Abstract
Circulating tumor cells (CTCs) enter the vasculature or lymphatic system after shedding from the primary tumor. CTCs may serve as “seed” cells for tumor metastasis. The utility of CTCs in clinical applications for sarcoma is not fully investigated, partly owing to the necessity for fresh blood samples and the lack of a CTC-specific antibody. To overcome these drawbacks, we developed a technique for sarcoma CTCs capture and detection using cryopreserved peripheral blood mononuclear cells (PBMCs) and our proprietary cell-surface vimentin (CSV) antibody 84-1, which is specific to tumor cells. This technique was validated by sarcoma cell spiking assay, matched CTCs comparison between fresh and cryopreserved PBMCs, and independent tumor markers in multiple types of sarcoma patient blood samples. The reproducibility was maximized when cryopreserved PBMCs were prepared from fresh blood samples within 2 hours of the blood draw. In summary, as far as we are aware, ours is the first report to capture and detect CTCs from cryopreserved PBMCs. Further validation in other types of tumor may help boost the feasibility and utility of CTC-based diagnosis in a centralized laboratory.
Keywords: Sarcoma, Circulating tumor cells, Cell surface vimentin, Cryopreservation, Peripheral blood mononuclear cells
1. Introduction
Sarcoma is a rare group of mesenchymal origin tumors, accounting for nearly 20% of pediatric malignancies and less than 2% of adult neoplasms [1, 2]. Despite the low incidence of sarcoma, it represents a much larger proportion in adolescents and young adults with high mortality rate due to late diagnosis and relapse. A potential new approach for the early detection of relapse is to capture the circulating tumor cells (CTCs) from peripheral blood of sarcoma patients who are under remission. CTCs are “seed” cells for tumor metastasis that are shed into the circulatory or lymphatic system from the primary tumor [3, 4]. These cells have attracted attention due to their potential role in early diagnosis and monitoring of therapeutic response to anti-cancer drugs [5–7]. At present, the CellSearch system is the only technique approved by the US Food and Drug Administration for the detection and enumeration of CTCs in metastatic breast, colorectal, and prostate cancers in the clinical setting [8–11]. CellSearch captures CTCs by utilizing the epithelial cell adhesion molecule (EpCAM) which, as its name suggests, is overexpressed only in epithelial cancer types [12, 13]. However, this marker is not effective in capturing CTCs originating from mesenchymal tumors such as sarcoma, and might even miss some of CTCs undergoing epithelial–mesenchymal transition (EMT) [12, 14]. Thus, a novel technique for accurately detecting CTCs from sarcoma patients’ peripheral blood is quite necessary.
Previously, we have reported that cell-surface vimentin (CSV) is a marker unique to different types of tumor cells [15–17]. By utilizing CSV as a specific target, we captured and enumerated mesenchymal-derived CTCs and EMT-like CTCs from fresh blood samples of patients bearing different types of cancer with high sensitivity and specificity [14, 18]. However, to the best of our knowledge, the current CTCs capture techniques require fresh blood samples [19]. A reliable and reproducible cytometric technique for the enumeration of CTCs from cryopreserved samples is still lacking. Fresh samples have to be processed within 72 hours as collection to maintain the reproducibility [20]. Transportation from multiple laboratories is not only expensive but also may affect the reproducibility of CTCs measurement. All the above barriers limit the application of current CTCs isolation techniques for large multiple-center trials. To boost CTCs assay utility, cryopreserved sample-based CTCs capture should be investigated.
In the current study, we investigated an assay for capturing CTCs from cryopreserved peripheral blood mononuclear cells (PBMCs) from patients with various types of sarcoma using the tumor specific CSV antibody 84-1. The new isolation step can be highly time limiting, which prevents large numbers of samples being processed on the same day. Such a technology will boost the feasibility and utility of CTC-based diagnosis and therapeutic treatment monitoring in large multiple-center trials.
2. Materials and Methods
2.1 Patient eligibility and recruitment
Patients with metastatic cancer disease were consented in the Department of Laboratory Medicine and Sarcoma Center at The University of Texas MD Anderson Cancer Center. Blood was drawn either before or at least 7 days after intravenous therapy. Blood samples from healthy donors were obtained from Gulf Coast Blood Center in Houston, Texas. The healthy donors had no known disease or infection at the time of blood draw and no history of malignant disease. The study was approved by our institutional review board (Protocol: PA13-0353 and LAB06-0581). Informed and written consent was obtained from all the patients involved in this project.
2.2 Blood collection and cryopreserved PBMC preparation
Freshly drawn blood samples from patients were collected from patients in 10-mL BD Vacutainer tubes with K2 EDTA (BD Diagnostics Franklin Lakes, New Jersey) (Fig. 1A). A total of 500 mL of fresh blood from healthy donors was also collected. Blood was processed into 50 mL of Buffy Coat, which contained mononuclear cells and neutrophils, as well as smaller numbers of contaminating red blood cells, plasma, and platelets (Fig. 1B). All the blood samples were processed within 2 hours of blood collection for best results. Obtained blood was pooled into 50-mL conical tubes and diluted with room-temperature phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS). Next, the diluted blood was layered carefully over 15 mL of Ficoll-Paque PLUS density gradient medium (Ficoll Paque Premium, GE Healthcare, Pittsburgh, PA) in a 50-mL SepMate™ tube (StemCell™ Technologies, Vancouver, Canada) while continuously pipetting the diluted blood down the side of the tube (Fig. 1C).
