Abstract
Increasing antibiotic resistance and beneficial effects of host microbiota has motivated the search for anti-infective agents that attenuate bacterial virulence rather than growth. For example, we discovered that specific flavonoids such as baicalein and quercetin from traditional medicinal plant extracts could attenuate Salmonella enterica serovar Typhimurium type III protein secretion and invasion of host cells. Here, we show epigallocatechin-3-gallate from green tea extracts also inhibits the activity of S. Typhimurium type III protein effectors and significantly reduces bacterial invasion into host cells. These results reveal additional dietary plant metabolites that can attenuate bacterial virulence and infection of host cells.
Graphical Abstract
1. Introduction
The discovery of specific microbial virulence mechanisms has motivated the search for anti-infective agents that attenuate bacterial pathogenesis mechanisms instead of growth.1, 2 For example, the essential function of type III secretion systems (T3SSs) in Gram-negative bacteria virulence has highlighted these protein translocation organelles as prime targets for next-generation anti-infectives.2–9 In the absence of T3SSs, bacterial pathogens such as Salmonella enterica serovar Typhimurium (S. Typhimurium) cannot inject “effector” proteins into host cells to manipulate signaling pathways for invasion and intracellular replication.6, 10 Beyond Salmonella, T3SSs are also crucial for the pathogenesis of other Gram-negative bacterial pathogens including Shigella, Pseudomonas, enteropathogenic Escherichia coli (EPEC), enterohemorrhagic Escherichia coli (EHEC), Chlamydia and Yersinia.4, 5, 11
To identify T3SS inhibitors, we have explored traditional medicinal extracts and discovered that specific flavonoids could attenuate S. Typhimurium invasion of epithelial cells through covalent inactivation of the Salmonella Pathogenicity Island -1 (SPI-1) T3SS substrates and effectors.12 Based on these initial findings, we explored additional plant metabolites from other medicinal and dietary sources with proposed anti-infective activities towards Gram-negative bacterial pathogens. Here we show that epigallocatechin-3-gallate (EGCG), a major metabolite from green tea,17 a previously reported inhibitor of α-synuclein amyloid formation13, 14 and hepatitis C viral entry15, 16, also effectively inhibits S. Typhimurium T3SS and invasion of host cells.
2. Results and discussion
2.1 Analysis of polyphenolic catechins on S. Typhimurium type III protein secretion
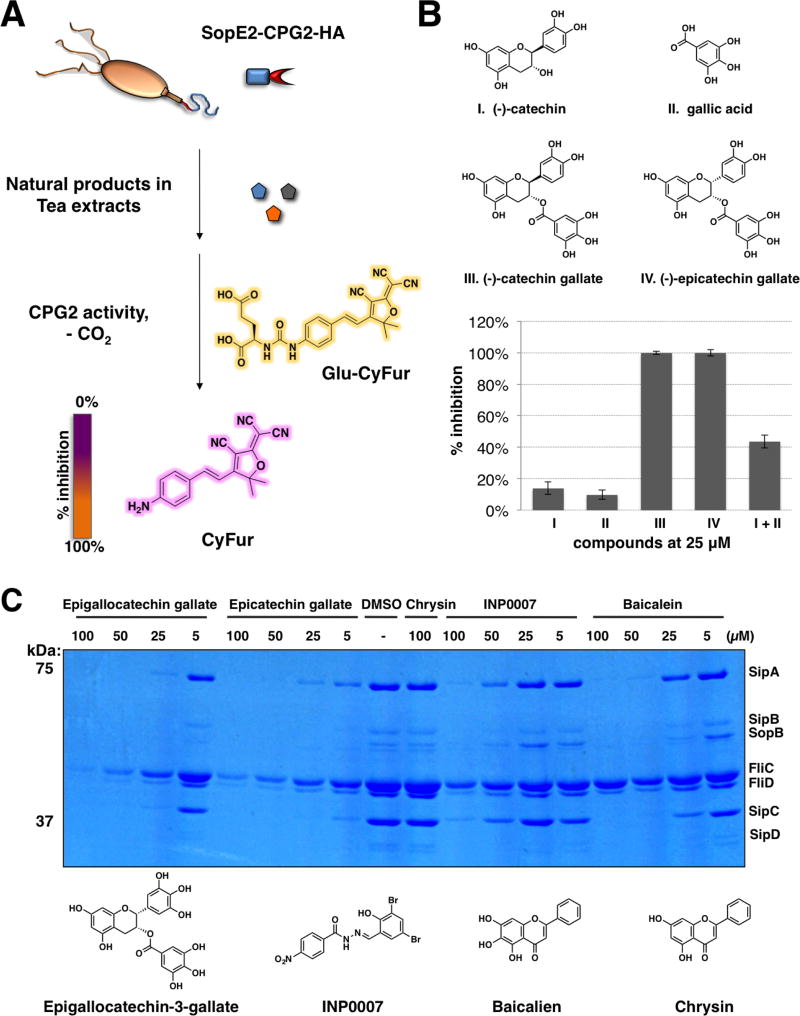
To explore other anti-infective plant metabolites, we used a sensitive two-component enzymatic reporter system, SopE2-CPG2-HA:Glu-CyFur, for monitoring type III protein secretion in S. Typhimurium previously developed in our laboratory.18 This two-component assay takes advantage of the unique enzyme activity of carboxypeptidase G2 (CPG2) that when attached to the C-terminus of a known S. Typhimurium bacterial effector (SopE2) can rapidly and specifically report