Abstract
Yersinia outer protein J (YopJ) family of bacterial effectors depends on a novel acetyltransferase domain to acetylate signaling proteins from plant and animal hosts. However, the underlying mechanism is unclear. Here, we report the crystal structures of PopP2, a YopJ effector produced by the plant pathogen Ralstonia solanacearum, in complex with inositol hexaphosphate (IP6), acetyl-coenzyme A (AcCoA), and/or substrate RRS1-RWRKY. PopP2 recognizes the WRKYGQK motif of RRS1-RWRKY to position a targeted lysine in the active site for acetylation. Importantly, the PopP2 – RRS1-RWRKY association is allosterically regulated by IP6 binding, suggestive of a previously unidentified role of the eukaryote-specific cofactor in substrate interaction. Furthermore, we provide evidence for the reaction intermediate of PopP2-mediated acetylation, an acetyl-cysteine covalent adduct, lending direct support to the “Ping-Pong”-like catalytic mechanism proposed for YopJ effectors. Together, our study provides critical mechanistic insights into the virulence activity of YopJ class of acetyltransferases.
Introduction
The co-evolutionary arms race between pathogens and hosts has cultivated multiple lines of invasion and counter-invasion schemes. The first line of host defense involves the recognition of microbe-associated molecular patterns (MAMPs), which effectively restricts the colonization of the majority of potential pathogens1,2. To counteract this MAMP-triggered immunity (MTI), successful pathogens employ secretion systems to inject virulence proteins, collectively known as effectors, into the host cells3,4. Effectors secreted by the bacterial type III secretion system (T3SEs) play a central role in the pathogenesis of Gram-negative bacteria5,6. Once translocated into the host cells, T3SEs manipulate specific host cellular processes, thereby promoting bacterial invasion and proliferation7. On the other hand, plant hosts have developed the nucleotide-binding/leucine-rich-repeat receptors (NLRs) to sense the activities of T3SEs, which consequently initiates a robust innate immune response termed effector-triggered immunity (ETI)3.
The YopJ superfamily of T3SEs define a broad class of novel acetyltransferases, with members produced from both animal and plant pathogens8,9. YopJ effectors produced by mammalian pathogens, including YopJ from Yersinia spp., AvrA from Salmonella enterica and VopA from Vibrio parahaemolyticus, acetylate specific serine, threonine and/or lysine residues in mitogen-activated protein kinase kinases (MAPKKs) to inhibit enzymatic activation10–13, or ATP binding14, leading to suppressed inflammatory response10–14. YopJ effectors produced by plant pathogens are more diversified on targeting host proteins8. Among them, PopP2 produced by the root-infecting bacterium Ralstonia solanacearum acetylates a conserved lysine residue in the heptad sequence WRKYGQK found in the defense-related WRKY transcription factors to inhibit their DNA binding activity15,16,. As such, PopP2 dampens the activation of defense genes in plant hosts. As a counter strategy, resistant plants evolved a chimeric NLR known as RRS1-R (Resistance to Ralstonia solanacearum 1), which incorporates a C-terminal DNA-binding WRKY domain as a “trap”. Acetylation of the WRKY domain of RRS1-R (RRS1-RWRKY) by PopP2 activates ETI and confers resistance to R. solanacearum15,16. By contrast, another YopJ homologue HopZ1a from Pseudomonas syringae modifies multiple substrates including tubulin17 and the Jasmonate ZIM domain (JAZ) protein18.
YopJ effectors bear no resemblance with other acetyltransferases. Instead, they depend on a catalytic core that is homologous to ubiquitin-like proteases (ULPs), with a conserved His/Cys/Glu or His/Cys/Asp catalytic triad19,20. Furthermore, the activities of YopJ family effectors are regulated by a host cofactor inositol-hexakisphosphate (IP6)20–23, which is highly abundant in eukaryotes but absent in bacteria. Our recent study revealed that HopZ1a evolved a regulatory domain that, together with the ULP-like catalytic core, forms the IP6- and AcCoA-binding pockets. IP6 binding induces a conformational transition of HopZ1a to permit the formation of an AcCoA-binding pocket, thereby enhancing the acetylation reaction allosterically20. However, the mechanism by which YopJ effectors bind and acetylate substrates remains unclear.
In this study, we determined the crystal structures of PopP2 in complex with IP6, AcCoA and/or RRS1-RWRKY. The complex structures define the interaction interface required for the acetylation of RRS1-R and possibly WRKY transcription factors by PopP2. Importantly, the interaction of PopP2 with RRS1-RWRKY is allosterically regulated by IP6 and AcCoA, establishing a link between its cofactor binding and substrate recognition. Moreover, we characterized the structural details of the acetyl-cysteine reaction intermediate, which lends the long-sought-after evidence for the previously proposed “Ping-Pong” route of catalysis24. Together, these studies provide key mechanistic insights into the virulence activity of PopP2 and other YopJ family effectors.
Result
Overall structures of PopP2 in complex with IP6, AcCoA and/or RRS1-RWRKY
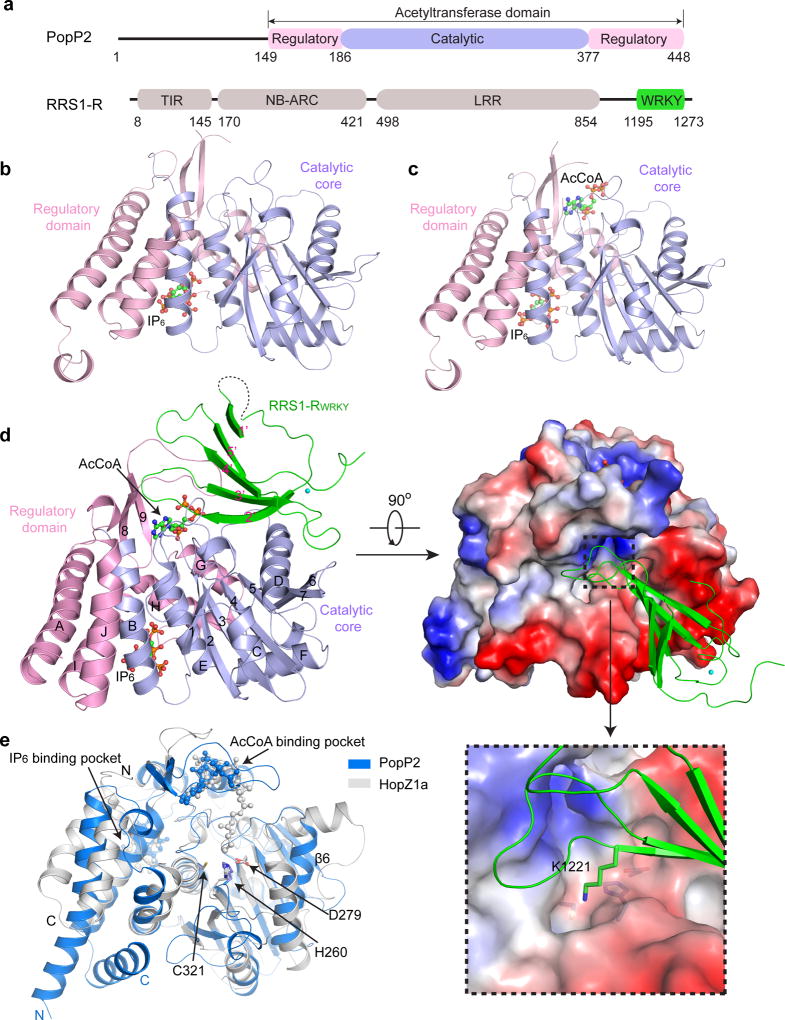
To understand how PopP2 binds and acetylates host substrates, we solved the crystal structures of the acetyltransferase domain of PopP2 (PopP2WT, residues 149–488) or its catalytic mutant C321A (PopP2C321A) in complex with IP6, AcCoA and/or the RRS1-RWRKY domain (residues 1195–1273 of RRS1-R) (Fig. 1a–d, Supplementary Table 1). The structures of the PopP2WT – IP6, PopP2C321A – IP6 – AcCoA and PopP2WT – IP6 – AcCoA – RRS1-RWRKY complexes reveal that PopP2 is comprised of a catalytic core assuming a ULP-like fold packed against a regulatory domain formed by the N- and C-terminal flanking sequences (Fig. 1b–d). The closely packed catalytic core and regulatory domain together form one IP6-binding pocket (Fig 1b), one AcCoA-binding cleft (Fig 1c), and one surface groove cradling the RRS1-RWRKY domain (Fig 1d), similar to what was observed for HopZ1a (Fig. 1e). Structural comparison of all three PopP2 complexes reveals no substantial conformational change in PopP2, except for a disorder-to-order transition of loopβ8β9 upon its binding to RRS1-RWRKY (discussed in detail below) (Fig. 1b–d).
