Abstract
Uncovering the mechanisms by which single-stranded binding proteins both protect and expose single-stranded DNA has important implications for our understanding of DNA replication and repair. A new study serves up a master class in developing a full kinetic model for one such protein, mtSSB, showing how DNA can be reeled in and set free to control accessibility.
Introduction
Separation of the double helix to form single-stranded DNA (ssDNA) is needed for DNA replication, recombination, and repair. However, exposure of single-stranded DNA can also lead to damage or breaks from a myriad of chemical and enzymatic sources. The critical importance of protecting ssDNA from damage has led to the evolution of proteins conserved across species that bind and sequester ssDNA (1). In this regard, single-stranded binding proteins (SSBs)3 are preeminent in the protection and utilization of nucleic acids. To borrow a phrase from the fishing world, SSBs facilitate a “catch and release” program: catching ssDNA to stabilize it and then releasing it to appropriate proteins involved in DNA metabolism. How can SSB proteins protect ssDNA while simultaneously providing access to it? In this issue of JBC, Qian and Johnson (2) take an important step toward answering this question by providing a quantitative view of the binding of human mitochondrial SSB to ssDNA.
The work from Qian and Johnson builds on the healthy literature surrounding the Escherichia coli SSB (EcoSSB). The EcoSSB homotetramer exhibits two major modes of binding to ssDNA, termed SSB35 and SSB65 (3). For EcoSSB35, a single tetramer occludes up to 35 nucleotides (nt) through binding of two of the four protein subunits. The EcoSSB65 tetramer occludes 65 nt by wrapping the ssDNA around all four subunits. The binding modes are dependent on the salt concentration, with lower salt favoring EcoSSB35. SSBs bind strongly to ssDNA; however, the interaction is highly dynamic as illustrated by the ability of EcoSSB to diffuse on ssDNA (4). An important structural feature of EcoSSB is the C-terminal, 9-amino acid sequence that interacts with many different proteins, serving as an assembly point for protein complexes that function in DNA metabolism (5).
The human mitochondrial SSB (mtSSB) has significant sequence homology to EcoSSB. Evolutionarily, the presence of mitochondria in eukaryotes is thought to have arisen through endosymbiosis, whereby mitochondria are derived from an early endocytic event. The circular genome and homology of several proteins, including mtSSB, provides some of the evidence in support of this theory. However, characterization of the mitochondrial DNA replication proteins has lagged behind prokaryotic and eukaryotic replication systems. The work of Qian and Johnson is noteworthy because mtDNA replication mechanisms are actively debated (e.g. asynchronous unidirectional versus strand-coupled and/or RITOLS (RNA incorporation throughout the lagging strand) mechanisms, organism-specific differences, identity of the mitochondrial primase, mechanisms of damage repair, and bypass) and mtSSB participates in most if not all aspects of mitochondrial DNA metabolism (6). Thus, determining whether mtSSB behaves like its E. coli counterpart or substantially diverges from it could similarly enable inferences as to whether mtDNA mechanisms more generally follow prokaryotic or eukaryotic strategies.
In the current work, Qian and Johnson (2) combine a variety of quantitative biochemical approaches, including DNA footprinting, fluorescence anisotropy, ITC, and stopped-flow experiments to measure thermodynamic and kinetic properties of mtSSB. With each experiment, data were initially analyzed by fitting to appropriate equations to determine observed rate constants, which allowed the authors to evaluate specific steps such as whether DNA binding and wrapping occurred as kinetically separable events and whether DNA exchange required full release of the initially bound DNA or just fraying of one end. However, some of the data were not well-described by traditional equation-based data–fitting. Directly fitting the data to appropriate kinetic schemes, along with variation of specific kinetic parameters where appropriate, led to improved descriptions of the data. It was the use of this “scheme-based” data analysis with real-time statistical evaluation that allowed the authors to produce a unifying description of the whole system (Fig. 1). The Johnson lab has been at the forefront of making quantitative kinetic/thermodynamic data more accessible to biochemists and molecular biologists (7), and the work presented in this issue of the JBC nicely illustrates the power of their data-fitting tools.
Figure 1.