Figure 1. Isolation of human peripheral blood mononuclear cells (PBMCs) and selection of cell-surface vimentin (CSV)-positive circulating tumor cells (CTCs).
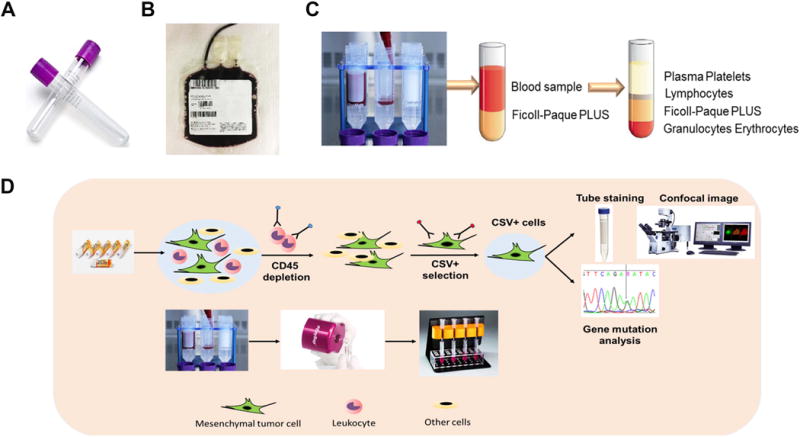
(A) Vacutainers used to collect the blood. (B) Buffy coat containing mononuclear cells, neutrophils, smaller numbers of contaminating red blood cells, plasma and platelets. (C) Separation of blood cell types in the SepMate™ tubes. (D) Schematic representation of 84-1+/CD45- CTCs selection, enumeration and analysis.
The samples were centrifuged at 1200 × g for 10 minutes at room temperature with the brake on. After density gradient centrifugation, differential migration of cells during centrifugation resulted in the formation of layers containing different cell types. The bottom layer contained erythrocytes and granulocytes. PBMCs could be found together with other low-density slowly sedimenting particles (e.g., platelets) at the interface between the plasma and the Ficoll-Paque layer (Fig. 1C). The PBMCs were harvested by pouring the top layer, transferred to a new tube and then washed twice with PBS containing 2% FBS at room temperature. The cell pellet was resuspended with medium at room temperature. After cell counting, PBMCs were cryopreserved at 6 × 106 (6 million, nearly 6 mL of whole blood) cells per 1 mL and placed inside a Nalgene® Mr. Frosty® Cryo 1°C Freezing Container (Thermo Fisher Scientific, Waltham, MA) container at −80°C for 48 hours, and the cells were later transferred to liquid nitrogen until further experiments were performed.
2.3 84-1+ cell selection
The selection and validation methods used to isolate and enrich 84-1+ cells were previously described [15]. Briefly, an antibody against human Fc receptor (Miltenyi Biotec, Auburn, CA) was added to the PBMCs cell suspension to reduce nonspecific binding. CD45+ immune cells were then depleted using an EasySep human CD45 depletion kit (StemCell™ Technologies) according to the manufacturer’s recommendation. Later, the CD45- cell fractions from the blood were labeled with the 84-1 anti-vimentin antibody, followed by the addition of mouse IgG-binding microbeads (Miltenyi Biotec) to the mixture after incubation. The labeled cells were then extracted using the magnetic column according to the manufacturer’s protocol (Miltenyi Biotec). The 84-1+ and CD45- cells obtained were CSV+ CTCs ready for further analysis (Fig. 1D).
2.4 Immunofluorescence imaging
For immunofluorescence, the cell pellet was mixed with MACS buffer (Miltenyi Biotec) and stained with 84-1 antibody (1:200) in tube at room temperature for 1 hour. Cells were cytospun onto Polysine™ microscope adhension slides (Thermo Fisher Scientific) by Cytofuge (Iris, Westwood, MA). Cells were fixed with 4% paraformaldehyde (Fisher Scientific) and blocked in blocking buffer (1% FBS in PBS) for 1 hour. For the staining of other markers, such as CD45 (1:100; Abcam, Cambridge, MA), α-SMA (1:100; Abcam), CD117 (1:100; Cell Signaling Technology, Danvers, MA), and MDM2 (1:100; Santa Cruz Biotechnology Santa Cruz, CA), the cells were incubated with primary antibody overnight in a cold room followed by permeabilization in PBS (pH 7.4)/0.2% NP40 (Sigma Aldrich) for 20 minutes. Next, the slides were washed with PBS three times and stained with Alexa Fluor-555 secondary antibody (1:100; Invitrogen, Carlsbad, CA) for CD45, Alexa Fluor-647 (1:100; Invitrogen) for 84-1, and Sytox Green (1:200; Invitrogen) for nuclei staining for 1 hour at room temperature. Then the slides were washed with PBS three times and mounted in Slow fade antifade (Invitrogen). All slides were visualized by Zeiss LSM 510 confocal microscope using LSM 5 3.2 image capture and analysis software (Carl Zeiss, Thornwood, NY).
2.5 Spiking assay
First, all cells used for the spiking assay were subjected to 84-1+ expression selection one day before the analysis. After being labeled with CFSE tracking dye (eBioscience, San Diego, CA), the cells were diluted to the required count numbers. For sensitivity analysis, 0, 5, or 25 LM7 cells (see below) labeled with CFSE were spiked into 6 million PBMCs (nearly isolated from 6 mL of whole blood). PBMCs with spiking cells were cryopreserved with an initial 1 mL of prepared ice-cold freezing media (90% FBS and 10% DMSO). Spiking assays were performed in triplicate to test the reproducibility of the cell recovery rate using this method.
