Abstract
Multiple myeloma (MM) is a plasma cell malignancy characterized by molecular and clinical heterogeneity. The outcome of the disease has been dramatically improved with the advent of new drugs in the past few years. In this context of increasing therapeutic options, important challenges are to accurately evaluate patients’ prognosis and to predict sensitivity to specific treatments and drug combinations. Transcriptomic studies have largely contributed to decipher MM complexity, characterizing MM sub-groups featured by different outcomes. Micro-arrays and more recently RNA sequencing allows evaluation of expression of coding and non-coding genes, alternate splicing events and mutations as well as novel transcriptome modifiers providing new information regarding myeloma biology, prognostication and therapy. In this review, we discuss the role and impact of gene expression profiling studies in myeloma.
Introduction
Multiple myeloma (MM) results from a multi-step transformation of normal to malignant plasma cells (1). The distinct stages of the disease include monoclonal gammopathy of undetermined significance (MGUS), smoldering MM (SMM), symptomatic MM and extramedullary disease or plasma cell leukemia. MM is clinically heterogeneous with a spectrum of symptoms that can comprise bone lesions, extramedullary locations, renal failure, hypercalcemia, cytopenia, and short or long survival (2,3). This heterogeneity is supported by distinct molecular and cytogenetic profiles. Based on karyotype, MM is divided into 2 groups: Hyperdiploid MM (HDMM) and non-hyperdiploid MM (NHDMM) which is associated with recurrent translocations (40% of MM) involving the immunoglobulin heavy chain (IGH) locus and different partners (mainly CCND1, cMAF, and MMSET) (1,4). Transcriptomic studies have provided important information regarding pathways and genes involved in myelomagenesis, distinguishing 8 to 10 subgroups of MM patients with different clinical and biological patterns (4–9). Initially array-based and more recently RNA-sequencing-based, gene expression profile (GEP) studies constitute a reliable prognostic tool that has been independently validated by different MM cooperative groups in different clinical trials. However, in daily clinical practice, no consensus has evolved to integrate GEP in MM care. In this review, we will discuss how GEP can be integrated in the clinical practice by identifying high and low risk patients at each step of the disease, predicting treatment sensitivity, identifying targetable pathways and specific mutations to guide personalized medicine.
GEP and Molecular Heterogeneity in Myeloma
The MM transcriptome has been evaluated using various different cohorts of patients (6–9). Differentially expressed genes have been identified between MM and normal plasma cells and also between different phases and stages of the disease. Deregulation of the Cyclin D family (CCND1, CCND2 and CCND3) appeared to be a universal characteristic of MM cells affecting the vast majority of patients from the early MGUS stage (5). The mechanisms involved in Cyclin D deregulation are multiple and include the translocation of CCND1 or CCND3 with the IgH gene in the t(11;14) and the t(6;14), specific Cyclin D amplifications, and trisomies and other cytogenetics events that indirectly contribute to over-expression of CCND genes. In particular, CCND2 is overexpressed in t(4;14) and t(14;16) MM (6,7). These observations allowed classification of MM in 8 subgroups in the translocation cyclin D (TC) classification (5). Further studies have reported other molecular subgroups independent of Cyclin D deregulation and associated with other clinical and phenotypical features. For exemple, a Low-Bone subgroup, that includes MM patients with lower or no bone lesions and underexpresses Dickkopf WNT Signaling Pathway Inhibitor 1 (DKK1) or the proliferative subgroup which harbors overexpression of numerous cell cycle– and proliferation-related genes with a significantly higher gene expression–defined proliferation index group have been identified (8). Overall, GEP highlights an important molecular heterogeneity in MM (Table 1). More than 500 genes have a significant variation between the different subgroups (5). Cytogenetic alterations, mainly hyperdiploidy and translocations involving IgH are highly correlated with molecular subgroups clusters. For example, t(4;14) which leads to the over-expression of the histone methyl transferase Multiple Myeloma SET Domain (MMSET), is associated with a specific gene profile in part due to MMSET activity (10). More globally, HDMM and NHDMM can be identified based on GEP (11).
Table 1. Molecular classification of multiple myeloma.
Gene expression profiling of MM samples allowed identifying 11 sub-groups have been identified correlated with clinical outcome. CD1 and CD2 are clustering together and related to IgH translocation with CCND1 or CCND3 (t(11;14) and t(6;14)). LB corresponds to low-bone disease and is associated with low DKK1 expression. PR and CTA are clustering together and featured by high proliferation index but CTA sub-groups is featured by better outcome and higher Cancer Testis Antigen. MS relates to IgH-MMSET translocation t(4;14) and MF to IgH-MAF translocation t(14;16). HY subgroup is related to hyperdiploid samples whereas NFKB is related to a subgroup of patients with high activation of this pathway. The PRL3 subgroup is characterized by the over-expression of the tyrosine phosphatase PRL3. Finally, the myeloid subgroup is controversial and it is associated with low infiltration by plasma cell in the bone marrow suggesting contamination of the samples (7, 8).
