Abstract
The mechanisms by which human immunodeficiency virus (HIV) circumvents and coopts cellular machinery to replicate and persist in cells are not fully understood. HIV accessory proteins play key roles in the HIV life cycle by altering host pathways that are often dependent on post-translational modifications (PTMs). Thus, the identification of HIV accessory protein host targets and their PTM status is critical to fully understand how HIV invades, avoids detection and replicates to spread infection. To date, a comprehensive characterization of HIV accessory protein host targets and modulation of their PTM status does not exist. The significant gap in knowledge regarding the identity and PTMs of HIV host targets is due, in part, to technological limitations. Here, we applied current mass spectrometry techniques to define mechanisms of viral protein action by identifying host proteins whose abundance is affected by the accessory protein Vpr and the corresponding modulation of down-stream signaling pathways, specifically those regulated by phosphorylation. By utilizing a novel, inducible HIV-1 CD4+ T-cell model system expressing either the wild type or a vpr-negative viral genome, we overcame challenges associated with synchronization and infection-levels present in other models. We report identification and abundance dynamics of over 7000 proteins and 28,000 phospho-peptides. Consistent with Vpr's ability to impair cell-cycle progression, we observed Vpr-mediated modulation of spindle and centromere proteins, as well as Aurora kinase A and cyclin-dependent kinase 4 (CDK4). Unexpectedly, we observed evidence of Vpr-mediated modulation of the activity of serine/arginine-rich protein-specific kinases (SRPKs), suggesting a possible role for Vpr in the regulation of RNA splicing. This study presents a new experimental system and provides a data-resource that lays the foundation for validating host proteins and phosphorylation-pathways affected by HIV-1 and its accessory protein Vpr.
In 2015, 40 million people were living with human immunodeficiency virus (HIV)1 worldwide, although 1 in 8 individuals (particularly those from medically underserved populations) were unaware of their infection (1). Currently, antiretroviral drugs can control HIV replication and increase survival (2). However, current therapies come at substantial financial cost (3), and they are not curative. Moreover, if HIV patients are not medically compliant, drug-resistance and treatment-failure can ensue (4). Thus, new and sustainable approaches are needed to treat HIV. The mechanisms by which HIV circumvents cellular defenses and coopts cellular machinery to replicate and persist are not fully known; elucidating these mechanisms could lead to the identification of new drug-targets.
HIV is a retrovirus containing nine protein-encoding genes (5). These gene products include structural proteins, essential regulatory proteins, and so-called accessory proteins (6). The accessory proteins are attractive targets for novel therapeutics, because they provide evasion of innate and adaptive immune responses and are important for viral persistence, replication, and transmission (6). The accessory proteins Vif, Vpr and Vpu direct host proteins that interfere with viral replication to a degradation pathway by hijacking the host ubiquitin-proteasome system (6). For example, Vpu recruits the SCF/CRL1 E3 ubiquitin ligase complex to degrade the HIV receptor CD4, which would otherwise inhibit the infectivity of newly produced virus particles (7). Several host targets for Vif and Vpu have been identified, including the so-called “restriction factors” of the APOBEC family and BST-2 (6, 8), but more might exist. The known cellular targets of Vpr have not defined a clear role for this gene product in viral replication or pathogenesis, but its ability to induce the degradation of the cellular proteins uracil DNA glycosylase (UNG), the endonuclease MUS81, and the DNA helicase HLTF suggest that modulation of DNA-repair is somehow important to HIV replication (9–12).
In general, HIV accessory proteins affect diverse pathways, from altering membrane protein trafficking (Vpu and Nef) to modulating the cell cycle (Vpr) (6). These host pathways are highly dependent on post-translational modifications (PTMs) such as ubiquitination and phosphorylation. Many studies have utilized genomics and transcriptomic approaches to study the interaction of HIV with host cells. However, the PTM status of proteins cannot be predicted from DNA or RNA sequence alone. The identification of HIV accessory protein targets and of the PTMs that accessory proteins induce seems critical to fully understand how HIV avoids immune-detection and replicates to spread infection.
