w/v) glucose, 2% agar).
2.4.2.2 Protocol
Plate the strains on YPD plates as small patches and place them in the 30°C incubator for 2 days.
Replicate the YPD plates onto the methanol medium plates and incubate at 30°C for another 2 days.
Replicate the cells from the methanol plates to the SD plates and incubate them at 30°C (~16 h).
-
Overlay the SD plates with 10 mL of the AOX activity mixture (50 mM Tris–HCl, pH 8.0, 0.5 mg/mL ABTS, 1 mg/mL digitonin, 0.3% low melt agarose, 6 IU/mL peroxidase, 1% methanol).
Note: peroxidase and methanol are added in the last step after other components of the mixture are boiled and cooled to 40°C.
Keep the mixture plates at room temperature for 30 min and incubate them with the medium side up at 30°C till the strains are stained green.
3. CONCLUDING REMARKS
In summary, we have described different ways to monitor pexophagy in yeast. Each of them has its own advantages. Biochemical analysis of thiolase or AOX degradation and thiolase–GFP processing is less likely to be biased and can be easily handled for many samples. It can also be quantitative. Microscopic observation of BFP–SKL localization facilitates tracking pexophagy at different stages (Sakai, Koller, Rangell, Keller, & Subramani, 1998). The AOX plate assay allows high-throughput screening of genes required for pexophagy (Stasyk, Nazarko, & Sibirny, 2008). Additionally, however, it is possible that the gene of interest might affect general autophagy and hence pexophagy. Atg8 lipidation and GFP-Atg8 cleavage assays can be employed to exclude this possibility (Cheong & Klionsky, 2008). The above assays are based on different aspects of pexophagy and we recommend using at least two different assays to monitor pexophagy. Similar assays, except the AOX degradation and plate assays, can also be applied to S. cerevisiae because oleate can also induce peroxisomes in this organism.
Since autophagy is a highly conserved cellular process from lower to higher eukaryotes, we assume that this is also true for pexophagy. In fact, the earliest description of pexophagy came from mammalian cells where peroxisomes were detected in the lysosomes (De Duve & Baudhuin, 1966). Though the molecular mechanisms of pexophagy are well established in yeast, little is known in mammalian cells. This is largely due to the challenging detection of the pexophagy process in higher organisms. The pexophagy studies in yeast would definitely help uncover the mysteries of pexophagy in mammalian cells.
Acknowledgments
We thank Dr. Jean-Claude Farré for careful and critical reading of the manuscript. This work was supported by NIH grant GM 069373 to S.S.
References
- Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods in Enzymology. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- De Duve C, Baudhuin P. Peroxisomes (microbodies and related particles) Physiological Reviews. 1966;46:323–357. doi: 10.1152/physrev.1966.46.2.323. [DOI] [PubMed] [Google Scholar]
- Fahimi HD, Reinicke A, Sujatta M, Yokota S, Ozel M, Hartig F, et al. The short- and long-term effects of bezafibrate in the rat. Annals of the New York Academy of Sciences. 1982;386:111–135. doi: 10.1111/j.1749-6632.1982.tb21410.x. [DOI] [PubMed] [Google Scholar]
- Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Reports. 2013;14:441–449. doi: 10.1038/embor.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Developmental Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré JC, Subramani S. Mechanistic insights into selective autophagy pathways: Lessons from yeast, Nature Reviews. Molecular Cell Biology. 2016;17(9):537–552. doi: 10.1038/nrm.2016.74. http://dx.doi.org/10.1038/nrm.2016.74. Epub 2016 Jul 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. The Journal of Cell Biology. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, McCollum D, Spong AP, Heyman JA, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Developmental Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Ma C, Agrawal G, Subramani S. Peroxisome assembly: Matrix and membrane protein biogenesis. The Journal of Cell Biology. 2011;193:7–16. doi: 10.1083/jcb.201010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. The EMBO Journal. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko TY, Ozeki K, Till A, Ramakrishnan G, Lotfi P, Yan M, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. The Journal of Cell Biology. 2014;204:541–557. doi: 10.1083/jcb.201307050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: Identification of specific steps and morphological intermediates. The Journal of Cell Biology. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasyk OV, Nazarko TY, Sibirny AA. Methods of plate pexophagy monitoring and positive selection for ATG gene cloning in yeasts. Methods in Enzymology. 2008;451:229–239. doi: 10.1016/S0076-6879(08)03216-3. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Dunn WA., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. Journal of Cell Science. 1995;108(Pt. 1):25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. The Journal of Cell Biology. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S. Quantitative immunocytochemical studies on differential induction of serine: Pyruvate aminotransferase in mitochondria and peroxisomes of rat liver cells by administration of glucagon or di-(2-ethylhexyl)phthalate. Histochemistry. 1986;85:145–155. doi: 10.1007/BF00491762. [DOI] [PubMed] [Google Scholar]
- Yokota S. Formation of autophagosomes during degradation of excess peroxisomes induced by administration of dioctyl phthalate. European Journal of Cell Biology. 1993;61:67–80. [PubMed] [Google Scholar]
- Yurimoto H, Oku M, Sakai Y. Yeast methylotrophy: Metabolism, gene regulation and peroxisome homeostasis. International Journal of Microbiology. 2011;2011:101298. doi: 10.1155/2011/101298. [DOI] [PMC free article] [PubMed] [Google Scholar]