| Subgroup | CD1 | CD2 | MF | MS | LB | PR | HY | CTA | NFKB | PRL3 | Myeloid |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetic | t(11;14) |
t(11;14) t(6;14) |
t(14;16) t(14;20) |
t(4;14) 1q gain |
1q gain | HD | 1q gain | HD | HD | ||
| CCND expression | CCND1 | CCND1 CCND3 |
CCND2 | CCND2 | CCND1 CCND2 |
CCND2 CCND1 |
CCND1 | CCND1 CCND2 |
CCND1 CCND2 |
CCND2 |
CCND1 CCND2 |
| High-expressed genes | INHBE ETV1 MACROD2 |
cd79a cd20 |
IL6R c-MAF MAFB |
MMSET FGFR3 PBX1 |
EDN1 IL6R SMAD1 |
CCNB1 MCM2 CDC2 BIRC5 CCNB2 AURKA |
TRAIL DKK1 CCR5 |
Cancer Testis Antigen AURKA |
CD40 BCL10 IL8 |
SOX3 PTP4A3 PTPRZ1 |
CD163 CA1 LIZ |
|
Low-expressed genes |
CD9 NOTCH2NL |
CCND2 | DKK1 CCND1 |
CCND1 DUSP2 SYK PAX5 |
DKK1 STAT1 STAT2 |
CXCR4 CD27 |
CCND2 CD52 TAGLN2 CKS1B OPN3 |
MALAT1 | TRAF3 CCR2 MAT2A |
CD44 DUSP6 |
PRMT1 DUSP5 SMAD7 |
|
Clinical feature/ prognosis |
Low risk | Low risk | High risk | High risk | Low risk Low rate of bone lesions |
High risk | Low risk | High risk | Low risk | Low risk | Low risk Low rate of plasma cell in the bone marrow |
| Frequency | 4–9% | 11–17% | 6–10% | 15–17% | 12–17% | 11% | 26–32% | 7% | 11% | 2–3% | 12% |
GEP and Myeloma Pathogenesis
In order to investigate the molecular basis of myelomagenesis, several studies have reported GEP at the different stages of the disease, comparing normal plasma cells, with plasma cells from MGUS, smoldering MM, newly-diagnosed symptomatic MM, relapsed MM and cells from patients with plasma cell leukemia (PCL). A large study of 877 patients has confirmed that MGUS plasma cells share similarities with MM and relapse MM but has identified genes and pathways that are activated lately during MM progression. These activated pathways include cMYC, E2F activation and chromosomal instability-defined GEP signature representing a risk of progression to MM if present at MGUS or SMM stage (12). Other groups have reported enrichment in antiapoptotic, NF-kB, DNA repair and cytokines-signaling pathway related genes in MM cells in comparison with MGUS cells (13). Importantly, impact of microenvironment on gene profile of the MM cells has been performed revealing activation of crucial pathways such as NF-kB, Notch and Ras, and genes affecting proliferation, survival, cell cycle regulators/activation (14).
GEP and Prognostication
Ability to investigate complete transcriptomic profile of MM cell provided an unique opportunity to investigate whether the disease behavior can be predicted based GEP. Clinical trials and long-follow-up of MM patients allowed evaluating the ability of GEP to predict prognosis in different cohorts. Number of reports, using distinct approaches, have identified gene expression signatures capable of predicting EFS and OS in MM. Most of these studies have identified GEP signature as an independent prognostic factor (Table 2). Some groups have used a biological approach relative to specific features of MM cells. Chromosome instability signature (15), centrosome index signature, and cell death signature (16) were defined based on genomic instability features, whereas a 7-gene prognostic signature was developed from MM cell lines study (17,18). Other signatures like the 15-gene prognostic signature or the proliferation signature have also been reported (19). Other groups have developed a GEP signature using a simple correlation between GEP with overall survival of MM patients in distinct cohorts. The HOVON-65/GMMG-HD4 clinical trial researchers (20), the Intergroupe Francophone du Myelome 99 clinical trial (9), and UAMS researchers (21) reported a 92, 15 and 70 genes signature respectively, able to identify poor outcome in independent cohorts. Importantly, only very few or no genes overlap between these signatures suggesting that each signature doesn’t incorporate all high risk patients and also highlights the redundancy in the system. In an attempt to simplify GEP use in clinical practice and to define a unique tool, a combination of existing prognostic signatures have been recently reported defining a single reliable signature that might be used to predict outcome in MM at diagnosis and relapse (22).
Table 2. Summary of main reported prognosis signatures allowing identification of high risk multiple myeloma patients.
No single gene is common across the 8 signatures.
| Signature | Number of genes | Number of common genes with 70-gene signature | Number of common genes with 92-gene signature | Clinical significance |
|---|---|---|---|---|
| UAMS (21) | 70 genes | 70 genes | 2 genes (BIRC5, LTBP1) | Identifies low and high risk patients at diagnosis and relapse. The high risk patients overexpress genes mapping at 1q21, 1q22, 1q43–q44 and 8q21–8q24 regions. |
| HOVON-65/GMMG-HD4 (EMC92) (20) | 92 genes | 2 genes (BIRC5, LTBP1) | 92 genes | An independant prognosis marker identifying high risk patients at diagnosis and relapse. |
| Intergroupe Francophone du Myelome (9) | 15 genes | None | 1 gene (FAM49A) | Identifies high risk patients featured by overexpression of genes involved in cell cycle. |
| Chromosome instability signature (CINGECS) (15) | 214 genes | 7 genes | 15 genes | Signature is based on copy-number alterations identified by aCGH.. It alows separating MM patients in 4 groups : low, 2 intermediates and high risk group. |
| Centrosome index signature (CNTI) (16) | 4 genes | None | None | An independant prognosis factor that identifies high risk patients featured by higher sensitivity to aurora kinase inhibitor. |
| Cell death signature (18) | 6 genes | None | None | Based on the presence of genomic deletions involving cell death genes, it identifies low and high risk patients. |
| 7-gene prognostic signature HMCL 6-gene signature for non t(4;14) patients (17) | 7 genes 6 genes |
None None |
None None |
Based on MM cell lines, identify low, intermediate and high risk MM patients and can discriminate low and high risk patients within molecular sub-groups, especially in non t(4 ;14). |
| Proliferation signature (19) | 50 genes | 3 genes (BIRC5, ASPM, CKS1B) | 6 genes (ESPL1, MCM6, NCAPG, SPAG5, ZWINT, BIRC5) | Identifies 3 groups of MM associated with high, intermediate and low risk. It correlates with chromosomal aberrations (amp(1q) and del(13p)) and molecular subgroups. |
| Number of overlapping genes | None |
Abbreviation: aCGH, array comparative genomic hybridization; HMCL: human myeloma cell lines.
Interestingly, GEP signature has also been used in early stages of MM or in plasma cell leukemia patients. Investigators from UAMS has reported that 70-genes signature and its derivative are able to predict outcome in context of MGUS and SMM (23). In the context of PCL, in a cohort of 21 patients, a 27 gene expression signature was identified as an independent prognostic factor (24).