Figure 1. Crystal structures of PopP2 in complex with IP6, AcCoA and/or RRS1-RWRKY.
a, Domain architectures of PopP2 and RRS1-R, with individual domains colored differently. TIR, Toll/interleukin1 receptor; NB, nucleotide binding; ARC, Apaf-1, R-protein and CED-4; LRR, leucine-rich repeats. Similar color schemes are used in other figures unless otherwise indicated. b, Crystal structure of PopP2WT in complex with IP6. c, Crystal structure of PopP2C321A in complex with IP6 and AcCoA. d, Ribbon and surface representations of the PopP2WT-IP6-AcCoA-RRS1-RWRKY complex. The α–helices and β-strands in PopP2 are labeled from A to J and 1 to 9, respectively. The β-strands in RRS1-RWRKY are labeled from 1’ to 5’. The IP6 and AcCoA molecules are shown in ball-and-stick representation. The zinc ion is shown as a cyan sphere. The active site of PopP2 and the target lysine K1221 in RRS1-RWRKY are shown in an expanded view. e, Structural alignment of PopP2 and HopZ1a, colored in blue and grey, respectively. The catalytic triad (H260/D279/C321 in PopP2 or H150/E170/C216 in HopZ1a) is shown in stick representation.
Despite with only ~30% sequence identity (Supplementary Fig. 1), PopP2 and HopZ1a superimpose well in their catalytic core and the IP6- and AcCoA-binding pockets (Fig. 1e), giving a root-mean-square-deviation of 2.0 Å over the backbone of 224 residues. Their major structural difference lies in the linker sequence preceding the last β strand of the ULP fold, which forms a strand (β6) in PopP2 but assumes two helices in HopZ1a, as well as the N- and C-terminal helices that constitute one half of the regulatory domain (Fig. 1e). Another notable structural difference arises from their active sites: The active site of PopP2 is surrounded by several bulky hydrophobic residues (L191, L281 and F318), and T229 and S259, forming a deep cavity harboring the target lysine K1221; by contrast, the active site of HopZ1a is surrounded by two serines (S87 and S213), resulting in a shallow concave (Supplementary Fig. 2). Such distinct active site environments may explain why PopP2 prefers lysine for acetylation whereas HopZ1a acetylates serine and threonine residues21.
The RRS1-RWRKY domain is comprised of a five-stranded antiparallel β-sheet stabilized by a zinc cluster, reminiscent of what was observed for other WRKY domains25. The positively charged β2’- and β3’-strands of RRS1-RWRKY penetrate into an acidic surface groove of PopP2, with the side chain of K1221 inserting into the active site (Fig. 1d and Supplementary Fig. 3). Note that each asymmetric unit of the crystal contains two PopP2WT–IP6–AcCoA–RRS1-RWRKY complexes, crystal packing of which resulted in a displaced β1’-strand from one of the RRS1-RWRKY molecules and reduced electron density for the loop connecting β1’ and β2’ (Supplementary Fig. 4). For the purpose of clarity, our structural analysis only focuses on the intact region of the complex.
IP6 regulation of the AcCoA binding of PopP2
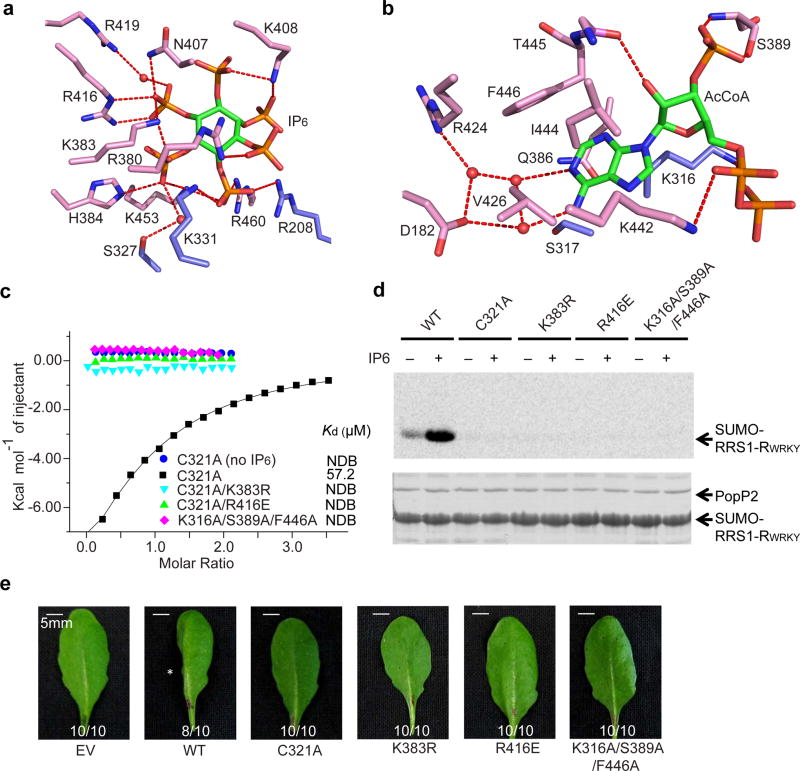
The IP6- and AcCoA-binding pockets of PopP2 are located distantly at the interface of the regulatory domain and the catalytic core (Fig. 1c). The PopP2 – IP6 association is mediated by a cluster of polar residues (R208, S327, K331, K383, H384, N407, K408, R416, R419, K453 and R460) and the six phosphate groups of IP6 through hydrogen bonding and/or electrostatic interactions (Fig. 2a). Note that the previously identified auto-acetylation site of PopP2, K38326, is unmodified in the crystal structure. On the opposite side, the AcCoA molecule is embedded in a surface cleft extending from the regulatory domain toward the catalytic triad of PopP2, with the pantetheine arm of AcCoA untraceable, likely due to its high flexibility (Fig. 1c,d). The adenine moiety of AcCoA is buried in a hydrophobic pocket surrounded by the residues of both the catalytic core (K316, and S317) and the regulatory domain (Q386, V426, I444 and F446); while the 2’-hydroxyl, 3’-phosphate and 5’-pyrophosphate groups are hydrogen bonded to T445, S389 and K442, respectively (Fig. 2b). In the presence of RRS1-RWRKY, we observed additional hydrogen bonds formed between the 5’-pyrophosphate and PopP2 S432 on the loopβ8β9 (Supplementary Fig. 5), suggesting a stabilization of the PopP2 – RRS1-RWRKY interaction by AcCoA.