2.6 LM7 cell line and culture conditions
Human osteosarcoma cell line LM7 was kindly provided by Dr. Eugenie S. Kleinerman (MD Anderson). LM7 cells were cultured in DMEM/F12 (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS and 10 U/mL penicillin and streptomycin (Life Technologies, Carlsbad, CA) in an atmosphere of 5% CO2 at 37°C. Cells were subcultured every 2–3 days and harvested in the logarithmic phase of growth. Cell viability was assessed using the trypan blue dye (Life Technologies) assay. Live cells with viability greater than 98% were used for spiking assay experiments.
2.7 Statistical analysis
The average spiking cell recovery data were expressed as the mean ± standard deviation and representative results were from at least three independent experiments. The correlation between spiking cell count and recovered cell count, as well as reproducibility of CTC measurements, was assessed using liner regression and Bland-Altman analysis. For Bland-Altman analysis, the error of each CTC count was determined by the difference in the CTC count between two samples divided by the mean CTC count of both samples, and these errors were plotted on a graph. All statistical analyses were performed using GraphPad Prism software. P values less than 0.05 were considered significant.
3. Results
3.1 The modified technique enables capture of CTCs from cryopreserved PBMCs
We previously established a method in which enriched CSV-positive CTCs were stained with 84-1 antibody after fixation for immunofluorescence imaging [14]. However, this method was not a feasible approach for cryopreserved PBMC samples. CSV-positive CTCs, captured from cryopreserved samples, stained positive for 84-1 antibody both on the cell surface (CSV) and in the cytoplasm (intracellular vimentin; Fig. 2A). Our previous published results showed that CSV was specific to cancer cells and did not appear on normal immune cells occasionally co-captured with CTCs when using fresh blood samples. However, normal CD45+ immune cells were also stained positively with CSV antibody when newly thawed PBMCs were used (Fig. 2B). It appeared that the thawed cells were too fragile to be membrane-permeabilized during spinning and fixation with paraformaldehyde. Thus, it was not feasible to accurately measure CSV-positive CTCs from cryopreserved samples via the previous staining method, although the CTCs were generally large (>10 μm). Plus, there tended to be more contamination with normal blood cells when cryopreserved samples were used, which made counting difficult. Therefore, the current CTCs staining technique must be modified to reduce non-specific staining in CTCs analysis.
Figure 2. Modified technique for CSV marker staining in captured CTCs from osteosarcoma patient and normal PBMCs that have been cryopreserved and thawed.

(A–B) Immunofluorescent staining for 84-1 and CD45 antibody in both captured CTCs and normal immune cells after fixation. (C–D) Immunofluorescent staining for 84-1 and CD45 in both captured CTCs and normal immune cells before fixation (non-permeabilized cells). CSV (84-1, green), CD45 (red) and nuclear stain (blue). Scale indicates 10 μm.
To solve this problem, we stained the captured CTCs from newly thawed osteosarcoma patient samples with 84-1 antibody before spinning the cells onto slides and fixing the cells. Compared with cells stained after spinning on slides and fixation, those stained prior to spinning on slides and fixation showed specific staining of vimentin on the tumor cell surface (Fig. 2C). Additionally, normal CD45+ immune cells were not observed with CSV staining in the newly thawed PBMCs (Fig. 2D). These results indicated that staining cells with 84-1 CSV antibody prior to spinning on slides and fixation holds the key for specifically detecting CSV on CTCs captured from cryopreserved and thawed PBMCs. This assay also improved the quality of CTCs staining in fresh blood samples (data not shown).
3.2 The modified technique is highly sensitive according to the cell spiking assay
To examine the sensitivity of the modified technique for enumerating CTCs in cryopreserved PBMC samples, we performed the spiking assay using cryopreserved PBMCs from healthy donors. Different concentrations of CFSE-labeled LM7 cells were spiked into 1 mL of PBMCs (isolated from 6 mL of blood). Some of the fresh samples spiked with LM7 cells were processed immediately using the CSV-positive CTC isolation protocol described above (schematic representation in Fig. 1D), and the rest were cryopreserved and processed 1 week later. The live recovered cells in the thawed samples were subjected to immunofluorescence staining with the 84-1 antibody, using the modified technique.
Confocal microscopy revealed that CFSE-labeled cells captured from cryopreserved PBMCs using the modified staining technique described above were as easily detectable as those in the fresh samples (Fig. 3A). The recovery rates were >75% in both the fresh and cryopreserved samples (Fig. 3B, D). As expected, the coefficient of variation for cryopreserved samples increased as the number of spiked cells decreased, ranging from 5.26% at 25 spiked cells to 13.3% at 5 spiked cells (Fig. 3D). Linear regression analysis yielded a positive correlation between the number of detected LM7 cells and the number of LM7 cells spiked in both of the fresh and cryopreserved PBMCs (P<0.05; Fig. 3C, E). These results indicated high sensitivity for CSV-positive CTCs capture in both cryopreserved samples and fresh samples using the modified technique.
Figure 3. Spiking assay results.