on type III protein secretion through cleavage of fluorogenic substrates (Glu-CyFur) (Fig 1A).18 Amongst the plant metabolites we explored, polyphenolic catechins such as catechin gallate (CG) and epicatechin gallate (ECG) completely inhibited SopE2-CPG2-HA reporter activity, whereas catechin and gallic acid had less than 20% inhibitory activity at 25 µM (Fig. 1B). In addition, co-incubation of catechin with gallic acid showed marginal improvement in the inhibitory activities when compared to either catechin or gallic acid alone (Fig. 1B). These data suggest the catechin core must be covalently coupled with gallic acid for optimal inhibitory activity against the T3SS-dependent SopE2-CPG2-HA reporter activity.
Figure 1. Tea extracts inhibit SPI-1 T3SS.
(A) Scheme for SopE2-CPG2-HA:Glu-CyFur reporter system. SopE2-CPG2-HA (SPI-1 T3SS) adopts enzymatic activity of carboxypeptidase G2 (CPG2) that when fused to the C-terminus of SopE2, a known S. Typhimurium T3SS bacterial effector, can be secreted and used for monitoring type III protein secretion via cleavage of fluorogenic substrates (Glu-CyFur). (B) Structures of catechin, gallic acid, catechin gallate, and epicatechin gallate. (C) Dose-dependent effect of EGCG, epicatechin gallate, chrysin, INP0007, and baicalein on the levels of SPI-1 T3SS secreted proteins (SipA, SipB, SopB, SipC, and SipD) and flagella components in S. Typhimurium growth media.
2.2 Epigallocatechin gallate inhibits secretion of S. Typhimurium SPI-1 T3SS substrates in vitro
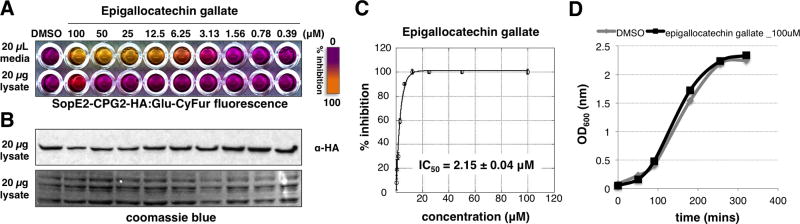
As epigallocatechin gallate (EGCG) is the major polyphenolic components among the tea derived catechins, we compared the activities of EGCG and ECG with previously reported T3SS inhibitors baicalein12 and INP000719 on endogenously secreted SPI-1 T3SS effectors from the growth media of S. Typhimurium (Fig. 1C). With chrysin as the inactive control, all active compounds treated S. Typhimurium growth media exhibited dose-dependent reduction of SPI-1 T3SS substrates, such as SipA, SipB, SopB, SipC and SipD (Fig. 1C). In addition, both EGCG and ECG also exhibited a pronounced effect on the level of FliC and FliD, two proteins associated with bacterial flagella. These SPI-1 T3SS components are key virulence factors for pathogenesis and successful invasion of the host cells.20–25 In general, both EGCG and ECG exerted stronger effects in reducing endogenous SPI-1 T3SS substrate levels compared to INP0007 and baicalein. EGCG effectively attenuated the level of SopE2-CPG2-HA in a dose-dependent manner with an IC50 of 2.15 µM (Fig. 2A-C). Moreover, fluorescence and western blot analysis of the bacterial lysates from EGCG-treated S. Typhimurium indicated that the expression of the SopE2-CPG2-HA was not impaired (Fig. 2A, B). To alleviate the concern of bacterial toxicity by EGCG, we showed that EGCG did not affect S. Typhimurium growth at 100 µM (Fig. 2D). Taken together, these data show that EGCG is a non-bactericidal and potent plant-derived metabolite that inhibits SPI-1 T3SS substrates.