Figure 2. IP6-dependent acetyltransferase activity of PopP2.
a, Close-up view of the PopP2 – IP6 interaction. b, Close-up view of the PopP2 – AcCoA interaction. In a and b, the ligands are shown as green sticks, the hydrogen bonds are depicted as red dashed lines, and the water molecules are shown as red spheres. c, ITC binding curves of PopP2 mutants with AcCoA in the presence or absence of IP6. d, In vitro acetylation activity of wild-type (WT) or mutant PopP2. The acetylated proteins were detected by autoradiography (top), and the protein amount is indicated by Coomassie blue staining (bottom). e, Cell death-triggering activity of PopP2 in Arabidopsis eco. Nd-1. The asterisk indicates the cell death symptom triggered by wild-type PopP2. Ratios indicate number of leaves with indicated phenotype/total number of inoculated leaves in each treatment. Experiments in d and e were repeated twice and once, respectively, with consistent results.
We then performed Nuclear Magnetic Resonance (NMR) spectroscopy and Isothermal Titration Calorimetry (ITC) on the catalytic mutant PopP2C321A to evaluate the effect of IP6 binding on the PopP2 – CoA interaction. First, it is apparent that the addition of IP6 led to appearance of additional NMR signals in the aliphatic proton region of PopP2C321, suggestive of IP6-induced structural ordering (Supplementary Fig. 6a). Consistently, ITC analysis revealed that the presence of IP6 enhanced the binding of PopP2C321A to CoA from an undetectable level to a dissociation constant (Kd) of 57.2 µM. However, such an interaction was severely impaired when mutations of the IP6-binding sites (K383R or R416E) or AcCoA-binding sites (K316A/S389A/F446A) were introduced (Fig. 2c and Supplementary Table 2).
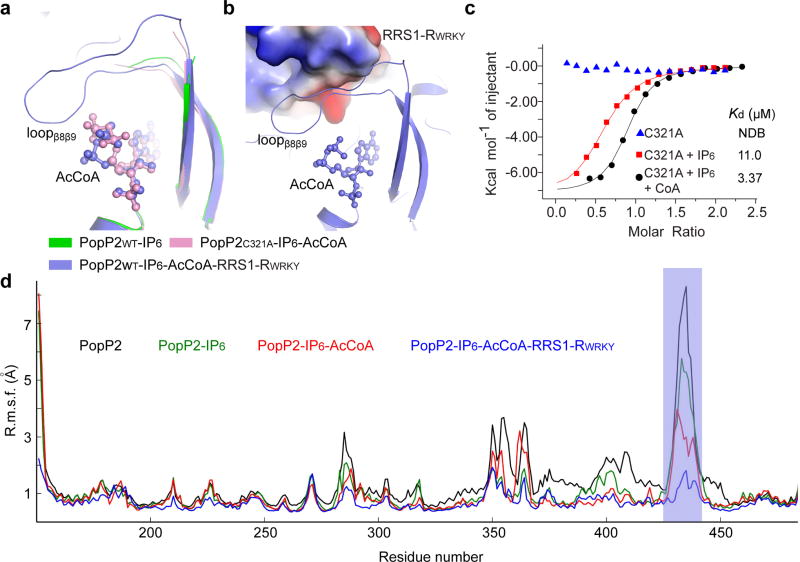
Furthermore, In vitro acetylation assay revealed that the presence of IP6 significantly increased the PopP2-mediated acetylation of RRS1-RWRKY, whereas the IP6- and AcCoA-binding mutations nearly or completely abolished the acetyltransferase activity (Fig.2d and Supplementary Fig. 6b,c). To test the activity of the PopP2 proteins in planta, PopP2 was fused to the N-terminal 79 residues of another T3SEs effector AvrRpt2, which serves as an artificial effector delivery system by bacterial pathogen Pseudomonas syringae pv. tomato (Pto)27. As expected, Pto strain D28E expressing wild-type PopP2 triggered programmed cell death in resistant Arabidopsis thaliana plants (eco. Nd-1), which produces RRS1-R (Fig. 2e). By contrast, PtoD28E expressing the mutants of PopP2 all failed to trigger cell death (Fig. 2e), confirming that these mutations resulted in the loss of biological function of PopP2. Note that these mutants showed similar protein stability (Supplementary Fig. 6d). Together, these studies suggest that IP6 enhances the activity of PopP2 by increasing its binding with AcCoA, a mechanism similar to that observed for HopZ1a.
