This is a response to a letter by Richarme (1).
We thank Dr. Richarme for the opportunity to clarify the points raised in his letter. We address the points relating to our data in the order in which they are raised.
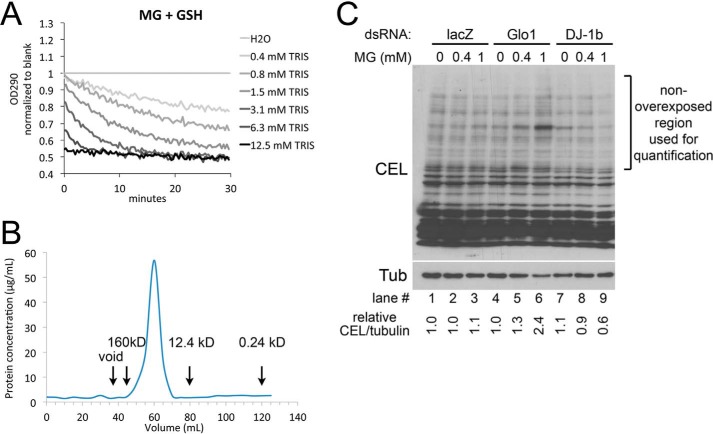
In our paper (2), we find that Tris buffer alone is able to reproduce the drop in A290 described in Ref. 3, whereas neither Drosophila DJ-1β nor human DJ-1, when dialyzed into PBS, causes this drop in A290. Dr. Richarme points out that we did not specify the final concentration of Tris used in our assays. We set up our assay to be similar to the conditions described in Richarme et al. (3). According to the “Experimental Procedures” of this paper, “DJ-1 was purified as described previously (28),” and according to the “Experimental Procedures” of Ref. 28 (shown below as Ref. 4), the DJ-1 homolog, YajL, was “dialyzed for 2 h against 30 mm Tris, pH 8.” Hence, in our initial assays, we dialyzed our DJ-1 proteins into 20 mm Tris, 150 mm NaCl, yielding a final concentration of 1 mm Tris in our deglycation assay. Thus, our final deglycation reaction assays are: 50 mm sodium phosphate buffer, pH 7.0, 2 mm methylglyoxal, 2 mm reduced glutathione or N-acetylcysteine, and either 4 μm DJ-1 or 1.0 mm Tris, pH 8.0. We provide here in Fig. 1A a titration curve of Tris in the deglycation assay, which shows that even 0.4 mm Tris can cause this drop.
Figure 1.
Additional data supporting our original publication. A, effect of various Tris concentrations on the deglycation assayed as in Ref. 3, measured as the drop in absorption at 290 nm, which detects the hemithioacetal formed by MG reacting with glutathione. B, gel filtration of the HIS-hDJ-1 protein sample used in our paper (2), using a HiPrep 16/60 Sephacryl S-200 HR column, reveals that the protein is not aggregated. Eluates were collected from the column, and the protein concentration in each eluate was measured using the micro BCATM Protein Assay Kit from Thermo Fisher. Aggregated protein would elute with the void volume under 40 ml. Rough estimates for elution volumes of three reference proteins are indicated. C, addition of 1 mm methylglyoxal to S2 cells results in increased levels of the MG adduct CEL only in Glo1 knockdown cells but not in DJ-1β knockdown cells. S2 cells were treated with indicated MG concentrations for 24 h prior to lysis and immunoblotting.
Dr. Richarme claims that in our study “DJ-1 samples displayed massive protein aggregation and potential protein inactivation.” We expressed our HIS-tagged proteins in Escherichia coli and provided in Fig. 3A in Ref. 2 a Coomassie-stained SDS-PAGE gel showing the various steps of the purification procedure. As is usual in such a setup, not all the recombinant protein expressed in the bacteria is soluble. After lysing the bacteria, insoluble proteins as well as non-lysed bacteria are pelleted by centrifugation, yielding the sample we termed “insoluble,” whereas, following standard procedure, the HIS-tagged protein is then purified from the soluble fraction by affinity chromatography. We assume Dr. Richarme's comment relates to the fact that there is recombinant HIS-DJ-1 in the insoluble bacterial pellet; however, this is not the fraction used for subsequent purification steps. Indeed, our purified DJ-1 is soluble, and we have no indication that it is aggregated. We have now analyzed our sample by gel filtration on a HiPrep 16/60 Sephacryl S-200 HR column and quantified protein concentration from the various fractions using the micro BCATM Protein Assay Kit (Thermo Fisher) (Fig. 1B). This shows there is almost no aggregated protein in our prep, as this would elute at the void volume of the column. That said, we cannot exclude that for some other reason, unbeknownst to us, both our dDJ-1β and hDJ-1 proteins are inactive.