Figure 2. Epigallocatechin gallate (EGCG) inhibits SPI-1 T3SS.
(A) Dose-dependent activity of EGCG on SPI-1 T3SS (SopE2-CPG2-HA) reporter in S. Typhimurium growth media and cell lysate. (B) Western blot analysis of SopE2-CPG2-HA levels in cell lysate. (C) IC50 value of EGCG measured with the SopE2-CPG2-HA reporter fluorescence assay. Mean ± s.d., n = 3. (D) S. Typhimurim growth curve with 100 µM of EGCG.
2.3 Epigallocatechin gallate inhibits SPI-1 T3SS-mediated invasion of host cells
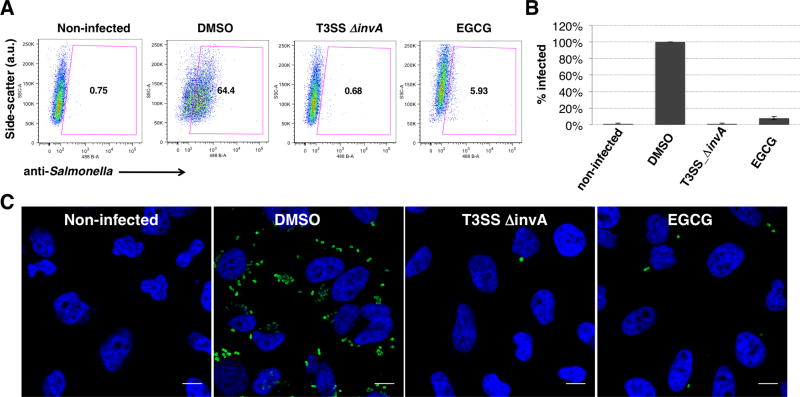
We then investigated the invasion of S. Typhimurium into HeLa cells with the presence of EGCG. Ability of S. Typhimurium to invade epithelial cells is dependent on the SPI-1 effector and translocation proteins, SipA, SipB, SipC, SopE, SopE2, and SopB, which trigger bacterial internalization by stimulating the Rho-family GTPases Rac1 and RhoG.26 We evaluated S. Typhimurium invasion into HeLa cells by flow cytometry and immunofluorescence imaging using anti-Salmonella antibody staining. S. Typhimurium grown in the presence of EGCG showed significant reduction in its ability to invade the cultured HeLa cells (Fig. 3A). The inhibitory effect of EGCG at 100 μM was similar to that of a non-invasive S. Typhimurium strain with an insertion mutation in the invA gene 27–29 (Fig. 3A,B). The inhibition of bacterial invasion could also be readily observed by immunofluorescence analysis of intracellular S. Typhimurium (Fig 3C). Collectively, our results demonstrated that EGCG did not affect bacterial growth but could potently inhibit SPI-1 T3SS-dependent S. Typhimurium invasion of host cells.
Figure 3. EGCG inhibits SPI-1 T3SS-mediated bacterial invasion of epithelial cells.
(A) Flow cytometry analysis of EGCG (100 μM) on S. Typhimurium invasion of HeLa cells judged by anti-S. Typhimurium antibody staining. MOI = 10, 30 minute infection. Experiment was done in triplicate and similar results were seen in two independent runs. (B) Quantification of the invasion studies and values are normalized to those of DMSO treated. (C) Immunofluorescence analysis of intracellular S. Typhimurium in HeLa cells with 100 μM EGCG. Scale bar = 10 μm.
3. Conclusion
The flavonoid EGCG is most abundant and bioactive catechin in green tea17 with diverse pharmacological activities in mammalian cells,13, 14, 30 and viruses,15, 16 but its effects on Gram-negative bacteria T3SS-mediated virulence have not been described. Here, we demonstrate that EGCG targets bacterial virulence with potent inhibitory activity in blocking SPI-1 T3SS-dependent S. Typhimurium invasion of host cells. As EGCG also contains polyphenolic functionality similar to baicalein and quercetin, the uptake and reactivity of these plant metabolites may covalently react with T3SS substrates and inactivate these secreted bacterial protein effectors to attenuate pathogen virulence pathways. These results suggest that specific plant metabolites amongst medicinal extracts and dietary sources (i.e. green tea) may attenuate the virulence mechanisms of microbial pathogens.
4. Experimental
4.1 General
Chemical compounds were purchased from commercial suppliers and were used without further purifications.