Structural basis for the PopP2 – RRS1-RWRKY interaction
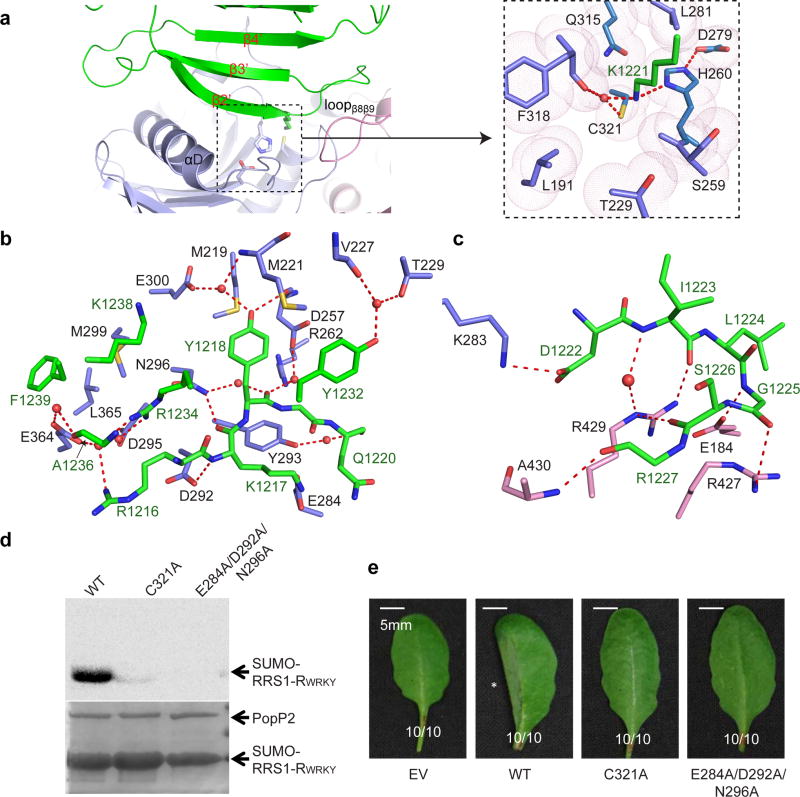
The association of RRS1-RWRKY with PopP2 is mediated by the αD-helix and three loops (loopβ1αC, loopβ2β3 and loopβ8β9) of PopP2, and the β2’-β3’ segment of RRS1-RWRKY (Fig 1d and Fig 3a). Notably, the strictly conserved WRKYGQK motif of RRS1-RWRKY (Supplementary Fig. 7a) is recognized by PopP2 through a number of hydrogen bonds and van der Waals contacts (Fig. 3b). Along the β2’-strand, the side chain of RRS1-RWRKY R1216 donates a water-mediated hydrogen bond to PopP2 E364; the backbone and side chain of RRS1-RWRKY K1217 form hydrogen bonds with PopP2 D292 and N296, and E284, respectively; the aromatic ring of RRS1-RWRKY Y1218 inserts into the cavity formed by PopP2 M219, M221 and D257, with its backbone and side chain engaging water-mediated hydrogen bonds with PopP2 D257, R262 and N296, and M221, D257 and E300, respectively (Fig. 3b); and the backbone of RRS1-RWRKY Q1220 forms a water-mediated hydrogen bond with PopP2 Y293. Most notably, the side chain of RRS1-RWRKY K1221, the target acetylation site, inserts into the catalytic pocket formed by PopP2 L191, S259, L281, Q315 and F318, with its side chain interacting with PopP2 H260, F318 and C321 through direct or water-mediated hydrogen bonds (Fig. 3a). Mutation of RRS1-RWRKY K1221 into glutamine, mimic of acetylated lysine, led to abolished DNA binding activity of RRS1-RWRKY (Supplementary Fig. 7b), supporting previous findings that PopP2-mediated acetylation disrupts the DNA binding activities of WRKY transcription factors15,16. C-terminal to the WRKYGQK motif, the loopβ2’β3’ stacks against two loops (loopβ5αE and loopβ8β9) of PopP2, with hydrogen bond interactions involving RRS1-RWRKY D1222, I1223, G1225, S1226 and R1227 and PopP2 E184, R427, R429 and A430 (Fig. 3c). Additional intermolecular contacts are mediated by the β3’-strand of RRS1-RWRKY (Y1232, R1234, A1236, K1238 and F1239), which makes hydrogen bonding or van der Waals contacts with loopβ1αC (V227 and T229), αD (D295, N296 and M299) and loop β6β7 (E364 and L365) of PopP2 (Fig. 3b).
Figure 3. The PopP2 – RRS1-RWRKY interaction.
a, The interface between PopP2 and RRS1-RWRKY, with the catalytic site shown in expanded view. b, Close-up views of the interactions of PopP2 with β2, β3 and Loopβ3β4 of RRS1-RWRKY. c, Close-up views of the interactions of PopP2 with loopβ2β3 of RRS1-RWRKY. Hydrogen bonds are depicted as dashed lines, and water molecules are shown as red spheres. d, In vitro acetylation activity of wild-type (WT) or mutant PopP2. The acetylated proteins were detected by autoradiography, and the protein amount is indicated by Coomassie blue staining. e, Cell death-triggering ability of PopP2 in Arabidopsis eco. Nd-1. The asterisk indicates the cell death symptom triggered by wild-type PopP2. Ratios indicate number of leaves with indicated phenotype/total number of inoculated leaves in each treatment. Experiments in d and e were repeated twice and once, respectively, with consistent results.
Interestingly, the majority of the PopP2-interacting residues is strictly conserved within the WRKY domains, and also required for DNA binding (Supplementary Fig 7a)28. We therefore tested whether PopP2 binding would directly affect the DNA binding of RRS1-RWRKY using electrophoretic mobility shift assay (EMSA). We observed that increasing amount of PopP2 led to a modest reduction in the DNA binding of RRS1-RWRKY (Supplementary Fig. 7c), suggesting that the PopP2 interaction contributes to the decreased DNA binding activity of WRKY domains.
To evaluate how the interaction with the WRKY domain affects the virulence activity of PopP2, we generated an E284A/D292A/N296A triple mutant, which eliminates the PopP2–RRS1-RWRKY binding (Supplementary Fig. 8 and Supplementary Table 2). Although this mutant retains the activities of CoA-binding (Supplementary Table 2) and auto-acetylation (Supplementary Fig. 9a), it was no longer able to acetylate RRS1-RWRKY in vitro, regardless of the presence of IP6 (Fig. 3d), and failed to induce cell death in the resistant Arabidopsis plants (Fig. 3e). These data suggest that the RRS1-RWRKY-interacting region of PopP2 is essential for its biological function in planta.
Distinct substrate binding strategies between YopJ effectors and ULPs
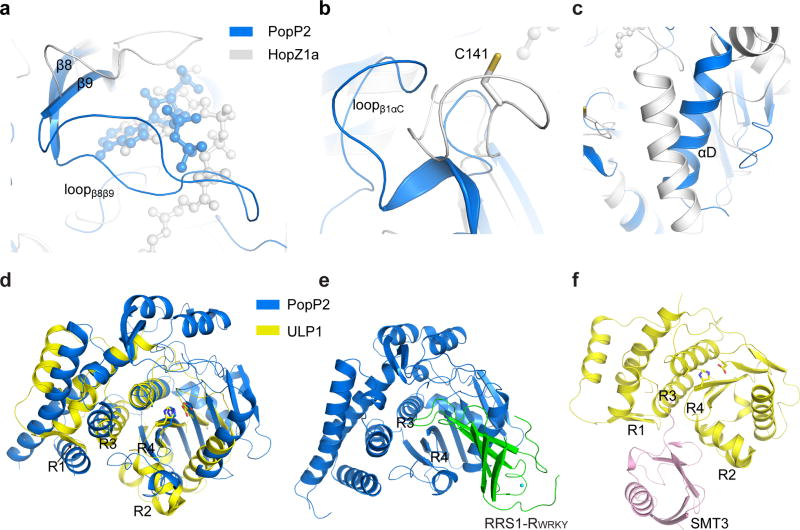
To assess how YopJ family of effectors evolves into a class of acetyltransferases with diverse substrate specificities, we compared the substrate binding sites of PopP2 with the corresponding regions of HopZ1a. Notably, the WRKY-binding sites of PopP2 (loopβ1αC, αD and loopβ8β9) are largely preserved in HopZ1a, albeit with distinct conformations (Fig 4a–c), implying that this region may also underlie the substrate specificity of HopZ1a. Consistently, our previous study revealed that introduction of a mutation (C141K) to the HopZ1a region, equivalent to loopβ1αC of PopP2 (Fig 4b), leads to altered allelic specificity of HopZ1b, an allele evolved from a HopZ1a–like ancestor29, suggesting that alteration in substrate binding might be an important mechanism for the evolution of YopJ effectors.