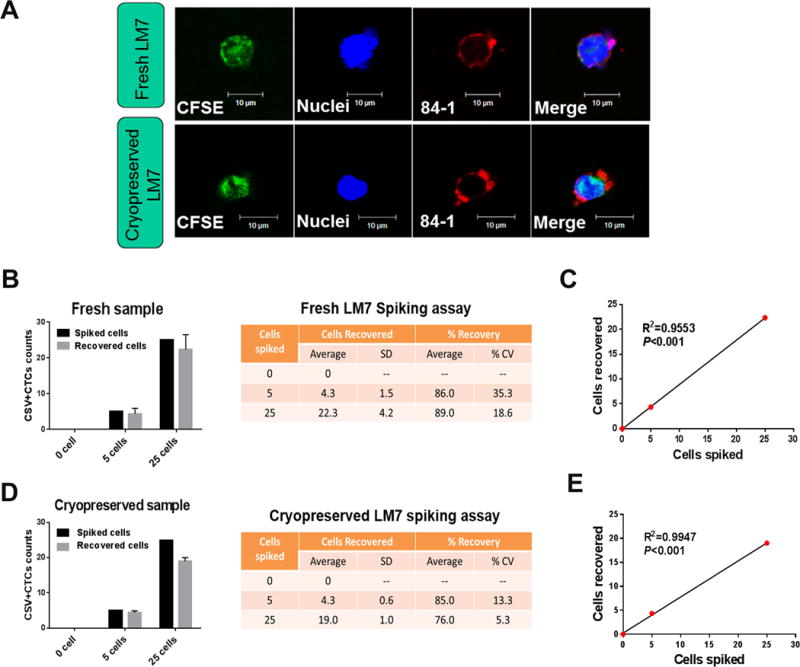
(A) Micrographs showing CFSE-labeled LM7 cells in the fresh and cryopreserved peripheral blood mononuclear cells from healthy donors. Graphs and tables show the recovery rates for each concentration of spiked cells in the fresh samples (B) and cryopreserved samples (D). The mean results are from at least three independent experiments (error bars indicate standard deviation). Correlations of CTCs counts between the numbers of recovered cells and spiked cells in the fresh samples (C) and cryopreserved samples (E). CSV (84-1, red), tracker dye CFSE (green) and nuclear stain (blue). Scale indicates 10 μm. SD, standard deviation; CV, coefficient of variation.
3.3 CTCs capture from cryopreserved PBMCs using the modified technique is reproducible in patient blood samples across duplicate tubes
To measure the reproducibility of CTCs capture with the modified technique, we collected six blood samples from metastatic osteosarcoma patients. Each sample was aliquoted into two tubes and stored in liquid nitrogen for up to 2 to 3 years before analysis. Vimentin was specifically detected on the cell surface in these 12 cryopreserved samples using the modified staining technique (Fig. 4A). Regression analysis indicated a slope of 1.33 and a correlation coefficient (R2) of 0.934 (P=0.002) across duplicate tubes (Fig. 4B). The data from all samples with an average duplicate CTCs counts (n=12) were analyzed using a Bland-Altman plot (Fig. 4C). The error of each CTC enumeration was represented by the difference in the CTCs counts between tube1 and tube2 divided by the average of both CTCs counts. The largest differences in the average CTCs counts were located within the range of mean difference ± 1.96 standard deviation (SD; −7.12 to 12.78), and no value was out of the range.
Figure 4. Comparison of CTCs counts between matched samples.
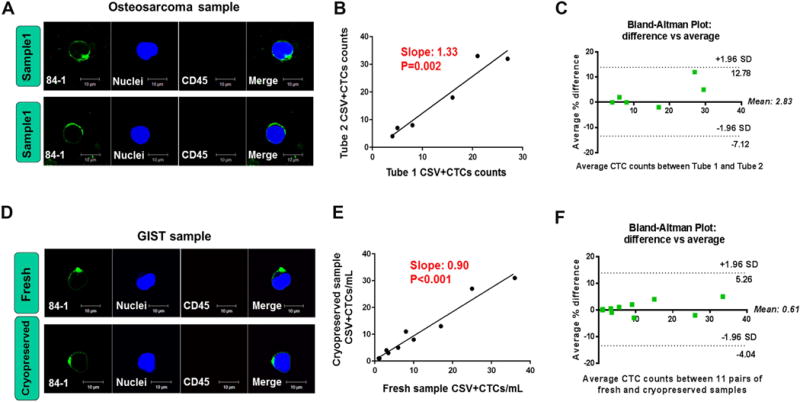
(A–C) Duplicate samples of osteosarcoma PBMCs (N=6) that had been cryopreserved for 2 to 3 years were measured. (D–F) Matched fresh and cryopreserved samples from multiple types of metastatic sarcoma were compared (N=11). (A, D) Immunofluorescent staining for cell surface vimentin and CD45. (B, E) Correlations between CTCs counts for matched samples. (C, F) Bland-Altman plots of the average differences between the matched samples. CSV (84-1, green), CD45 (red), nuclear stain (blue). GIST, Gastrointestinal stromal tumor. Scale indicates 10 μm.