4.2 Type III secretion assay
S. Typhimurium strain expressing SopE2-CPG2-HA fusion protein18 and the T3SS-defective invA mutant31 have been previously described. All LB media used were made from BD Difco™ LB (Luria-Bertani) Broth Miller, which contains 10 g/L NaCl. S. Typhimurium containing SopE2-CPG2-HA plasmid was grown overnight at 37 °C in LB containing 50 μg/mL ampicillin and was diluted by 1:30. In each well of the sterile Nunc 96 deep-well plates (2 mL), 400 µL of the diluted culture was then grown for 4 hours in the presence of compounds at indicated concentrations with DMSO or H2O as controls. All added volumes of inhibitors or DMSO were <1% of the total volume in each experiment. After 4 hours, the OD600 was measured to confirm comparable bacterial growth. 250 µL of each sample was transferred into a 96- well plate and bacterial cells were spun down at 5,250 g for 6 minutes. 20 μL of supernatant from each sample was then transferred into a 384- well costar black-bottom plate. 80 μL of CPG2 buffer (50 mM Tris, 0.1 mM ZnCl2, pH 7.4) containing 0.1% Brij-97 to aid in solubility of CyFur18 and 10 μM Glu-CyFur was added, and fluorescence readings (λex= 563 nm, λem= 610 nm with no cutoff) were started immediately and monitored as kinetic readings for 2 hours. Absorbance and fluorescence data were collected on SpectraMax M2 multi-detection reader (Molecular Devices). The percent inhibition by chemical inhibitors was determined by calculating the rates of change in fluorescence of the CPG2-expressing strain in the presence of compound in comparison to the DMSO or H2O control. IC50 values of the inhibitors were then determined using KaleigaGraph version 4.1 and fitting to the exponential decay equation: y = m1 + m2*exp(-m3*x).
4.3 Coomassie staining of secreted proteins
1:30 dilutions of overnight cultures of S. Typhimurium were grown in LB for 4 h in the absence or presence of compounds at indicated concentrations. Secreted proteins from 1 mL culture were precipitated overnight with a final concentration of 10% TCA at 4 °C. Secreted effectors were pelleted at 14,000 rpm for 30 minutes and washed with 250 µL ice-chilled acetone. This procedure was repeated 2 times and the precipitates were allowed to dry for 15 minutes before the addition of Laemmli buffer. Secreted proteins were then separated by 4–20% SDS-PAGE and stained with SimplyBlue™ SafeStain (Invitrogen). Gels were imaged with ChemiDoc™ XRS+ System and Image Lab™ Software (Bio-Rad).
4.4 Cell culture, bacterial infection, flow cytometry and immunofluoresence
HeLa cells were cultured in 12-well plates in DMEM supplemented with 10% FBS (Gemini Bio-Products) at 37 °C in a humidified incubator with an atmosphere of 5% CO2. S. Typhimurium strains IR715, and the invA deletion mutant were grown overnight in LB at 37 °C in a shaker set at 250 rpm. Bacteria were diluted 1/33 in LB containing DMSO or inhibitors, and were grown for additional 4 hours. Optical density readings were used to determine an MOI of 10 and bacteria was added to cells in a total volume of 500 uL DMEM/10% FBS containing inhibitors or DMSO as a control. Plates containing cells and bacteria were then centrifuged at room temperature for 5 min at 1000×g. Plates were then transferred back to the 37 °C incubator for 30 min to allow infection to proceed. Cells were then washed three times with room temperature PBS containing 100 ug/mL gentamycin and were then incubated with DMEM/10% FBS containing 100 ug/mL gentamycin and inhibitors or DMSO as a control at 37 °C for an additional 30 min. Cells were then washed an additional three times with room temperature PBS containing 100 ug/mL gentamycin to remove any remaining extracellular bacteria. Cells were trypsinized and then fixed with ice cold 3.7% paraformaldehyde in PBS for 10 minutes followed by permeabilization with ice cold 0.2% saponin in PBS for 10 min. Cells were then blocked with ice cold 2% FBS in PBS for 10 min. All antibody stainings and washes were performed with ice-cold 0.2% saponin in PBS. Cells were stained for bacterial antigens with anti-Salmonella rabbit serum (Biodesign International, 1/250 dilution) for 1 h and washed three times. Goat anti-rabbit secondary antibody conjugated to AlexaFluor 488 (Invitrogen) was used at a 1/1000 dilution for 30 min. Cells were then washed 3 times. Flow cytometry was performed using a Becton Dickinson LSRII machine and FlowJo software was used for analysis. 30,000 cells were analyzed for each sample. Cellular debris was eliminated from analysis of all samples based on forward and side scatter measurements and never constituted more than 10% of collected events. Gates drawn to illustrate infected cells were based on a value close to 0% for non-infected samples. For microscopy, HeLa cells were grown on glass coverslips. Infections and staining were performed using the same protocol as described above for flow cytometry. For the imaging the extracellular S. Typhimurium, the scrapped-harvest HeLa cells were not permeablized and there is no gentamycin added. Cells were treated with TOPRO-3 (1/1000 dilution, Invitrogen) as a final step to stain nuclei and were mounted using Prolong Gold Antifade Reagent (Invitrogen).