Figure 4. Structural comparison of PopP2WT – RRS1-RWRKY, HopZ1a and ULP1 – SMT3.
a–c, Structural overlay of the substrate binding sites of PopP2 (blue) with the corresponding regions in HopZ1a (grey). d, Overlaid structures of PopP2 (blue) and ULP1 (PDB 1EUV, yellow), with the catalytic triads shown in stick representation. e–f, Crystal structures of the PopP2WT – IP6 – AcCoA – RRS1-RWRKY e and ULP1 – SMT3 f complexes in the same orientation as shown in d. The substrate binding regions of ULP1, and the R3- and R4-equivalent regions in PopP2, are labeled.
Next, we performed structural comparison between PopP2WT – IP6 – AcCoA –RRS1-RWRKY and one of the closely related enzyme-substrate complexes, the ULP1-SMT3 complex30 (Fig. 4d–f). ULPs contain four conserved regions for recognition of SUMO- or ubiquitin-like substrates (R1 and R2) or guiding the C-terminus of the substrate toward the catalytic site (R3 and R4)31 (Fig 4d–f and Supplementary Fig. 1). By comparison, although PopP2 and ULP1 contain similar catalytic sites (R3 and R4), the substrate-recognition regions in ULP1 are either absent (in the case of R1) or structurally divergent (in the case of R2) in PopP2 (Fig. 4d). Notably, the R2-correponding region of PopP2 not only blocks R3, but also contributes to the formation of a surface groove right on top of the catalytic center, resulting in the distinct substrate-recognition site for PopP2 (Fig. 1d and Fig. 4e). In fact, the R1- and R2-corresponding regions are also structurally varied in HopZ1a (Fig. 1e), further highlighting the functional divergence between YopJ effectors and ULPs.
IP6 regulation of the PopP2 – WRKY interaction
The observation that the loopβ8-β9 of PopP2 is involved in binding to both AcCoA and RRS1-RWRKY (Fig. 1d) suggests a role of this region in linking cofactor binding to substrate binding. Indeed, structural comparison of the PopP2WT – IP6, PopP2C321A – IP6 – AcCoA and PopP2WT – IP6 – AcCoA – RRS1-RWRKY complexes reveals that the β8-β9 segment of PopP2 is partly stabilized by AcCoA binding, and becomes even more ordered upon RRS1-RWRKY binding (Fig. 5a,b). These observations, together with the observation that the loopβ8-β9-equivalent region in HopZ1a undergoes an IP6-induced structural ordering20, prompts us to investigate whether the IP6 or AcCoA binding regulates the PopP2 – RRS1-RWRKY association. Indeed, whereas the binding affinity of IP6-free PopP2 for RRS1-RWRKY was undetectable, addition of IP6 increased the binding to a Kd of 11.0 µM, and another ~3.3-fold increase when CoA was also added (Fig. 5c and Supplementary Table 2). By contrast, the IP6-binding deficient mutations (K383R and R416A) largely abolished the interaction (Supplementary Fig. 8 and Supplementary Table 2). Consistently, molecular dynamics (MD) simulations revealed that the root mean square fluctuation (RMSF) of the atomic fluctuations of PopP2 loopβ8β9 undergoes a gradual reduction upon binding to IP6, IP6 and AcCoA, or IP6, AcCoA and RRS1-RWRKY, suggesting that IP6 and AcCoA regulate the interaction of PopP2 with RRS1-RWRKY by inducing structural ordering of loopβ8-β9 (Fig. 5d). Taken together, these data establish that the IP6 binding not only allosterically stimulates the formation of AcCoA-binding pocket, but also regulates the substrate binding of PopP2.
Figure 5. IP6-stimulated binding between PopP2 with RRS1-RWRKY.
a, Close-up view of the conformational change of the β8-β9 segment of PopP2 in different complexes. AcCoA is shown in ball-and-stick representation. b, The structure of loopβ8β9 in the PopP2WT– IP6 – AcCoA – RRS1-RWRKY complex. The PopP2-binding site of RRS1-RWRKY is shown in electrostatic surface representation. c, ITC binding curves of PopP2 with RRS1-RWRKY in the presence or absence of IP6 and/or CoA. d, RMSF of atomic fluctuations of PopP2, free or in complex with IP6, AcCoA and/or RRS1-RWRKY. The region corresponding to loopβ8β9 is shaded in blue.
Structure of a PopP2 – AcCoA covalent adduct
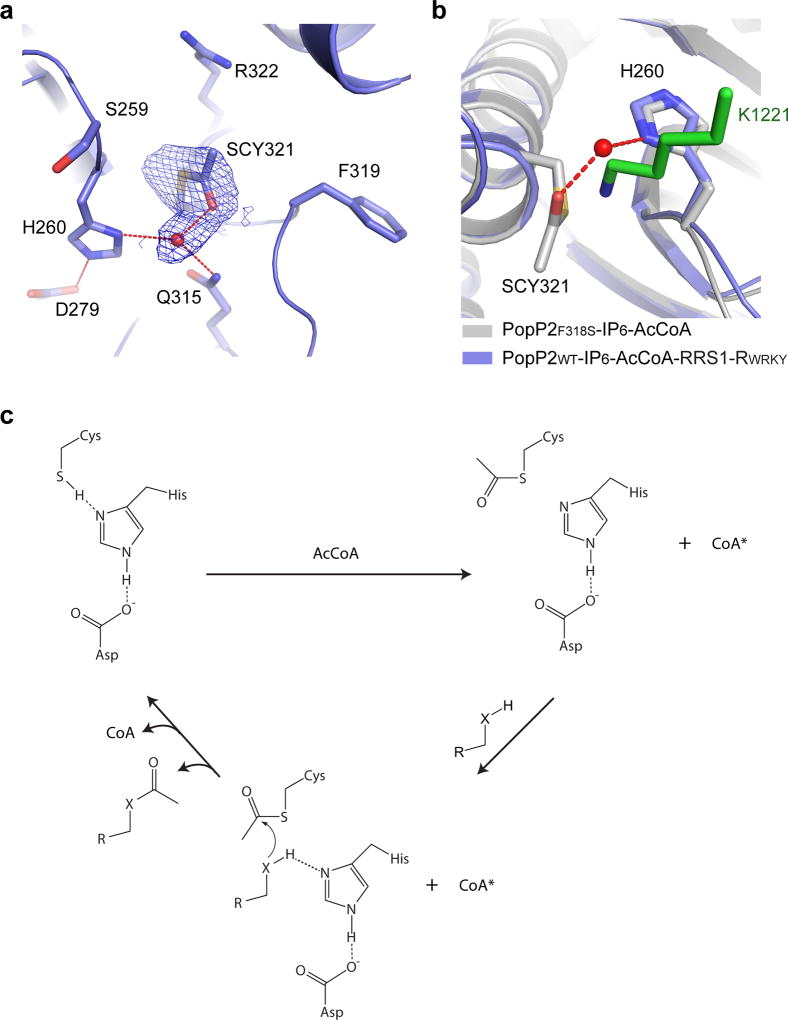
Given the fact that YopJ effectors contain a ULP-like catalytic triad, it has been proposed that they acetylate substrates via a “Ping-Pong” route of catalysis12, but not the “direct transfer” mechanism adopted by the GCN5-related N-acetyltransferase (GNAT) or the MYST family of acetyltransferases32–34. The “Ping-Pong” model predicts the presence of an acetyl-cysteine intermediate on the reaction pathway, generated by nucleophilic attack of the catalytic cysteine on AcCoA, which then reacts with the target residue to create an acetylated product12,35. To provide experimental support for this model, we captured the reaction intermediate between PopP2 and AcCoA by soaking AcCoA into the crystals of the IP6-bound, enzymatically active mutant PopP2F318S (Supplementary Fig. 9b). In comparison with wild-type PopP2, this mutant yielded a higher-quality crystal structure.