To validate this cryopreserved sample-based and modified staining technique further, we also enumerated CTCs in 11 pairs of fresh and cryopreserved PBMCs from patients with various types of metastatic sarcoma to confirm the reproducibility. The blood sample was divided into two tubes after PBMCs isolation. CTCs enumeration using the modified technique was performed immediately in one tube and after 1 week of cryopreservation at −80°C in the other. We did not observe any obvious effect of the freeze-thaw procedure on morphologic features or biomarker staining in the matched samples (Fig. 4D). Positive correlation of CTCs counts was shown between the fresh and cryopreserved PBMCs from the same patient (P <0.001; Fig. 4E). Furthermore, duplicate measurements showed very little variability in CTCs counts; the largest differences in the average CTCs counts were located within the range of mean difference ± 1.96 standard deviation (SD) and no values were outside of the range according to Bland-Altman analysis (Fig. 4F). The error of each CTC measurement was represented by the difference in the CTCs counts between fresh and cryopreserved samples divided by the average of both CTCs counts. These results collectively confirmed that the CTCs counts obtained using the modified technique were reproducible.
3.4 Time lapse between collection and cryopreservation of PBMCs affects the reproducibility of CTCs capture
To determine whether time lapse between collection and cryopreservation of PBMCs affects the reproducibility of CTC measurement, we compared the number of CTCs in both fresh and cryopreserved samples for different time lapses between collection and cryopreservation of PBMCs in seven metastatic sarcoma samples and six epithelial cancer samples. As shown in Fig. 5A, 2 hours may be the maximum allowable delay time for cryopreservation because CTCs captured later than this were stained positively with vimentin both on the cell surface and in the cytoplasm in the newly thawed samples. From this time point on, intracellular vimentin began to leak out and was stained with 84-1 owing to the broken membrane after thawing in most of the CTCs isolated from aged blood samples. Compared with newly thawed samples, fresh samples stored at room temperature for 2h, 24h or 48h did not affect the morphology or the reproducibility of CTCs measurement (Fig. 5A, B). However, with the delayed time lapse on: 24 hours and 48 hours before cryopreservation of PBMCs, CTC numbers noticeably decreased compared to the group of time-lapse less than 2 hours (Fig. 5C). Additionally, CTCs counts between fresh blood and stored overnight blood prior to cryopreservation of PBMCs were highly variable in both metastatic sarcoma and epithelial cancer (Supplementary Fig. 1). These results indicated that waiting longer than 2 hours before cryopreserving PBMCs affected the reproducibility of CTCs capture and detection.
Figure 5. Comparison of CTCs counts within different time lapse before cryopreserving PBMCs.
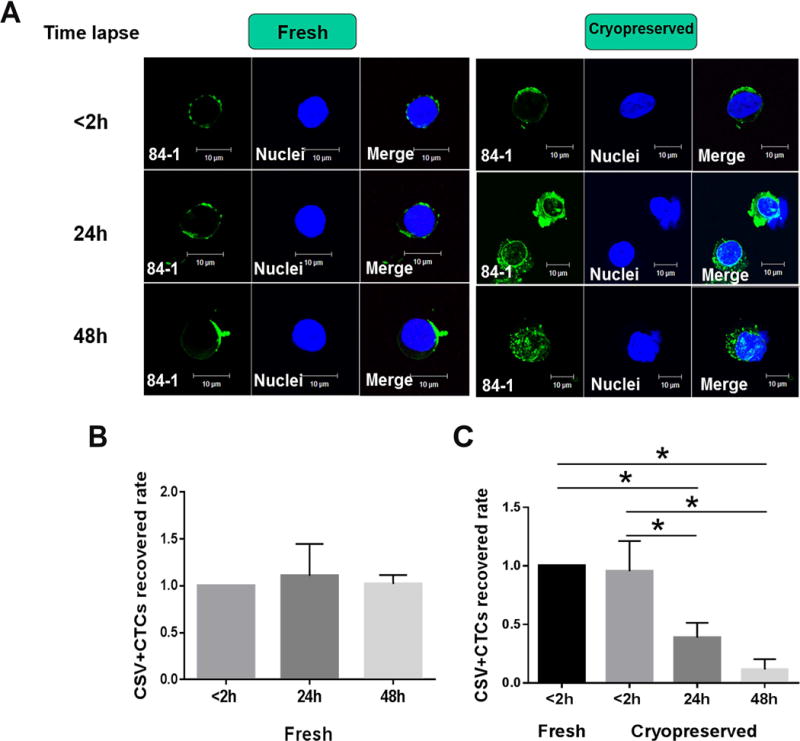
(A) Micrographs showing cell surface vimentin staining in the fresh and cryopreserved samples with different time lapse before cryopreserving PBMCs: less than 2 h, 24h and 48h. Graph shows the results of CTCs counts among fresh sarcoma samples (B) and cryopreserved ones (C) with different time lapse before isolating PBMCs: less than 2h, 24h and 48h. CSV (84-1, green), nuclear stain (blue). GIST, Gastrointestinal stromal tumor. Scale indicates 10 μm. The group of time-lapse less than 2 hours was used as the reference.* P<0.05
3.5 The modified technique detects CTCs that express independent tumor markers
To further verify that the isolated CTCs from cryopreserved PBMCs were of cancerous origin, we further characterized the cells captured from patients with sarcoma by independent specific marker staining. As shown in Fig. 6, the isolated CSV-positive CTCs were validated by the presence of specific mesenchymal markers, namely α-SMA for osteosarcoma, α-SMA and MDM2 for myxoid liposarcoma, CD117 for gastrointestinal stromal tumor, across samples of different types of sarcoma. These molecular characterization results verified that CTCs captured from cryopreserved samples using the modified staining technique originated from sarcoma.
Figure 6. Independent tumor markers staining against captured CTCs using modified technique.