Profiling Transcriptome Modifiers
Initially transcript of the gene was generally measured by its overall level, using array-based methods to reflect overall gene expression profile. However, it is now clear that number of intermediate molecules process the initial RNA transcript and eventually modify its level, sequence or characteristics to significantly affect the eventual protein being produced. These transcriptome-modifiers play an important role in development of malignancies including myeloma and affect both the initial tumorigenesis and eventual tumor cell behavior. RNA sequencing has progressively replaced micro-array based studies as, besides being highly sensitivity and specificity, it allows measurement of various transcripts and their isoforms along with identification of majority of the transcriptome modifiers as well as mutations. These new data from RNA-sequencing have been generated and will be integrated into GEP to improve prediction of the outcome in the near future.
Of these modifiers, non-coding RNA are particularly studied in MM since reports have already shown that micro-RNA participate to myelomagenesis, segregate with MM subgroups, and can be used to predict outcome or complete response to auto-transplant. MiR17 and miR886-5p have been identified as a strong prognostic marker in a cohort of 163 newly diagnosed patients from the MRC Myeloma IX l trial for example (25). An increasing literature is now describing how microRNAs play an important role in MM and characterize distinct MM subgroups (26). For example, miR-126 promotes cMYC overexpression in t(4;14) MM (27), and miRs-192, -194, and -215 deregulate p53 and MDM2 in a subgroup of MM, contributing to a poorer outcome (28,29). Very interestingly, Expression of circulating microRNAs, which are easily accessible has been evaluated and may represent a good prognostic biomarkers in MM (30). Furthermore, treatments that can restore miRNAs (in case of tumor suppressor miRNA) or inhibit miRNAs (in case of oncogene miRNA) are being developed and may constitute a major therapeutic option in the future (31,32).
Long noncoding RNAs (lncRNA) are also being carefully evaluated in MM. Our group with others, is currently identifying important deregulation of lncRNAs expression and its impact on clinical outcome (33).
Alternate splicing is a critical post-transcriptional event that tremendously increases the transcript repertoire affecting number of cellular processes including cell growth and survival. It has been recognized as important mediators of malignant phenotype and the understanding of the alternate splicing events will contribute in the next future to better establish prognosis in MM. Some reports have shown that splicing events affecting specific genes as hyaluronan synthase 1 (HAS1) (34,35) or deleted in colorectal carcinoma gene (DCC) occur recurrently in MM (36), or that a strategy targeting the splicing of X-box binding protein 1 (XBP1) increases sensitivity of MM cells to proteasome inhibitor. Pilot investigations by our group as well as others have identified significant number of spliced isoforms in myeloma in comparison to normal plasma cells with both functional consequence as well as prognostic implications.
Interestingly, the ability to detect/depict mutations at the RNA level is becoming well documented. DNA –based studies in MM, including mainly whole exome sequencing, have highlighted the mutational landscape of the disease, which includes few recurrent mutations (NRAS, KRAS, TP53, DIS3, and FAM46C). NFkB and ERK pathways are the most reccurent affected pathways, with mutations in 43 % and 17 % of MM patients respectively (37–39).
Although only certain mutations have a clear impact on prognosis (TP53, ATR, and ATM), the ability to detect those mutations at the RNA level (40) can now be used to identify mutations impacting outcome that can be integrated in the future models predicting prognosis in MM.
Finally, next generation sequencing provides the ability to perform single cell studies. This method, illustrated by the drop-seq technology (41), allows identification of the clonal heterogeneity as well as evaluation of the transcriptome in the context of the microenvironment. The initial data regarding single cell transcriptome measurement suggest exciting applications (42) including integration into a new GEP signature (43).
Clonal Heterogeneity and GEP
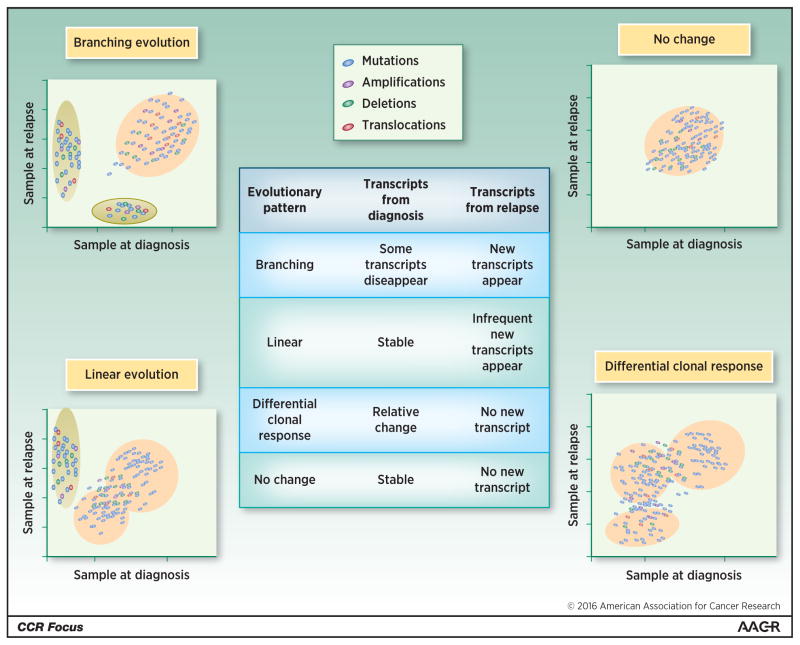
Intraclonal heterogeneity is an important feature of cancer that has been shown in MM (44,45). It refers to the presence of tumoral cells sharing most of its genomic alterations with subtle differences in mutations, copy number abnormalities and chromosome rearrangements including translocations between different clones. In MM cells, the evaluation of Ig gene rearrangement by next-generation sequencing is particularly useful. Our group has performed deep sequencing of the IgH gene at diagnosis and relapse in a large cohort of patients highlighting the complexity of the clonal and sub-clonal architecture of the disease (46). However, only few reports have described the clonal evolution in MM. Four patterns have been observed (Fig. 1) (38,47). The change in sub clonal abundance will also be reflected in changes in GEP. For example a linear evolution may not significantly impact overall GEP, on the other hand branching evolution may be reflected as decrease in expression of genes representing clones which have disappeared and appearance of genes from newly formed clones. Evaluation of GEP as yet is not sufficient to study or recognize neither evolutionary pattern, nor to estimate sub clonal content, which remains major limitation of studies utilizing GEP. Further investigations are necessary to evaluate the influence of therapies on clonal evolution. As a consequence of the clonal heterogeneity and clonal evolution, the genome and the transcriptome vary across the sub-clones and over the time, requiring new methods and iterative investigations. The ability to evaluate transcriptome at a single cell level might be necessary in order to determine the true impact of intraclonal heterogeneity on GEP and to identify potential marker of sensitivity or resistance to specific drugs (42,48).