Specific and important roles for host-phosphorylation during HIV replication have been demonstrated (13). From the moment that HIV binds to its receptor, host phosphorylation signaling cascades are initiated (13, 14), rendering the cell an optimal host for the virus. A hallmark of HIV replication in dividing cells is delay in the G2/M phase of the cell cycle, which is caused by Vpr (15). The cell cycle is tightly regulated by phosphorylation events (16). Thus, we hypothesized that phospho-proteomic analysis would provide insight into how Vpr affects the cell cycle.
In this study, we utilized a novel, inducible HIV-1 CD4+ T cell-based system that overcomes the challenges associated with cell synchronization and high multiplicity of infection (MOI) associated with traditional HIV infection models (17). Our objective was to comprehensively characterize the host proteome and phospho-proteome dynamics quantitatively in response to HIV gene expression in the presence and absence of the accessory protein Vpr. Our analysis incorporated a quantitative mass spectrometry approach in addition to phospho-peptide enrichment. Proteomic findings were validated by Western blotting in the induction-model and compared with a standard infection-model. We quantified over 28,000 phospho-peptides with statistical fidelity, complementing and extending the previously reported phospho-proteomics relating to HIV reported by Wojcechowskyj et al. (13), who identified 1700 phospho-sites utilizing the current technology at the time. These results validate our model system, identify novel potential targets of Vpr, and identify altered phospho-signaling events associated with HIV expression in the presence or absence of Vpr. We followed the dynamics of over 7000 proteins through time points post-induction of HIV gene expression. Numerous protein profiles, including those for APOBEC3C and PPP2 subunits aligned with a previous report (17). The combined proteomics and phospho-proteomics data indicated Vpr-dependent changes consistent with the alteration of cell cycle dynamics, such as modulation of the levels of CDK4 and Aurora kinase A, deregulation of spindle assembly/centromere related proteins, and modulation of phosphorylation of retinoblastoma 1. In addition, the phospho-proteomics data implicated Vpr in the regulation of splicing processes through modulation of SRPK. In general, this study shows that by using an inducible HIV-1 CD4+ T cell based system in combination with quantitative proteomic approaches, new mechanistic insights into the activities of accessory proteins can be achieved.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
All quantitative mass spectrometry experiments were performed in biological duplicate. All Western blottings were performed in biological triplicate. Significance for proteomics data and phospho-proteomics data was assessed by Student's t test; variance was assessed by an F-test to ensure the correct statistical assumptions were used. p values of p ≤ 0.05 were considered significant; adjustment for multiple comparisons was not included in the statistical analysis.
Cell Culture Model System
The T lymphocytic Jurkat Tet-On 3G cells expressing the Tet-On 3G tetracycline-controlled transactivator protein were obtained from Clontech Laboratories (Mountain View, CA). To aid the discovery and validation of HIV accessory protein events, Jurkat cells were modified to express replication-incompetent HIV-1 (derived from pNL4–3 with the V3 region of the R5-tropic 51–9 virus (18)), upon doxycycline (1 μg/ml) treatment. The tet-responsive replication-incompetent HIV was constructed through the deletion of a portion of the 3-prime Long Terminal Repeat U3 region of the virus and replacing the HIV promoter with a tet-responsive promoter (from pTRE-Tight, Clontech). The ΔVPR mutation was derived from pNL4–3-VprX (19) (a gift from Vincente Planelles). A puromycin resistance expression cassette was cloned into the plasmid backbone. The tet-inducible viral constructs were stably transfected into Jurkat Tet-On 3G cells following linearization with PvuI (New England Biolabs, Ipswich, MA) with Lipofectamine LTX with PLUS Reagent (Life Technologies, Carlsbad, CA) according to manufacturer's instructions. Transfected cells were selected with 200 μg/ml G418 plus 1 μg/ml puromycin and single cell clones were isolated by limiting dilution. The expression of HIV proteins in this system is tightly regulated and easily scalable, making it ideal for proteomics studies. Cell culture studies were approved by the University of California, San Diego institutional review board. Cell lines were cultured in RPMI 1640 (Gibco, Gaithersburg, MD) supplemented with penicillin/streptomycin and 10% Tet-free fetal bovine serum (Omega Scientific, Tarzana, CA), HIV gene expression was induced with 1 μg/ml doxycycline (SIGMA, Carlsbad, CA) and cells were harvested at 0, 3, 6, 12, and 24 h post-viral induction with doxycycline (Fig. 1).