Dr. Richarme points out that the lactate produced in the assays reported in his paper (3) implies a deglycase activity that cannot result from Tris (which can only shift the equilibrium of the hemithioacetal toward methylglyoxal formation). We are fully aware of this. In our deglycase assays, however, with dDJ-1β or hDJ-1 dialyzed in PBS (Figs. 3 and 4 in Ref. 2), we do not see any drop in levels of the hemithioacetal, detected by A290; hence, no lactate can possibly be produced. In our hands, DJ-1 has very little or no deglycase activity.
Dr. Richarme points out in Fig. 1E of our publication (2) that “DJ-1- and glyoxalase-deficient cells displayed similar increases in protein glycation levels.” The main point of this figure was to show that in the absence of Glo1, but not in the absence of DJ-1, the ability of cells to defend against a methylglyoxal (MG) challenge is impaired. This can be seen by the increased CEL signal in the Glo1 knockdown and Glo1/DJ-1 double-knockdown cells upon treatment with 1 mm MG, but not in the DJ-1 knockdown cells treated with 1 mm MG. We repeated this experiment several times, and this result was consistent and robust and is visually obvious. We assume Dr. Richarme is referring to the non-MG treated lanes. The signal in these lanes is variable. In the immunoblot we selected for the figure, indeed the signal in the DJ-1β knockdown + 0 mm MG lane is higher than the lacZ + 0 mm MG control lane. However, this is due to variability. For instance, lane 10, which is DJ-1β/Glo1 double-knockdown cells + 0 mm MG, should have at least as much signal as the DJ-1β-only knockdown cells (lane 7), but this is not the case, illustrating the fact that the baseline signal has some variability. We include as Fig. 1C another experiment of this kind, along with a quantification of the CEL signal normalized to tubulin. In this figure, one can see once again that cells with a Glo1 knockdown are impaired in their ability to resist the 1 mm MG challenge (lane 6), whereas the DJ-1β knockdown cells are not (lane 9). In this replicate, the CEL/tubulin quantification in the DJ-1β knockdown lanes, 7–9, is 1.1, 0.9, and 0.6 times the control lane (lacZ + 0 mm MG, lane 1), respectively (i.e. it is not elevated compared with the controls). In sum, from this figure and the figure in the original paper, we can only conclude that the absence of DJ-1β does not impair the ability of the cells to withstand an MG challenge. This is also corroborated clearly by the viability curves in Fig. 1B in our original paper, which show that knockdown of Glo1 makes cells more sensitive to MG whereas knockdown of DJ-1 does not.
Regarding the remaining claims in Dr. Richarme's letter, these refer to his own work and not to ours, so we cannot comment on them. Clearly there are some discrepancies between Dr. Richarme's findings and ours. Future work, also by others, will be important to shed more light on these issues.
Footnotes
The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1. Richarme G. (2017) Evidence against a role of DJ-1 in methylglyoxal detoxification. J. Biol. Chem. 292, 12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pfaff D. H., Fleming T., Nawroth P., and Teleman A. A. (2017) Evidence against a role for the Parkinsonism-associated protein DJ-1 in methylglyoxal detoxification. J. Biol. Chem. 292, 685–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richarme G., Mihoub M., Dairou J., Bui L. C., Leger T., and Lamouri A. (2015) Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J. Biol. Chem. 290, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kthiri F., Le H. T., Gautier V., Caldas T., Malki A., Landoulsi A., Bohn C., Bouloc P., and Richarme G. (2010) Protein aggregation in a mutant deficient in YajL, the bacterial homolog of the Parkinsonism-associated protein DJ-1. J. Biol. Chem. 285, 10328–10336 [DOI] [PMC free article] [PubMed] [Google Scholar]