Figure 1. Clonal heterogeneity and clonal evolution in multiple myeloma impacts gene expression profile.
The figure represents complex clonal content and clonal evolution in multiple myeloma. Four distinct evolutionary patterns have been identified and their impact on gene expression profile is presented. Branching evolution, which corresponds to the appearance of new clones expressing new genes while other clones disappear, affects GEP. Linear evolution is characterized by the emergence of one new clone that influences GEP. Differential clone response, featured by the modification of the relative proportion of the sub-clones, affects GEP. Stable clonality with no detectable change does not impact GEP.
GEP to Guide Therapy
Identification of high-risk patients
Amongst various applications for identifying high-risk patient, is to try and tailor therapy specifically to improve outcome in high-risk patients. To date the consensus from the International Myeloma Workshop recommends to assess MM risk at diagnosis, using the combination of 2 biological criteria determined by the international system staging (ISS) (beta2microglobulin and albumin levels) and 2 cytogenetic abnormalities (del(17p13) and t(4;14)). These criteria do not reflect the heterogeneity that exists within patients even at the time of diagnosis and the evolution of the risk category over time. Although the traditional risk categorization still has applicability in practice, the number of newer agents available for MM therapy can overcome these traditional high-risk features such as t(4;14). The newer genomic correlates, such as GEP, are now able to identify high-risk population even with utilization of combination newer agent therapies. Moreover, the combination of gene expression, splicing events, and mutations should soon provide an even better tool to predict prognosis in MM that can be combined with other clinico-biological parameters.
GEP in combination with ISS
A recent study has evaluated GEP in combination with other prognosis marker in MM including cytogenetic alterations and ISS score. This study evaluated different GEP signature and showed that the combination of GEP with ISS is a powerful prognostic tool that significantly improves risk stratification (49). Identifying high risk patients remains an important task to try and tailor therapy in future discussed by Landgren and Rajkumar in this CCR Focus section (50). Currently, no specific therapy is indicated especially for the high-risk patients, there is increasing emphasis on including multi-agent therapy as consolidation and maintenance including longer term use of both proteasome inhibitor and immunomodulatory agent. High dose melphalan followed by autologous stem cell transplant (ASCT) seems to be the best consolidation therapy to date (51).
Prediction of treatment response
GEP has also been evaluated to predict complete response (CR) to different treatment as well. CR is an independent prognosis factor and is an indirect marker of overall survival (52). A specific GEP signature has been identified In context of 3 drugs combinations (VTD) in newly diagnosed patients (53), in context of high dose therapy (54), another in context of Imids/dexamethaosne and double auto-transplant (55) and another at relapse, in context of bortezomib based-regimen (56). However, a prediction model study comparing different dataset have shown that GEP alone is not efficient to predict CR in different datasets (57).
This study utilized various methods to develop a response predictive model; even with the best GEP-based CR predictive model accuracy was between 56% to 78% across four different datasets. The ability to predict CR was not affected by different platforms used to measure GEP, or treatment regimens used or newly-diagnosed or relapsed patients. This study highlights the fact that it may be necessary to incorporate multiple other genomic correlates in such response predicting model in future.
Personalized therapy selection
Based on GEP, the activation or deregulation of certain pathways can be monitored and provide important information to select a therapy. For example, the presence of high DKK1 level, which is also predictive of bone lesions can be exploited for the use of anti-DKK1 drug (58,59), or the evaluation of the ratio BCL2 /MCL1 level can predict the sensitivity to BH3 mimetic drugs (60). Alternatively, combining the information regarding gene expression and mutation expression is very important to consider personalized medicine (61). The detection of specific expressed mutations such as BRAF V600E can indicate the use of BRAF inhibitor such as vemurafenib (62,63), or mutations activating the MAPK pathway can provide rationale for the use of MEK inhibitors such as trametinib (64). Other specific and targetable mutations such as SF3B1, FGFR3, ATM/ATR, IDH1/2, and CCND1 as well as RAS/RAF, NFkB pathway–related genes have been reported in myeloma. These mutations can be targeted by appropriate inhibitors.
Some mutations can also be evaluated to predict drug sensitivity. Preliminary data showed that the presence of NRAS mutations in relapsed MM is associated with lower response to bortezomib (65) or in contrast, that the presence of IRF4 mutations is associated with higher sensitivity to immuno-modulatory agents (37). These data need to be confirmed in distinct clinical trials but may constitute important results.
The identification of specific micro-RNA expression profile can also be exploited to inform therapy. Several microRNAs are being investigated as therapeutic targets with potential for development of small molecules that affect micro-RNA function.
Similarly, GEP has been utilized to predict resistance to individual agents with a view that one can avoid toxicity of agents which are not likely to work. Using number of B-cell lines including multiple myeloma cell lines, a microarray-based GEP signature was developed to predict melphalan resistance. Although the signature was able to predict sensitivity versus resistance in cell lines, its application to patient remains to be confirmed (66,67). Interestingly, a pharmacogenomic analysis of global GEP of myeloma cells retrieved from patients with myeloma, certain time after administration of various therapeutic agents have been performed (68,69). Prognostic information was obtained from GEP of purified plasma cells 2 days after administration of thalidomide and dexamethasone or bortezomib to newly-diagnosed myeloma patients. An 80-gene signature was identified following bortezomib administration allowing improving patients’ risk stratification (69).
From therapeutic as well as prognostic end points, it is also important to consider continues evolution of genome which happens spontaneously as well as under the influence of microenvironment, epigenomic changes or therapy. Therefore, evaluation of GEP at a single time point may not be adequate. The evolution of GEP from diagnosis, response and relapse should be investigated to theoretically provide important information for potential selection of the most appropriate therapy.