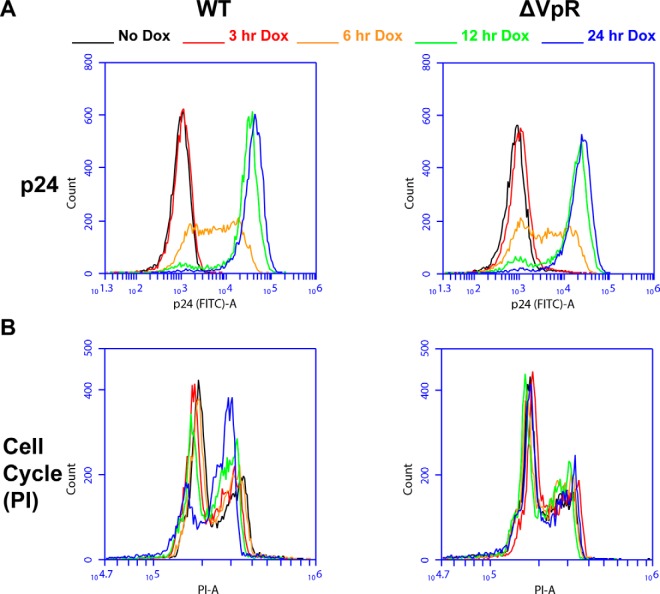
Fig. 1.
- Characterization of the Jurkat WT and ΔVPR cell lines following induction of HIV with doxycycline treatment. A, Time course of HIV-1 Gag/p24 expression in WT and ΔVPR cells. Conditions are color coded as above for 0, 3, 6, 12, and 24 h postdoxycycline induction of HIV expression. Histograms show FITC (intracellular Gag/p24) intensity for live gated WT (left) and ΔVPR (left) cells. B, The cell cycle block (or not) in these cells. WT (left) and ΔVPR (right) cells were labeled with PI and gated on live single cells. Histograms show PI-labeled DNA content with the lower peak representing cells in G0/G1 and the higher peak representing cells at G2/M phase.
Flow Cytometry
For confirmation of HIV (Gag/p24) expression by FACS, harvested cells were fixed and permeabilized (20) using the BD (San Diego, CA) Cytofix/Cytoperm kit according to the manufacturer's instructions, followed by staining with fluorescein isothiocyanate (FITC)-conjugated anti-HIV-1 p24 antibody (KC57; Beckman Coulter, Brea, CA), washing and analysis by flow cytometry. For cell cycle analysis, cells were harvested, washed with PBS, then ethanol fixed for 6–24 h. After fixation, cells were washed, then resuspended in Propidium Iodide (PI)/RNase Staining Solution (Cell Signaling, Danvers, MA, #4087) and incubated for 15 min at room temperature in the dark prior to analysis of PI fluorescence using a BD Accuri C6 flow cytometer.