The overall structure of the PopP2F318S – IP6 – AcCoA complex is identical to that of the PopP2C321A – IP6 – AcCoA complex (Fig. 6a), except for the presence of an acetyl group covalently attached to the side chain of C321 (SCY321), surrounded by residues S259, H260, Q315, F319 and R322 (Fig. 6a). A water molecule, situated in a position that is otherwise occupied by the side chain amino group of the target lysine, bridges the acetyl group with H260 of the catalytic triad (Fig. 6a, b). Meanwhile, the water molecule also receives a hydrogen bond from another highly conserved residue, Q315 (Fig. 6a and Supplementary Fig. 1). The presence of a target lysine would presumably lead to eviction of this water molecule (Fig. 6b). Subsequently, hydrogen bonding with both H260 and the acetyl-cysteine adduct would permit the target lysine to initiate a nucleophilic attack of the acetyl-cysteine, resulting in substrate acetylation. Consistently, mutation of H260 into alanine leads to abolished enzymatic activity of PopP2 as well as PopP2-triggered cell death in Arabidopsis (Supplementary Fig. 9b,c), reinforcing the notion that the H260-equivalent residue serves as the catalytic base in YopJ effector-mediated acetylation.
Figure 6. A model for enzymatic catalysis of YopJ effectors.
a, Close-up view of the catalytic site in the crystal structure of PopP2F318S – IP6 – AcCoA, with the 2Fo-Fc omit map of the acetyl-cysteine (SCY321) contoured at 1.0σ level. The water molecule is shown as a red sphere and the hydrogen bonds are shown as dashed lines. b, The active site of PopP2F318S – IP6 – AcCoA overlaid with that of PopP2WT– IP6 – AcCoA– RRS1-RWRKY. The target lysine is shown in green stick. c, A model for the catalytic steps of YopJ effector-mediated acetylation. *The adenosine moiety of CoA remains bound to PopP2, whereas the pantetheine arm is released from the catalytic center. “X” denotes an NH2 group or oxygen atom.
Discussion
Enzyme-substrate interaction of PopP2 determines the outcome of pathogen invasion
The molecular interactions between bacterial effectors and host targets critically influence the outcome of pathogenic infections. Although PopP2 has evolved to target multiple WRKY transcription factors to suppress MTI, the incorporation of a decoy WRKY domain into the C-terminus of RRS1-R in resistant Arabidopsis plants successfully translates the virulence activity of PopP2 into defense activation15,16. This study provides the structural basis for the recognition of the conserved WRKYGQK motif by PopP2, which guides the insertion of the target lysine into the active site pocket. Strikingly, the IP6-induced structural ordering of loopβ8β9 enhances both AcCoA and WRKY bindings, providing a novel mechanism by which IP6 regulates the basal activity of PopP2. Given the structural similarity between PopP2 and HopZ1a, it is conceivable that such an IP6 regulation may help ensure efficient host-specific substrate recognition of both PopP2 and HopZ1a in plant cells.
Structural evolution and target recognition of YopJ effectors
YopJ effectors modify a diverse array of host proteins to promote pathogenesis8. Structural analysis of the PopP2WT – IP6 – AcCoA – RRS1-RWRKY complex suggests that YopJ effectors have evolved a distinct substrate-recognition strategy from that of ULPs. Furthermore, it provides insight into how YopJ effectors achieve distinct substrate specificities. In particular, the WRKY-binding sites of PopP2 (e.g. αD-helix), despite with structural homology with the corresponding region of HopZ1a, manifest substantial conformational and sequence divergence from other YopJ effectors (Supplementary Fig. 1), implying that the substrate binding sites of YopJ effectors are under strong selective pressure during arms race with specific hosts. Taken together, this study provides a framework for further understanding of the effector-target interaction of other YopJ effectors.
YopJ family effectors share a novel acetyltransferase mechanism
The finding that YopJ effectors contain a conserved ULP-like catalytic core challenges the current structure-function paradigm of acetyltransferases. Here, structural characterization of the acetyl-cysteine reaction intermediate of PopP2 lends a key support to a previous notion that YopJ effector-mediated acetylation mechanistically resembles the proteolysis process mediated by cysteine proteases24; that is, YopJ effectors employ a “Ping-Pong”-like route of catalysis involving sequential binding of AcCoA and protein substrates. It is also worth pointing out that the presence of the PopP2 – IP6 – AcCoA – RRS1-RWRKY quaternary complex also suggests that the substrate binding of YopJ effector may precede a complete release of the product CoA (Fig. 6c), which deviates from the classic “Ping-Pong” model.
Materials and Methods
Protein Preparation
The DNA sequences encoding PopP2, except for its N-terminal 148 residues that are predicted to be structurally disordered and responsible for transferring the effector into host cells by Ralstonia solanacearum, and RRS1-RWRKY (residues 1195–1273) were inserted in frame with a hexahistidine-SUMO tag in the pRSF-Duet vector. The recombinant proteins of PopP2 or RRS1-RWRKY were overexpressed in BL21(DE3) RIL cell strain overnight by the addition of 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 20°C when the cell density reached an OD600 of 0.8. For Se-Met labeling, cells cultured in M9 minimum medium to mid-log phase were induced for 16 hours after the addition of 50 mg/L of each of the following amino acids: lysine, phenylalanine, threonine, isoleucine, leucine, valine and L-selenomethionine. Cells were lysed in buffer containing 50 mM Tris-HCl (pH 8.0), 1 M NaCl, 25 mM imidazole, 0.5 mM DTT and 1 mM PMSF. The soluble fraction was subjected to a Nickel column. For purification of PopP2, the eluted fusion proteins were cleaved by ULP1, followed by further purification using hydrophobic interaction chromatography (Phenyl HP, GE Healthcare). Peak fractions were collected, concentrated and subsequently applied to Superdex 200 16/600 (GE Healthcare) pre-equilibrated with a buffer containing 25 mM Tris (pH 7.5), 200 mM NaCl, 5 mM IP6, 2 mM DTT and 5% glycerol. Purified PopP2 samples were stored in −80°C at a concentration of ~50 mg/mL for future use. For purification of RRS1-RWRKY, the RRS1-RWRKY protein in fusion with the SUMO tag was cleaved by ULP1. Subsequently, the RRS1-RWRKY protein was purified through a Heparin column (GE Healthcare) followed by size exclusion chromatography on a Superdex 200 16/600 column pre-equilibrated with a buffer containing 25 mM Tris-HCl (pH 7.5), 100 mM NaCl and 2 mM DTT.