Supplementary Material
Acknowledgments
This work was supported by NIH-NICCH grant R01AT007671 and Lerner Trust to H.C.H.
References
- 1.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430(6996):242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9(2):117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 3.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306(5698):1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galan JE. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature. 2006;441(7093):637–640. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 5.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4(11):811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 6.Kubori T, Matsushima Y, Nakamura D, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280(5363):602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 7.Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clin Microbiol Rev. 2007;20(4):535–549. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3(9):541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 9.Tsou LK, Dossa PD, Hang HC. Small molecules aimed at type III secretion systems to inhibit bacterial virulence. Medchemcomm. 2013;4(1):68–79. doi: 10.1039/C2MD20213A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6(1):53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 11.Izore T, Job V, Dessen A. Biogenesis, regulation, and targeting of the type III secretion system. Structure. 2011;19(5):603–612. doi: 10.1016/j.str.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Tsou LK, Lara-Tejero M, RoseFigura J, et al. Antibacterial Flavonoids from Medicinal Plants Covalently Inactivate Type III Protein Secretion Substrates. J Am Chem Soc. 2016;138(7):2209–2218. doi: 10.1021/jacs.5b11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bieschke J, Russ J, Friedrich RP, et al. EGCG remodels mature alpha-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc Natl Acad Sci U S A. 2010;107(17):7710–7715. doi: 10.1073/pnas.0910723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenzen N, Nielsen SB, Yoshimura Y, et al. How epigallocatechin gallate can inhibit alpha-synuclein oligomer toxicity in vitro. J Biol Chem. 2014;289(31):21299–21310. doi: 10.1074/jbc.M114.554667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciesek S, von Hahn T, Colpitts CC, et al. The green tea polyphenol, epigallocatechin-3-gallate, inhibits hepatitis C virus entry. Hepatology. 2011;54(6):1947–1955. doi: 10.1002/hep.24610. [DOI] [PubMed] [Google Scholar]
- 16.Calland N, Albecka A, Belouzard S, et al. (-)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology. 2012;55(3):720–729. doi: 10.1002/hep.24803. [DOI] [PubMed] [Google Scholar]
- 17.Yang CS, Wang ZY. Tea and cancer. J Natl Cancer Inst. 1993;85(13):1038–1049. doi: 10.1093/jnci/85.13.1038. [DOI] [PubMed] [Google Scholar]
- 18.Yount JS, Tsou LK, Dossa PD, Kullas AL, van der Velden AW, Hang HC. Visible fluorescence detection of type III protein secretion from bacterial pathogens. J Am Chem Soc. 2010;132(24):8244–8245. doi: 10.1021/ja102257v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. Small-molecule inhibitors specifically targeting type III secretion. Infect Immun. 2005;73(5):3104–3114. doi: 10.1128/IAI.73.5.3104-3114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collazo CM, Galan JE. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24(4):747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 21.Lilic M, Galkin VE, Orlova A, VanLoock MS, Egelman EH, Stebbins CE. Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms. Science. 2003;301(5641):1918–1921. doi: 10.1126/science.1088433. [DOI] [PubMed] [Google Scholar]
- 22.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77(7):2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galan JE. Interactions of Salmonella with host cells: encounters of the closest kind. Proc Natl Acad Sci U S A. 1998;95(24):14006–14008. doi: 10.1073/pnas.95.24.14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaniga K, Trollinger D, Galan JE. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177(24):7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou D, Mooseker MS, Galan JE. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc Natl Acad Sci U S A. 1999;96(18):10176–10181. doi: 10.1073/pnas.96.18.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93(5):815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 27.Galan JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A. 1989;86(16):6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galan JE, Curtiss R., 3rd Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun. 1990;58(6):1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galan JE, Curtiss R., 3rd Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59(9):2901–2908. doi: 10.1128/iai.59.9.2901-2908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambert JD. Does tea prevent cancer? Evidence from laboratory and human intervention studies. Am J Clin Nutr. 2013;98(6 Suppl):1667S–1675S. doi: 10.3945/ajcn.113.059352. [DOI] [PubMed] [Google Scholar]
- 31.Galan JE, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174(13):4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.