Limitation of GEP in Current Clinical Practice
Despite a significant amount of data, general adoption of GEP in clinical practice is neither observed nor recommended. Important obstacles still exist to application of gene expression profiling to general clinical practice. Although several distinct GEP signatures have been identified and a recent study has combined several of these signatures to generate a unique signature (70) no consensus has been adopted so far for universal application to all patient population. GEP remains an investigational tool and is not yet validated by the FDA. From clinical application point of view, the GEP data have been generated mostly in a context of specific treatments that includes thalidomide, lenalidomide and borteziomib with or without auto-transplant. Since the therapeutic landscape is largely evolving in MM, re-evaluations are needed for each new drug and/or combination. In particular the advent of new therapeutic classes such as antibody-based therapies (Elotuzumab, Daratumumab) and new Imids and proteasome inhibitors, as discussed elsewhere in this CCR Focus section (71,72), greatly modify the prognosis and may require new GEP studies and signatures (73–78). GEP has been used to date in a limited number of myeloma centers and mostly for research purposes. The development of simpler and faster methods should be considered. Simple quantitative PCR has been evaluated in a cohort of 157 newly diagnosed patients with good results (79). However, a consensus remains to be established to define the genes that should be evaluated. Most importantly, we believe that an integrated approach that includes, at the minimum, gene expression, mutational profile and microRNA expression will be required to allow a broader utilization of genomic data to guide both therapeutic selection as well as prognostication. Taking the current state-of-the-art to the next level, it will be necessary to understand the clinical impact of clonal content and evolution along with identification of sub-clonal variants and role of molecular mechanism driving such changes, on disease outcome. The current data that mutational load predicts outcome will need to be investigated for therapeutic purpose. These algorithms will also change with the advent of immunotherapeutic strategies which may have greater success in tumors with high number of mutations. Again, as demonstrated by our earlier study expression of mutations will need to be studied for eventual consideration in therapeutic decisions (61).
Future Direction
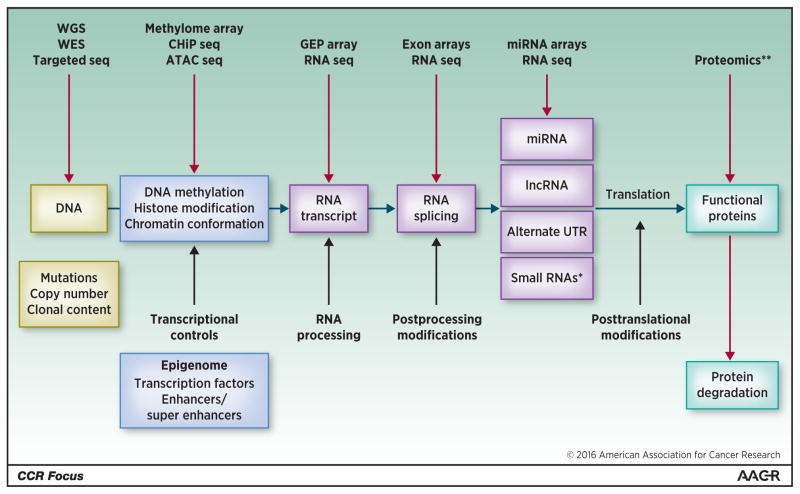
To improve upon tremendous progress made so far, newer high-throughput technologies are being incorporated. Array-based methodologies have given way to sequencing-based method, and newer bio-informatic methodologies are being developed to identify meaning from the large amount of data being generated (Fig. 2). Furthermore, integrative oncogenomic efforts are incorporating new markers such as mutations, splicing events, noncoding RNA, miRs to improve both predictive and prognostic markers. The personalized medicine based on the selection of a targeted therapy informed by the presence of a specific mutation or GEP signature is appealing. However, in future, the selection of patients will require consideration of dynamic evolution of the disease, coexistence of sub-clones, and the possibility of downstream activating mutations in the same pathway as it may be the case with KRAS and BRAF for the ERK pathway
Figure 2. Next generation highthroughput and sequencing technologies provides new markers for gene profiling.
Gene expression profile results from various processes (middle row) including post-translational leading to protein modification. Several genomic analysis technologies (upper row) are available to identify various genomic abnormalities to develop an integrated approach allowing understanding of the molecular pathogenesis of multiple myeloma, risk stratification and development of personalized medicine.
Alternate UTR, alternate untranslated region; ATAC seq, Assay for Transposase-Accessible Chromatin with high-throughput sequencing; GEP array, gene-expression profile array; lncRNA: long noncoding RNA; miRNA, microRNA; RNA seq, RNA sequencing.
*Small RNAs: small nucleolar RNA (snoRNAs), small nuclear RNA (U-RNA), small rDNA-derived RNA (srRNA).
**Protein modification such as phosphorylation, acetylation, ubiquitination, sumoylation, and glycosylation.
Adapted from Munshi and Avet-Loiseau (80).
To conclude, gene profiling studies provide important information regarding MM biology, and constitute a powerful tool to predict outcome and to guide therapy. The combination of mutational profile, splicing events, gene expression with ISS and cytogenetic may become a standard into MM care.