Crystallization and structure determination
The PopP2C321A – IP6 and PopP2WT – IP6 complexes, prepared by mixing of the corresponding proteins with IP6 in a molar ration of 1:10, was crystallized by hanging-drop diffusion method. Crystals appeared in a buffer containing 0.1 M HEPES (pH 7.5), 20% (v/v) PEG4000 and 10% (v/v) glycerol. The PopP2F318S – IP6 complex was crystallized in a buffer containing 0.1 mM imidazole (pH 8.0), 30% (v/v) PEG8000, 200 mM NaCl and 4% (v/v) isopropanol. The PopP2WT – IP6 – RRS1-RWRKY complex was prepared by mixing wild-type PopP2 and RRS1-RWRKY in a molar ratio of 1:1.1 in a buffer containing 25 mM Tris-HCl (pH 7.5), 250 mM NaCl, 50 mM Arg/Glu, 5% glycerol and 2 mM IP6, and crystallized in a buffer containing 0.1 mM Tris-HCl (pH 8.5), and 20% ethanol. The crystals for the PopP2C321A – IP6 – AcCoA, PopP2F318S – IP6 – AcCoA and PopP2WT – IP6 – AcCoA – RRS1-RWRKY complexes were obtained by soaking crystals in the corresponding crystallization buffer supplemented with 5 mM AcCoA for overnight before flash frozen in liquid nitrogen. Crystallization buffer was supplemented with glycerol or PEG400 to a final concentration of 15–20% (v/v) as cryo-protectant before flash freezing crystals in liquid nitrogen. Diffraction data were collected on beamline 5.0.1 and 5.0.2 at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory and 24-IDE NE-CAT beamline at the Advanced Photo Source (APS), Argonne National Laboratory. The data were indexed, integrated and scaled by HKL2000 package36 or XDS37. The structure of PopP2C321A–IP6 complex was solved by the Se-SAD method and the initial model was built by the Autosol module embedded in the PHENIX software package38. Iterative cycles of model rebuilding and refinement were performed using COOT39 and PHENIX, respectively. The structures of the PopP2WT – IP6, PopP2C321A – IP6 – AcCoA, PopP2F318S – IP6 – AcCoA and PopP2WT – IP6 – AcCoA – RRS1-RWRKY complexes were solved by molecular replacement using PHASER40 with PopP2C321A – IP6 and/or WRKY1 (PDB 2AYD) structure as searching model.
The statistics for data collection and structural refinement for the PopP2 complexes is summarized in Supplementary Table 1.
ITC measurements
IP6 and AcCoA were dissolved in water at an initial concentration of 100 mM (adjusted to pH 7.5 by 0.1 mM NaOH) and 30 mM, respectively, and diluted to 1 mM by the ITC buffer before the titration. Protein samples for the RRS1-RWRKY domain, wild-type, and mutant PopP2 were dialyzed against the ITC buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2 mM IP6 and 5% glycerol) at 4°C overnight. A MicroCal iTC200 system (GE Healthcare) was used to conduct the ITC measurements. A total of 17–20 injections with a spacing of 180 s and a reference power of 5 µcal/s were performed at 25°C. The ITC curves were processed with software ORIGEN (MicroCal) using one-site fitting model.
In vitro acetylation assay
In vitro acetylation assays were carried out to examine the acetyltransferase activity of PopP2. For [14C] labeling experiments, 4 µg PopP2 were incubated with [14C]-acetyl-CoA (55 µci/µmol) in 25 µL reactions (50 mM HEPES (pH 8.0), 150mM NaCl, 1 mM DTT, 1 mM PMSF, 10 mM sodium butyrate and 5% glycerol) at room temperature for an hour. 22.5 µg SUMO-RRS1-RWRKY was used as the substrates of PopP2. 10 nM IP6 and 30 µM AcCoA were added. The reactions were stopped by 4× Laemmli buffer and subjected to 12% SDS-PAGE. Acetylated proteins were detected after five days. After autoradiography, the protein gels were stained with Coomassie Blue as a loading control.
For immunoblot experiments, similar experiments were carried out except that 60µM 12C-acetyl-CoA was used instead. Acetylated proteins were detected using Ac-K antibody (Cell signaling, Ac-K-103). The protein gels were stained with Coomassie Blue as a loading control
Pseudomonas syringae infection assays
DNA sequence encoding N-terminal AvrRpt2 (residue 1–79) fused with truncated wild-type and mutant PopP2 carrying a C-terminal HA tag were cloned into the vector pUCP20tk. The genes are under the control of native avrRpt2 promoter. The plasmids were transformed into Pseudomonas syringae pv. tomato strain D28E (PtoD28E), which is an effector-less mutant of wild-type strain DC3000. The expression and stability of the chimeric proteins in PtoD28E was confirmed by immunoblotting using an anti-HA antibody (Roche Diagnostics) after the cells were induced in MG minimal medium for two days. Secretion of the chimeric proteins in plant cells during bacterial infection was not checked.
For hypersensitive response assay, P. syringae strains were grown on mannitol-glutamate (MG) agar at room temperature for two days. Leaves of five-week-old Arabidopsis thaliana plants (eco. Nd-1) were infiltrated with bacterial suspensions at OD600 = 0.4 (approximately 4×108 cfu/mL) in 10 mM MgSO4. The inoculated plants were transferred to a growth chamber (22°C and 16/8 light/dark regime, 90% humidity). Cell death symptoms were monitored at 24 hours post inoculation (hpi).
Molecular dynamics (MD) simulations
Protein structure was prepared with AmberTools1641. The pantetheine arm of AcCoA was interactively docked using YetiX42 using the solved crystal structure as template. Each complex was explicitly solvated in a rectangular box of TIP3P model water (at least 12 Å from the complex to avoid periodic artifacts from occurring) by using AMBER16 with the ff14SB and gaff2 force field for protein and ligands. Ligand partial charges were calculated by RESP fit with Gaussian09 using the B3LYP/6–31G* basis set after geometry optimization. The Zinc metal center parameters of the RRS1-RWRKY domain were processed with ZAFF parameters43. For each system, 200ns production MD simulations were performed for trajectory analysis. The last 50ns trajectory were subjected to RMSF analysis of C-alpha coordinates by cpptraj44.
Electrophoretic mobility shift assay
The samples were prepared by mixing various amounts of PopP2, RRS1-RWRKY domain and a 16 mer W-box DNA duplex (upper strand: CGCCTTTGACCAGCGC) in a total volume of 40 µL of solution containing 25 mM Tris·HCl (pH7.5), 200 mM NaCl and 5 mM DTT. For titration of PopP2, the sample mixture was supplemented with 2 mM IP6 and 1.5 mM AcCoA, and incubated on ice for 1 hr before gel electrophoresis. The samples were loaded onto 10% non-denaturing polyacrylamide gel and electrophoresed at 4°C in 0.25 × TBE buffer. The gel was stained by ethidium bromide and visualized under UV light.