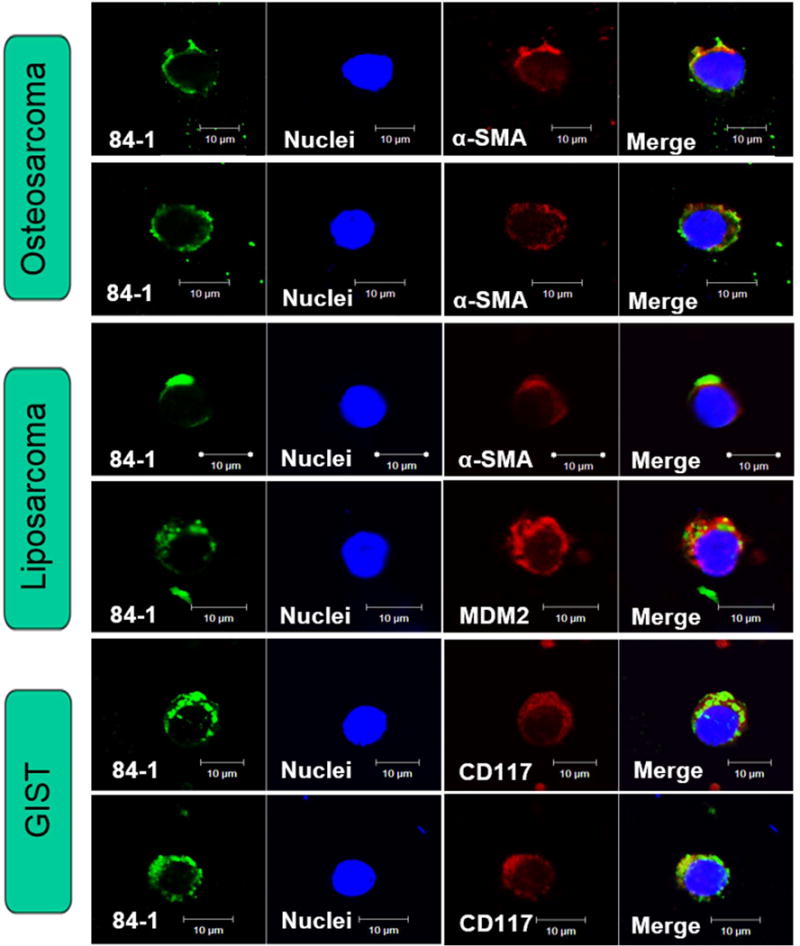
Captured CSV-positive CTCs validated by the presence of specific mesenchymal markers across 6 samples: α-SMA (red) for osteosarcoma, MDM2 (red) and α-SMA (red) for myxoid liposarcoma, and CD117 (red) for gastrointestinal stromal tumor (GIST). CSV (84-1, green), nuclear stain (blue). GIST, Gastrointestinal stromal tumor. Scale indicates 10 μm.
4. Discussion
In this study, we addressed a technological challenge faced by the entire CTCs research field: how to isolate CTCs from cryopreserved samples? As demonstrated by our findings, we firstly revised the cryopreserved PBMCs preparation for CTCs capture and then modified the established CTCs detection protocol, which was designed for fresh blood samples [15]. In short, we stained the captured CTCs with 84-1 antibody prior to spinning and fixation for immunofluorescent analysis. This is the first feasible technique for detecting CTCs from cryopreserved PBMCs in multiplies types of sarcoma tumors. The modified CSV staining time also improved CTCs detection quality for the fresh blood samples. A benefit of this new technology is that it will improve CTCs utility in clinical settings for sarcoma patients.
Current CTCs capture technologies are based on the properties of CTCs that distinguish them from normal immune cells, including physical properties (size, density, electric charges, and deformability) and biological properties (cell surface protein expression, viability, and invasive capacity) [21]. CellSearch, considered the gold standard for CTC detection methods, utilizes EpCAM, an epithelial-derived tumor marker that allows for the capture of CTCs [22, 23]. However, EpCAM-positive circulating epithelial cells have been observed in patients with benign colon diseases [24]. Moreover, carcinoma cells of mesenchymal origin or undergoing EMT cannot be detected using this technology [12]. Consequently, EpCAM cannot be considered a universal biomarker for isolating CTCs.
Physical properties of CTCs are different from normal immune cells: CTCs are larger and exhibit a more obvious nucleus-to-cytoplasm ratio with different nuclear morphology than normal immune cells [25]. Various approaches have been used to detect CTCs in blood samples using these physical properties. The microfluidic size-based CTCs capture system allows for the manipulation with relatively small volumes of biological fluids [26]. It has been reported that a higher purity and larger number of CTCs can be detected by the microfluidic system than the CellSearch system [27]. However, the main drawback of this method is the difficulty of eliminating immune cells with the same or larger size from CTCs. Although a wide range of techniques have been developed for isolating and characterizing CTCs, most of the reliable and reproducible methods for CTCs isolation require fresh blood samples and do not use a CTC-specific antibody. These limitations reduce the utility of these technologies in large multiple center clinical trials. To address these shortcomings, we used a tumor specific monoclonal antibody, 84-1, that was developed by our group. The 84-1 antibody showed high specificity for targeting CSV on both mesenchymal CTCs and EMT-CTCs but not in normal immune cells from fresh blood samples in patients with sarcoma, colon and prostate cancer [14, 15, 18]. Furthermore, enumeration of CTCs using CSV enrichment was shown to be a more reliable predictor of therapeutic outcome than CellSearch in metastatic breast cancer patients [16].