Acknowledgments
Grant Support
This work was supported by NIH grants PO1-155258 and P50-100707 (to N.C. Munshi and H. Avet-Loiseau), Department of Veterans Affairs Merit Review Award 1 I01BX001584-01 (to N.C. Munshi), and a Leukemia & Lymphoma Society Translational Research Program Award (to N.C. Munshi).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nature Reviews Cancer. 2012;12(5):335–48. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 2.Kyle RA, Gertz MA, Witzig TE, Lust JA, Lacy MQ, Dispenzieri A, et al. Review of 1027 patients with newly diagnosed multiple myeloma. Mayo Clinic Proceedings. 2003;78(1):21–33. doi: 10.4065/78.1.21. [DOI] [PubMed] [Google Scholar]
- 3.Palumbo A, Anderson K. Multiple myeloma. New England Journal of Medicine. 2011;364(11):1046–60. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 4.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9(4):313–25. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23(26):6333–8. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 6.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106(1):296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116(14):2543–53. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 8.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decaux O, Lode L, Magrangeas F, Charbonnel C, Gouraud W, Jezequel P, et al. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myelome. J Clin Oncol. 2008;26(29):4798–805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 10.Dring AM, Davies FE, Fenton JA, Roddam PL, Scott K, Gonzalez D, et al. A global expression-based analysis of the consequences of the t(4;14) translocation in myeloma. Clin Cancer Res. 2004;10(17):5692–701. doi: 10.1158/1078-0432.CCR-04-0467. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Wang X, Zheng H, Wang C, Minvielle S, Magrangeas F, et al. Classify hyperdiploidy status of multiple myeloma patients using gene expression profiles. PloS one. 2013;8(3):e58809. doi: 10.1371/journal.pone.0058809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anguiano A, Tuchman SA, Acharya C, Salter K, Gasparetto C, Zhan F, et al. Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J Clin Oncol. 2009;27(25):4197–203. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Corral L, Corchete LA, Sarasquete ME, Mateos MV, Garcia-Sanz R, Ferminan E, et al. Transcriptome analysis reveals molecular profiles associated with evolving steps of monoclonal gammopathies. Haematologica. 2014;99(8):1365–72. doi: 10.3324/haematol.2013.087809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nature medicine. 2010;16(4):483–9. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung TH, Mulligan G, Fonseca R, Chng WJ. A novel measure of chromosome instability can account for prognostic difference in multiple myeloma. PloS one. 2013;8(6):e66361. doi: 10.1371/journal.pone.0066361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chng WJ, Braggio E, Mulligan G, Bryant B, Remstein E, Valdez R, et al. The centrosome index is a powerful prognostic marker in myeloma and identifies a cohort of patients that might benefit from aurora kinase inhibition. Blood. 2008;111(3):1603–9. doi: 10.1182/blood-2007-06-097774. [DOI] [PubMed] [Google Scholar]
- 17.Moreaux J, Klein B, Bataille R, Descamps G, Maiga S, Hose D, et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica. 2011;96(4):574–82. doi: 10.3324/haematol.2010.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickens NJ, Walker BA, Leone PE, Johnson DC, Brito JL, Zeisig A, et al. Homozygous deletion mapping in myeloma samples identifies genes and an expression signature relevant to pathogenesis and outcome. Clin Cancer Res. 2010;16(6):1856–64. doi: 10.1158/1078-0432.CCR-09-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hose D, Reme T, Hielscher T, Moreaux J, Messner T, Seckinger A, et al. Proliferation is a central independent prognostic factor and target for personalized and risk-adapted treatment in multiple myeloma. Haematologica. 2011;96(1):87–95. doi: 10.3324/haematol.2010.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuiper R, Broyl A, de Knegt Y, van Vliet MH, van Beers EH, van der Holt B, et al. A gene expression signature for high-risk multiple myeloma. Leukemia. 2012;26(11):2406–13. doi: 10.1038/leu.2012.127. [DOI] [PubMed] [Google Scholar]
- 21.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276–84. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 22.Chng WJ, Chung TH, Kumar S, Usmani S, Munshi N, Avet-Loiseau H, et al. Gene signature combinations improve prognostic stratification of multiple myeloma patients. Leukemia. 2016;30(5):1071–8. doi: 10.1038/leu.2015.341. [DOI] [PubMed] [Google Scholar]
- 23.Khan R, Dhodapkar M, Rosenthal A, Heuck C, Papanikolaou X, Qu P, et al. Four genes predict high risk of progression from smoldering to symptomatic multiple myeloma (SWOG S0120) Haematologica. 2015;100(9):1214–21. doi: 10.3324/haematol.2015.124651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todoerti K, Agnelli L, Fabris S, Lionetti M, Tuana G, Mosca L, et al. Transcriptional characterization of a prospective series of primary plasma cell leukemia revealed signatures associated with tumor progression and poorer outcome. Clin Cancer Res. 2013;19(12):3247–58. doi: 10.1158/1078-0432.CCR-12-3461. [DOI] [PubMed] [Google Scholar]
- 25.Wu P, Agnelli L, Walker BA, Todoerti K, Lionetti M, Johnson DC, et al. Improved risk stratification in myeloma using a microRNA-based classifier. British Journal of Haematology. 2013;162(3):348–59. doi: 10.1111/bjh.12394. [DOI] [PubMed] [Google Scholar]
- 26.Corthals SL, Sun SM, Kuiper R, de Knegt Y, Broyl A, van der Holt B, et al. MicroRNA signatures characterize multiple myeloma patients. Leukemia. 2011;25(11):1784–9. doi: 10.1038/leu.2011.147. [DOI] [PubMed] [Google Scholar]