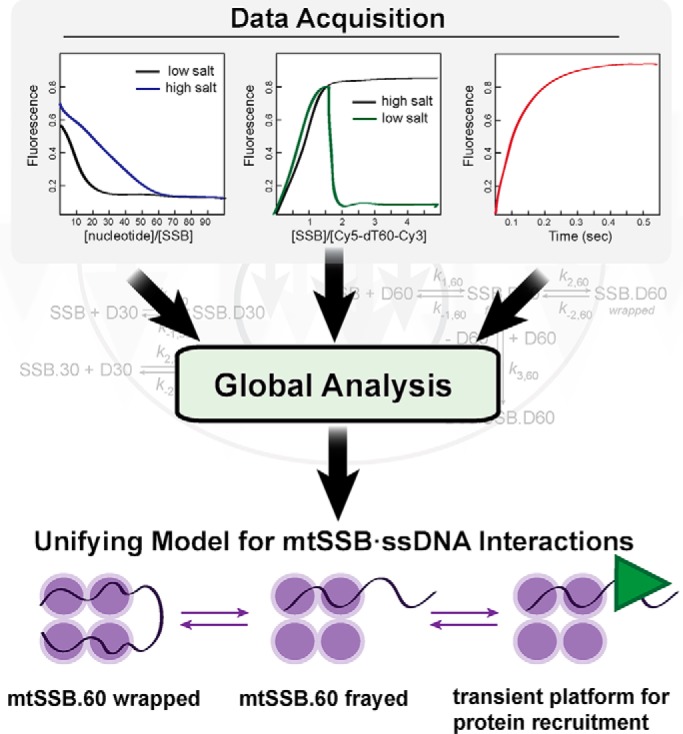
Global fitting of equilibrium and kinetic data establishes a “catch and release” mechanism of binding of ssDNA by human mtSSB.
The authors' model revealed many similarities and key differences between mtSSB and EcoSSB. For example, the mtSSB exhibits two modes of binding, termed mtSSB30 and mtSSB60 (Fig. 1), similarly to EcoSSB. Interestingly, mtSSB does not exhibit the negative cooperativity in the mtSSB30 binding mode that is observed with EcoSSB35 (8), perhaps explained by the lack of an acidic C-terminal tail on mtSSB, which mediates cooperative binding of EcoSSB to protein partners.
A key feature of both SSBs is the ability to exchange one strand of DNA for another. Strand exchange by mtSSB was attributed, in part, to the fact that wrapping of ssDNA around the tetramer is only moderately favorable. Thus, partial unwrapping of the ssDNA exposes one of the available protein-binding sites, which can re-wrap or bind to a different strand of ssDNA (Fig. 1). The exchange mechanism allows for transient exposure of ssDNA without complete dissociation of mtSSB, which may enable functions, such as diffusion along ssDNA and recruitment of other proteins.
Work from the Lohman lab (9) has shown that helicases can “push” SSB proteins along ssDNA through a mechanism whereby the translocation activity of the helicase strips ssDNA from the SSB. The free SSB-binding site binds to ssDNA downstream, resulting in net movement of the protein. The quantitative model put forth by Qian and Johnson indicates that unwrapping of ssDNA from the SSB can occur frequently based on the conclusion that ∼10% of the bound ssDNA can exist in the unwrapped state. The model further provides an opportunity for ssDNA to appear transiently, allowing other proteins in proximity to bind to the exposed ssDNA. Therefore, SSB proteins are able to catch ssDNA for protection and then release it so that DNA metabolic events can occur in a regulated manner. Release can occur spontaneously or due to the action of a motor protein such as a helicase.
The study from Qian and Johnson (2) is the first comprehensive, quantitative binding study of human mtSSB. Mitochondrial DNA replication requires at minimum a polymerase, helicase, and the SSB (6). Therefore, a detailed understanding of the binding modes for mtSSB is necessary to effectively study mtDNA replication in vitro, which is needed to understand the in vivo mechanism(s) of mtDNA replication. The stage is now set for reconstitution of other mitochondrial proteins with mtSSB to uncover the mechanisms of mtDNA metabolism.
This work was supported by National Institutes of Health Grants R01 CA183895 (to R. L. E.) and R35 GM122601 (to K. D. R). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SSB
- single-stranded DNA-binding protein
- EcoSSB
- E. coli SSB
- mtSSB
- mitochondrial SSB
- ITC
- isothermal titration calorimetry
- nt
- nucleotide(s).
References
- 1. Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., and Keck J. L. (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qian Y., and Johnson K. A. (2017) The human mitochondrial single-stranded DNA-binding protein displays distinct kinetics and thermodynamics of DNA binding and exchange. J. Biol. Chem. 292, 13068–13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lohman T. M., and Overman L. B. (1985) Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 260, 3594–3603 [PubMed] [Google Scholar]
- 4. Roy R., Kozlov A. G., Lohman T. M., and Ha T. (2009) SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461, 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antony E., Weiland E., Yuan Q., Manhart C. M., Nguyen B., Kozlov A. G., McHenry C. S., and Lohman T. M. (2013) Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 425, 4802–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ciesielski G. L., Oliveira M. T., and Kaguni L. S. (2016) Animal mitochondrial DNA replication. Enzymes 39, 255–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson K. A. (2013) A century of enzyme kinetic analysis, 1913 to 2013. FEBS Lett. 587, 2753–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lohman T. M., and Bujalowski W. (1988) Negative cooperativity within individual tetramers of Escherichia coli single strand binding protein is responsible for the transition between the (SSB)35 and (SSB)56 DNA binding modes. Biochemistry 27, 2260–2265 [DOI] [PubMed] [Google Scholar]
- 9. Sokoloski J. E., Kozlov A. G., Galletto R., and Lohman T. M. (2016) Chemo-mechanical pushing of proteins along single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 113, 6194–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]