In the current study, in addition to using CSV as a specific marker to detect CTCs in blood samples from patients with multiple types of sarcoma tumors, we sought to solve the technological challenge of how to efficiently capture CTCs in cryopreserved samples. First, we modified the cryopreserved PBMCs preparation protocol. Blood samples should be processed within less than 2 hours of blood collection for the best results. We compared the CTCs enumeration and morphology results at different time intervals between blood collection and PBMCs isolation: less than 2 hours, overnight, 24 hours and 48 hours. We found that longer elapsed time before cryopreservation resulted in lower reproducibility rate of CTC measurement, dirty background, and strange cell morphology. Second, we used the SepMate™-50 tube for isolation of PBMCs, which could eliminate the need for meticulously layering the blood onto the Ficoll-Paque™ Premium and allowed for fast and easy harvesting of the isolated PBMCs with a simple pour. Density gradient centrifugation eliminated unwanted cell pellet along with erythrocytes, leaving the PBMCs untouched and highly purified. Third, PBMCs were cryopreserved at 6 × 106 (6 million) cells per 1 mL for further experiments. Cell viability during freeze-thaw would be decreased if the cells were cryopreserved at a concentration of less than 6 million per 1 mL. Last but not least, we improved the technique for the capture and CSV-positive CTCs staining from cryopreserved blood samples by staining live cells in tube before fixation. This technique allowed the cell surface vimentin to remain intact so that 84-1 antibody binding was specific to the cell surface but not the cytoplasm.
Compared with a previous retrospective analysis in detection of tumor-associated cells from cryopreserved samples [28], this modified technique yields higher reproducibility for capturing CTCs in a relative large number of patients’ peripheral blood samples. In addition, all the samples were processed using the immunomagnetic CTC isolation technique, which was based on antibody-conjugated magnetic nanoparticles [29, 30]. This technique could attain high recovery and purity rates, as demonstrated by our previous data [15]. However, we only analyzed the immunomagnetic method platform using our modified technique in the current study. Whether this new procedure works with other antigen-independent cell capture techniques is unknown. Further study is required to determine whether our modified technology is compatible with other CTC detection and enumeration platforms.
It is noteworthy that this is the first technique demonstrated to be suitable for CTCs capture and enumeration from cryopreserved PBMC samples isolated from patients with different types of sarcoma tumor. The modified technology described here will enable bio-banking of samples to be processed in large numbers, reducing the effects of transportation of samples between laboratory locations. Most importantly, this technique allows for the wide use of CTC-based diagnosis and treatment in clinical settings.
Supplementary Material
Supplementary Figure 1. Comparison of CTCs counts isolated from fresh and overnight blood before cryopreserving PBMCs. Graph shows the counts of CTCs numbers isolated from fresh and overnight blood before cryopreserving PBMCs in multiplies types of cancer patients.
Highlights.
Cell-surface vimentin (CSV) is a specific marker to capture CTCs from patient cryopreserved PBMCs across sarcoma tumor types.
The novel technology is validated to specifically detect and isolate sarcoma CTCs from cryopreserved PBMCs.
This technique allows for the wide use of CTC-based diagnosis and treatment in clinical settings for sarcoma patients.
Acknowledgments
Not applicable.
Funding
This work was supported by grants from the National Institutes of Health to Dr. Shulin Li [NIH R01CA120895] and MD Anderson Institutional Research Grant to Dr. Qing H. Meng.
Abbreviations
- CTCs
Circulating tumor cells
- CSV
Cell-surface vimentin
- PBMCs
Peripheral blood mononuclear cells
- EpCAM
Epithelial cell adhesion molecule
- EMT
Epithelial–mesenchymal transition
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burningham Z, Hashibe M, Spector L, Schiffman JD. The epidemiology of sarcoma. Clinical sarcoma research. 2012;2:14. doi: 10.1186/2045-3329-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackall CL, Meltzer PS, Helman LJ. Focus on sarcomas. Cancer cell. 2002;2:175–178. doi: 10.1016/s1535-6108(02)00132-0. [DOI] [PubMed] [Google Scholar]
- 3.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clinical chemistry. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 4.Pantel K, Speicher M. The biology of circulating tumor cells. Oncogene. 2016;35:1216–1224. doi: 10.1038/onc.2015.192. [DOI] [PubMed] [Google Scholar]
- 5.Urtishak S, Alpaugh R, Weiner L, Swaby R. Clinical utility of circulating tumor cells: a role for monitoring response to therapy and drug development. Biomarkers in medicine. 2008;2:137–145. doi: 10.2217/17520363.2.2.137. [DOI] [PubMed] [Google Scholar]
- 6.Gorges TM, Pantel K. Circulating tumor cells as therapy-related biomarkers in cancer patients, Cancer Immunology. Immunotherapy. 2013;62:931–939. doi: 10.1007/s00262-012-1387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peeters DJ, Brouwer A, Van den Eynden GG, Rutten A, Onstenk W, Sieuwerts AM, Van Laere SJ, Huget P, Pauwels P, Peeters M. Circulating tumour cells and lung microvascular tumour cell retention in patients with metastatic breast and cervical cancer. Cancer letters. 2015;356:872–879. doi: 10.1016/j.canlet.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 8.Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clinical cancer research. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 9.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. New England Journal of Medicine. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 10.Cohen SJ, Alpaugh RK, Gross S, O’hara SM, Smirnov DA, Terstappen LW, Allard WJ, Bilbee M, Cheng JD, Hoffman JP. Isolation and characterization of circulating tumor cells in patients with metastatic colorectal cancer. Clinical colorectal cancer. 2006;6:125–132. doi: 10.3816/CCC.2006.n.029. [DOI] [PubMed] [Google Scholar]