- 27.Min DJ, Ezponda T, Kim MK, Will CM, Martinez-Garcia E, Popovic R, et al. MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC. Leukemia. 2013;27(3):686–94. doi: 10.1038/leu.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18(4):367–81. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(35):12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocci A, Hofmeister CC, Geyer S, Stiff A, Gambella M, Cascione L, et al. Circulating miRNA markers show promise as new prognosticators for multiple myeloma. Leukemia. 2014;28(9):1922–6. doi: 10.1038/leu.2014.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18(22):6260–70. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad N, Haider S, Jagannathan S, Anaissie E, Driscoll JJ. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia. 2014;28(4):732–8. doi: 10.1038/leu.2013.262. [DOI] [PubMed] [Google Scholar]
- 33.Samur M, Gulla A, Cleynen A, Magrangeas F, Minvielle S, Anderson KC, et al. Differentially expressed and prognostically significant lincrnas may impact immune system and tumor progression in multiple myeloma (MM) Blood. 2015;126:2989. [Google Scholar]
- 34.Adamia S, Reichert AA, Kuppusamy H, Kriangkum J, Ghosh A, Hodges JJ, et al. Inherited and acquired variations in the hyaluronan synthase 1 (HAS1) gene may contribute to disease progression in multiple myeloma and Waldenstrom macroglobulinemia. Blood. 2008;112(13):5111–21. doi: 10.1182/blood-2008-02-141770. [DOI] [PubMed] [Google Scholar]
- 35.Adamia S, Reiman T, Crainie M, Mant MJ, Belch AR, Pilarski LM. Intronic splicing of hyaluronan synthase 1 (HAS1): a biologically relevant indicator of poor outcome in multiple myeloma. Blood. 2005;105(12):4836–44. doi: 10.1182/blood-2004-10-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagoshi H, Taki T, Chinen Y, Tatekawa S, Tsukamoto T, Maegawa S, et al. Transcriptional dysregulation of the deleted in colorectal carcinoma gene in multiple myeloma and monoclonal gammopathy of undetermined significance. Genes, Chromosomes & Cancer. 2015;54(12):788–95. doi: 10.1002/gcc.22290. [DOI] [PubMed] [Google Scholar]
- 37.Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33(33):3911–20. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature Communications. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosen-Ansorena D, Bolli N, Samur MK, Magrangeas F, Minvielle S, Anderson KC, et al. Redefining mutational profiling using RNA-Seq: insight into the functional mutational landscape of multiple myeloma. Blood. 2015;126(23):837. [Google Scholar]
- 41.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitra AK, Mukherjee UK, Harding T, Jang JS, Stessman H, Li Y, et al. Single-cell analysis of targeted transcriptome predicts drug sensitivity of single cells within human myeloma tumors. Leukemia. 2016;30(5):1094–102. doi: 10.1038/leu.2015.361. [DOI] [PubMed] [Google Scholar]
- 43.Rashid N, Minvielle S, Magrangeas F, Samur MK, Cleynen A, Sperling A, et al. Alternative splicing is a frequent event and impacts clinical outcome in myeloma: a large RNA-Seq data analysis of newly-diagnosed myeloma patients. Blood. 2014;124:638. [Google Scholar]
- 44.Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120(5):1077–86. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- 45.Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28(2):384–90. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munshi NC, Minvielle S, Tai Y-T, Fulciniti M, Samur MK, Richardson PG, et al. Deep Igh sequencing identifies an ongoing somatic hypermutation process with complex and evolving clonal architecture in myeloma. Blood. 2015;126(23):21. [Google Scholar]
- 47.Weinhold N, Ashby C, Rasche L, Chavan SS, Stein C, Stephens OW, et al. Clonal selection and double hit events involving tumor suppressor genes underlie relapse from chemotherapy: myeloma as a model. Blood. 2016 Aug 11; doi: 10.1182/blood-2016-06-723007. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melchor L, Brioli A, Wardell CP, Murison A, Potter NE, Kaiser MF, et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28(8):1705–15. doi: 10.1038/leu.2014.13. [DOI] [PubMed] [Google Scholar]
- 49.Kuiper R, van Duin M, van Vliet MH, Broijl A, van der Holt B, El Jarari L, et al. Prediction of high- and low-risk multiple myeloma based on gene expression and the International Staging System. Blood. 2015;126(17):1996–2004. doi: 10.1182/blood-2015-05-644039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landgren O, Rajkumar SV. New developments in diagnosis, prognosis, and assessment of response in multiple myeloma. Clin Cancer Res. 2016;22:xxxx–xxxx. doi: 10.1158/1078-0432.CCR-16-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Attal M, Lauwers-Cances V, Hulin C, Facon T, Caillot D, Escoffre M, et al. Autologous transplantation for multiple myeloma in the era of new drugs: a phase III study of the Intergroupe Francophone Du Myelome (IFM/DFCI 2009 Trial) Blood. 2015;126(23):391. [Google Scholar]
- 52.Harousseau JL, Attal M, Avet-Loiseau H. The role of complete response in multiple myeloma. Blood. 2009;114(15):3139–46. doi: 10.1182/blood-2009-03-201053. [DOI] [PubMed] [Google Scholar]
- 53.Terragna C, Remondini D, Martello M, Zamagni E, Pantani L, Patriarca F, et al. The genetic and genomic background of multiple myeloma patients achieving complete response after induction therapy with bortezomib, thalidomide and dexamethasone (VTD) Oncotarget. 2016;7(9):9666–79. doi: 10.18632/oncotarget.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu P, Walker BA, Broyl A, Kaiser M, Johnson DC, Kuiper R, et al. A gene expression based predictor for high risk myeloma treated with intensive therapy and autologous stem cell rescue. Leukemia & Lymphoma. 2015;56(3):594–601. doi: 10.3109/10428194.2014.911863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terragna C, Renzulli M, Remondini D, Tagliafico E, Di Raimondo F, Patriarca F, et al. Correlation between eight-gene expression profiling and response to therapy of newly diagnosed multiple myeloma patients treated with thalidomide-dexamethasone incorporated into double autologous transplantation. Annals of Hematology. 2013;92(9):1271–80. doi: 10.1007/s00277-013-1757-6. [DOI] [PubMed] [Google Scholar]