Data availability
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes PDB 5W3T (PopP2WT – IP6), PDB 5W3Y (PopP2C321A – IP6 – AcCoA), PDB 5W40 (PopP2F318S-IP6-AcCoA) and PDB 5W3X (PopP2WT – IP6 – AcCoA – RRS1-RWRKY).
Supplementary Material
Acknowledgments
This work was supported by NIH (1R35GM119721), Kimmel Scholar Award from Sidney Kimmel Foundation for Cancer Research and March of Dimes Foundation (1-FY15-345) to J.S., and grants from US NSF (IOS#0847870) and the USDA Agriculture Experimental Station Funding (CA-R-PPA-5075-H) to W.M. We would like to thank staff members at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory for access to X-ray beamlines.
Footnotes
Author Contributions
Z-M. Z. and L. G. determined the crystal structures of PopP2 complexes and conducted ITC assays. J.S. and L. G. performed NMR analysis. K-W. M. and S.S. performed in vitro acetylation assays and in vivo functional analyses. Z.H. performed computational analysis. W.M and J.S. designed and organized the study, Z-M. Z., K-W. M., W.M. and J.S. prepared the manuscript.
Competing Financial Interests
The authors declare no competing financial interest.
References
- 1.Zipfel C. Plant pattern-recognition receptors. Trends Immunol. 2014;35:345–351. doi: 10.1016/j.it.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T. Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol. 2014;20:47–54. doi: 10.1016/j.pbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 4.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-Microbe Interactions: Shaping the Evolution of the Plant Immune Response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 2011;35:1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- 6.Büttner D. Behind the lines–actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 2016;6:894–937. doi: 10.1093/femsre/fuw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa TRD, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Micro. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 8.Ma K-W, Ma W. YopJ Family Effectors Promote Bacterial Infection through a Unique Acetyltransferase Activity. Microbiol. Mol. Biol. Rev. 2016;80:1011–1027. doi: 10.1128/MMBR.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis JD, et al. The YopJ superfamily in plant-associated bacteria. Mol. Plant. Pathol. 2011;12:928–937. doi: 10.1111/j.1364-3703.2011.00719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones RM, et al. Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell host & microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci U S A. 2006;103:18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 13.Paquette N, et al. Serine/threonine acetylation of TGFbeta-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc Natl Acad Sci U S A. 2012;109:12710–12715. doi: 10.1073/pnas.1008203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trosky JE, et al. VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J Biol Chem. 2007;282:34299–34305. doi: 10.1074/jbc.M706970200. [DOI] [PubMed] [Google Scholar]
- 15.Le Roux C, et al. A Receptor Pair with an Integrated Decoy Converts Pathogen Disabling of Transcription Factors to Immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 16.Sarris, Panagiotis F, et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 17.Lee AH-Y, et al. A Bacterial Acetyltransferase Destroys Plant Microtubule Networks and Blocks Secretion. PLoS Pathog. 2012;8:e1002523. doi: 10.1371/journal.ppat.1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang S, et al. Bacterial Effector Activates Jasmonate Signaling by Directly Targeting JAZ Transcriptional Repressors. PLoS Pathog. 2013;9:e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orth K, et al. Disruption of Signaling by Yersinia Effector YopJ, a Ubiquitin-Like Protein Protease. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z-M, et al. Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family. Nat. Struct. Mol. Biol. 2016;23:847–852. doi: 10.1038/nsmb.3279. [DOI] [PubMed] [Google Scholar]
- 21.Ma K-W, et al. Two serine residues in Pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor. New Phytol. 2015:1157–1168. doi: 10.1111/nph.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheong MS, et al. AvrBsT Acetylates Arabidopsis ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity. PLoS Pathog. 2014;10:e1003952. doi: 10.1371/journal.ppat.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittal R, Peak-Chew SY, Sade RS, Vallis Y, McMahon HT. The Acetyltransferase Activity of the Bacterial Toxin YopJ of Yersinia Is Activated by Eukaryotic Host Cell Inositol Hexakisphosphate. J. Biol. Chem. 2010;285:19927–19934. doi: 10.1074/jbc.M110.126581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee S, Hao Y-H, Orth K. A newly discovered post-translational modification – the acetylation of serine and threonine residues. Trends Biochem Sci. 2007;32:210–216. doi: 10.1016/j.tibs.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Duan M-R, et al. DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 2007;35:1145–1154. doi: 10.1093/nar/gkm001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tasset C, et al. Autoacetylation of the Ralstonia solanacearum Effector PopP2 Targets a Lysine Residue Essential for RRS1-R-Mediated Immunity in Arabidopsis. PLoS Pathog. 2010;6:e1001202. doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman DS, Greenberg JT. Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Molecular plant-microbe interactions : MPMI. 2001;14:145–155. doi: 10.1094/MPMI.2001.14.2.145. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki K, et al. Structural Basis for Sequence-specific DNA Recognition by an Arabidopsis WRKY Transcription Factor. J. Biol. Chem. 2012;287:7683–7691. doi: 10.1074/jbc.M111.279844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan RL, et al. Catalytic domain of the diversified Pseudomonas syringae type III effector HopZ1 determines the allelic specificity in plant hosts. Mol Microbiol. 2010;76:437–455. doi: 10.1111/j.1365-2958.2010.07107.x. [DOI] [PubMed] [Google Scholar]
- 30.Mossessova E, Lima CD. Ulp1-SUMO Crystal Structure and Genetic Analysis Reveal Conserved Interactions and a Regulatory Element Essential for Cell Growth in Yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 31.Pruneda, Jonathan N, et al. The Molecular Basis for Ubiquitin and Ubiquitin-like Specificities in Bacterial Effector Proteases. Mol. Cell. 2016;63:261–276. doi: 10.1016/j.molcel.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vetting MW, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Utley RT, Côté J. The MYST Family of Histone Acetyltransferases. Curr Top Microbiol Immunol. 2003;274:203–236. doi: 10.1007/978-3-642-55747-7_8. [DOI] [PubMed] [Google Scholar]
- 34.Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic Mechanism of a MYST Family Histone Acetyltransferase. Biochem. 2007;46:623–629. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cleland WW. Statistical Analysis of Enzyme Kinetic Data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case DA, Betz RM, Botello-Smith W, Cerutti DS, Cheatham I. T.E. AMBER 2016. University of California; San Francisco: 2016. [Google Scholar]
- 42.Rossato G, Ernst B, Smiesko M, Spreafico M, Vedani A. Probing Small-Molecule Binding to Cytochrome P450 2D6 and 2C9: An In Silico Protocol for Generating Toxicity Alerts. ChemMedChem. 2010;5:2088–2101. doi: 10.1002/cmdc.201000358. [DOI] [PubMed] [Google Scholar]
- 43.Peters MB, et al. Structural Survey of Zinc-Containing Proteins and Development of the Zinc AMBER Force Field (ZAFF) J. Chem. Theory Comput. 2010;6:2935–2947. doi: 10.1021/ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes PDB 5W3T (PopP2WT – IP6), PDB 5W3Y (PopP2C321A – IP6 – AcCoA), PDB 5W40 (PopP2F318S-IP6-AcCoA) and PDB 5W3X (PopP2WT – IP6 – AcCoA – RRS1-RWRKY).