- 11.Okegawa T, Nutahara K, Higashihara E. Prognostic significance of circulating tumor cells in patients with hormone refractory prostate cancer. The Journal of urology. 2009;181:1091–1097. doi: 10.1016/j.juro.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JW, Gratama JW, Sleijfer S, Foekens JA. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. Journal of the National Cancer Institute. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson DR, Dracopoli N, Petty BG, Compton C, Cristofanilli M, Deisseroth A, Hayes DF, Kapke G, Kumar P, Lee JS. Considerations in the development of circulating tumor cell technology for clinical use. Journal of translational medicine. 2012;10:138. doi: 10.1186/1479-5876-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satelli A, Mitra A, Brownlee Z, Xia X, Bellister S, Overman MJ, Kopetz S, Ellis LM, Meng QH, Li S. Epithelial–mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clinical Cancer Research. 2015;21:899–906. doi: 10.1158/1078-0432.CCR-14-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satelli A, Mitra A, Cutrera JJ, Devarie M, Xia X, Ingram DR, Dibra D, Somaiah N, Torres KE, Ravi V. Universal marker and detection tool for human sarcoma circulating tumor cells. Cancer research. 2014;74:1645–1650. doi: 10.1158/0008-5472.CAN-13-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satelli A, Brownlee Z, Mitra A, Meng QH, Li S. Circulating tumor cell enumeration using a combination of EpCAM and Cell-surface vimentin based methods for monitoring breast cancer therapeutic response. Clinical chemistry. 2015;61:259. doi: 10.1373/clinchem.2014.228122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cellular and molecular life sciences. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satelli A, Batth IS, Brownlee Z, Rojas C, Meng QH, Kopetz S, Li S. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Scientific Reports. 2016;6:28910. doi: 10.1038/srep28910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Mantel JM, Kojic N, Smith K, Chen P-i. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nature protocols. 2014;9:694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clinical Cancer Research. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 21.Gabriel MT, Calleja LR, Chalopin A, Ory B, Heymann D. Circulating tumor cells: a review of non-EpCAM-based approaches for cell enrichment and isolation. Clinical chemistry. 2016;62:571–581. doi: 10.1373/clinchem.2015.249706. [DOI] [PubMed] [Google Scholar]
- 22.Andreopoulou E, Yang LY, Rangel K, Reuben J, Hsu L, Krishnamurthy S, Valero V, Fritsche H, Cristofanilli M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. International journal of cancer. 2012;130:1590–1597. doi: 10.1002/ijc.26111. [DOI] [PubMed] [Google Scholar]
- 23.Hofman V, Ilie MI, Long E, Selva E, Bonnetaud C, Molina T, Venissac N, Mouroux J, Vielh P, Hofman P. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. International journal of cancer. 2011;129:1651–1660. doi: 10.1002/ijc.25819. [DOI] [PubMed] [Google Scholar]
- 24.Pantel K, Denève E, Nocca D, Coffy A, Vendrell J-P, Maudelonde T, Riethdorf S, Alix-Panabières C. Circulating epithelial cells in patients with benign colon diseases. Clinical chemistry. 2012;58:936–940. doi: 10.1373/clinchem.2011.175570. [DOI] [PubMed] [Google Scholar]
- 25.Bobek V, Gurlich R, Eliasova P, Kolostova K. Circulating tumor cells in pancreatic cancer patients: enrichment and cultivation. World Journal of Gastroenterology: WJG. 2014;20:17163. doi: 10.3748/wjg.v20.i45.17163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan SJ, Yobas L, Lee GYH, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomedical microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 27.Hughes AD, Mattison J, Western LT, Powderly JD, Greene BT, King MR. Microtube device for selectin-mediated capture of viable circulating tumor cells from blood. Clinical chemistry. 2012;58:846–853. doi: 10.1373/clinchem.2011.176669. [DOI] [PubMed] [Google Scholar]
- 28.Zhu P, Stanton ML, Castle EP, Joseph RW, Adams DL, Li S, Amstutz P, Tang CM, Ho TH. Detection of tumor-associated cells in cryopreserved peripheral blood mononuclear cell samples for retrospective analysis. Journal of Translational Medicine. 2016;14:198. doi: 10.1186/s12967-016-0953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pamme N. On-chip bioanalysis with magnetic particles. Current opinion in chemical biology. 2012;16:436–443. doi: 10.1016/j.cbpa.2012.05.181. [DOI] [PubMed] [Google Scholar]
- 30.Deng G, Herrler M, Burgess D, Manna E, Krag D, Burke JF. Enrichment with anti-cytokeratin alone or combined with anti-EpCAM antibodies significantly increases the sensitivity for circulating tumor cell detection in metastatic breast cancer patients. Breast Cancer Research. 2008;10:R69. doi: 10.1186/bcr2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Comparison of CTCs counts isolated from fresh and overnight blood before cryopreserving PBMCs. Graph shows the counts of CTCs numbers isolated from fresh and overnight blood before cryopreserving PBMCs in multiplies types of cancer patients.