- 56.Zhan F, Barlogie B, Mulligan G, Shaughnessy JD, Jr, Bryant B. High-risk myeloma: a gene expression based risk-stratification model for newly diagnosed multiple myeloma treated with high-dose therapy is predictive of outcome in relapsed disease treated with single-agent bortezomib or high-dose dexamethasone. Blood. 2008;111(2):968–9. doi: 10.1182/blood-2007-10-119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amin SB, Yip WK, Minvielle S, Broyl A, Li Y, Hanlon B, et al. Gene expression profile alone is inadequate in predicting complete response in multiple myeloma. Leukemia. 2014;28(11):2229–34. doi: 10.1038/leu.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. The New England Journal of Medicine. 2003;349(26):2483–94. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 59.Fulciniti M, Tassone P, Hideshima T, Vallet S, Nanjappa P, Ettenberg SA, et al. Anti-DKK1 mAb (BHQ880) as a potential therapeutic agent for multiple myeloma. Blood. 2009;114(2):371–9. doi: 10.1182/blood-2008-11-191577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Touzeau C, Dousset C, Le Gouill S, Sampath D, Leverson JD, Souers AJ, et al. The Bcl-2 specific BH3 mimetic ABT-199: a promising targeted therapy for t(11;14) multiple myeloma. Leukemia. 2014;28(1):210–2. doi: 10.1038/leu.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rashid NU, Sperling AS, Bolli N, Wedge DC, Van Loo P, Tai YT, et al. Differential and limited expression of mutant alleles in multiple myeloma. Blood. 2014;124(20):3110–7. doi: 10.1182/blood-2014-04-569327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharman JP, Chmielecki J, Morosini D, Palmer GA, Ross JS, Stephens PJ, et al. Vemurafenib response in 2 patients with posttransplant refractory BRAF V600E-mutated multiple myeloma. Clinical Lymphoma, Myeloma & Leukemia. 2014;14(5):e161–3. doi: 10.1016/j.clml.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 63.Andrulis M, Lehners N, Capper D, Penzel R, Heining C, Huellein J, et al. Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discovery. 2013;3(8):862–9. doi: 10.1158/2159-8290.CD-13-0014. [DOI] [PubMed] [Google Scholar]
- 64.Heuck CJ, Jethava Y, Khan R, van Rhee F, Zangari M, Chavan S, et al. Inhibiting MEK in MAPK pathway-activated myeloma. Leukemia. 2016;30(4):976–80. doi: 10.1038/leu.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mulligan G, Lichter DI, Di Bacco A, Blakemore SJ, Berger A, Koenig E, et al. Mutation of NRAS but not KRAS significantly reduces myeloma sensitivity to single-agent bortezomib therapy. Blood. 2014;123(5):632–9. doi: 10.1182/blood-2013-05-504340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boegsted M, Holst JM, Fogd K, Falgreen S, Sorensen S, Schmitz A, et al. Generation of a predictive melphalan resistance index by drug screen of B-cell cancer cell lines. PloS One. 2011;6(4):e19322. doi: 10.1371/journal.pone.0019322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bogsted M, Bilgrau AE, Wardell CP, Bertsch U, Schmitz A, Bodker JS, et al. Proof of the concept to use a malignant B cell line drug screen strategy for identification and weight of melphalan resistance genes in multiple myeloma. PloS One. 2013;8(12):e83252. doi: 10.1371/journal.pone.0083252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burington B, Barlogie B, Zhan F, Crowley J, Shaughnessy JD., Jr Tumor cell gene expression changes following short-term in vivo exposure to single agent chemotherapeutics are related to survival in multiple myeloma. Clin Cancer Res. 2008;14(15):4821–9. doi: 10.1158/1078-0432.CCR-07-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaughnessy JD, Jr, Qu P, Usmani S, Heuck CJ, Zhang Q, Zhou Y, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118(13):3512–24. doi: 10.1182/blood-2010-12-328252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chng WJ, Chung TH, Kumar S, Usmani S, Munshi N, Avet-Loiseau H, et al. Gene signature combinations improve prognostic stratification of multiple myeloma patients. Leukemia. 2016;30(5):1071–8. doi: 10.1038/leu.2015.341. [DOI] [PubMed] [Google Scholar]
- 71.Orlowski RZ, Lonial S. Integration of novel agents into the care of patients with multiple myeloma. Clin Cancer Res. 2016;22:xxxx–xxxx. doi: 10.1158/1078-0432.CCR-16-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar S, Aanderson KC. Immune therapies in myeloma. Clin Cancer Res. 2016;22:xxxx–xxxx. doi: 10.1158/1078-0432.CCR-16-0868. [DOI] [PubMed] [Google Scholar]
- 73.Avet-Loiseau H, Fonseca R, Siegel D, Dimopoulos MA, Spicka I, Masszi T, et al. Carfilzomib significantly improves the progression free survival of high-risk patients in multiple myeloma. Blood. 2016;128(9):1174–80. doi: 10.1182/blood-2016-03-707596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stewart AK, Rajkumar SV, Dimopoulos MA, Masszi T, Spicka I, Oriol A, et al. Carfilzomib, lenalidomide, and dexamethasone for relapsed multiple myeloma. The New England Journal of Medicine. 2015;372(2):142–52. doi: 10.1056/NEJMoa1411321. [DOI] [PubMed] [Google Scholar]
- 75.Jakubowiak A, Offidani M, Pegourie B, De La Rubia J, Garderet L, Laribi K, et al. Randomized phase 2 study: elotuzumab plus bortezomib/dexamethasone vs bortezomib/dexamethasone for relapsed/refractory MM. Blood. 2016;127(23):2833–40. doi: 10.1182/blood-2016-01-694604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lonial S, Dimopoulos M, Palumbo A, White D, Grosicki S, Spicka I, et al. Elotuzumab therapy for relapsed or refractory multiple myeloma. The New England Journal of Medicine. 2015;373(7):621–31. doi: 10.1056/NEJMoa1505654. [DOI] [PubMed] [Google Scholar]
- 77.Lokhorst HM, Plesner T, Laubach JP, Nahi H, Gimsing P, Hansson M, et al. Targeting CD38 with daratumumab monotherapy in multiple myeloma. The New England Journal of Medicine. 2015;373(13):1207–19. doi: 10.1056/NEJMoa1506348. [DOI] [PubMed] [Google Scholar]
- 78.Moreau P, Masszi T, Grzasko N, Bahlis NJ, Hansson M, Pour L, et al. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. The New England Journal of Medicine. 2016;374(17):1621–34. doi: 10.1056/NEJMoa1516282. [DOI] [PubMed] [Google Scholar]
- 79.Sarasquete ME, Martinez-Lopez J, Chillon MC, Alcoceba M, Corchete LA, Paiva B, et al. Evaluating gene expression profiling by quantitative polymerase chain reaction to develop a clinically feasible test for outcome prediction in multiple myeloma. British Journal of Haematology. 2013;163(2):223–34. doi: 10.1111/bjh.12519. [DOI] [PubMed] [Google Scholar]
- 80.Munshi NC, Avet-Loiseau H. Genomics in multiple myeloma. Clin Cancer Res. 2011;17(6):1234–42. doi: 10.1158/1078-0432.CCR-10-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]