Confirmatory Infections
To prepare virus, HEK-293 cells in 10-cm plates were transfected with pNL4–3 or pNL4–3-VprX with Lipofectamine2000 in Opti-MEM (Life Technologies) and after 4 h the culture media was replaced with 10 ml DMEM (Gibco) supplemented with penicillin/streptomycin and 10% fetal bovine serum. After 48 h, viral supernatants were harvested, cellular debris was pelleted, and clarified supernatants were concentrated by centrifugation at 4000 × g for 20 min at room temperature in Amicon Ultra-15 100K Centrifugal Filter Tubes (EMD Millipore, Temecula, CA). Viral infectivity was determined by infecting 25,000 HeLa P4-R5 cells (20–22)(acquired from the NIH AIDS Reagent Program from Dr. Nathaniel Landau) per well in a 48-well plate in duplicate with serial dilutions of the viral supernatants. The number of infectious centers per well was determined 2 days after infection, following fixation, staining, and counting of positive cells by computer-assisted image analysis as described previously (23). T cell lymphoblast SupT1 cells expressing human CCR5 (24) (SupT1-HuCCR5, a gift from James A. Hoxie) were infected at an MOI of 2 with 8 μg/ml polybrene (EMD Millipore) by spinoculation for 30 min at 800 × g at 32 °C, then plated. Cells were harvested after 6, 12, 24, and 48 h of infection.
Western Blotting Analysis
Cell lysates were prepared in Laemmli SDS-PAGE sample buffer and analyzed by immunoblot for GAPDH, HIV-1 Gag/p55, HIV-1 Vpr, VPRBP/DCAF1, PP2A (PPP2R5D), AURKA and CDK4. Samples were boiled for 10 min, then electrophoresed in a 12% Tris-HCl SDS-polyacrylamide gel, then electroblotted to a polyvinylidene difluoride (PVDF) membrane using the Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA). The membrane was blocked with 5% milk and PBS-Tween, then incubated with primary antibody overnight at 4 degrees C. Primary antibodies were diluted in 1% milk/PBS-T as follows: Mouse anti-GAPDH (GenTex, Zeeland, MI, GTX628408) 1:500, Mouse anti-HIV-1 gag (EMD Millipore, MAB880-A) 1:100, Rabbit anti-VPR antiserum (This reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Cat#11836, Anti-HIV-1 Vpr 1–50 aa Polyclonal from Dr. Jeffrey Kopp.) 1:500, Mouse anti-VPRBP/DCAF1 (Santa Cruz Biotechnology, Dallas, TX, sc-376850) 1:100, Mouse anti-PP2A/PPP2R5D (EMD Millipore, clone H5D12 (04–639)) 1:300, Mouse anti-Aurora A (Invitrogen, Carlsbad, CA, clone 35C1 (458900)) 1:250, and Mouse anti-CDK4 (Santa Cruz Biotechnology, (DCS-35) sc-23896) 1:1000. After washing, blots were incubated with 1:2000 Bio-Rad Immun-Star Goat Anti-Mouse-HRP Conjugate (#170–5047) or 1:3000 Bio-Rad Goat Anti-Rabbit IgG (H + L)-HRP Conjugate (#1706515) in 1% milk/PBS-T for 1.5 h at room temperature. After treatment with the Clarity Western ECL blotting substrate (Bio-Rad), chemiluminescence was detected using the ChemiDoc MP system (Bio-Rad).
Mass Spectrometry Based Proteomics. Cell Lysis and Protein Digestion
Cells were lysed in a buffer with a composition of 3% SDS, 75 mm NaCl, 1 mm NaF, 1 mm beta-glycerophosphate, 1 mm sodium orthovanadate, 10 mm sodium pyrophosphate, 1 mm PMSF and 1X Roche (Basel, Switzerland) Complete mini EDTA free protease inhibitors in 50 mm HEPES, pH8.5 (25). Lysates were passed through a 21-gauge needle 20 times and sonicated for 5 min to ensure full lysis. Debris was pelleted by centrifugation at 14,000 RPM for 5 min, with resultant supernatants used for downstream processing.
Protein disulfides were reduced with DTT and alkylated with iodoacetamide as previously described (26). Reduced/alkylated proteins were precipitated following a methanol-chloroform precipitation protocol (27). Precipitated proteins were re-suspended in 1 m urea in 50 mm HEPES, pH 8.5 for proteolytic digestion. Digestion was performed in a two-step process, where proteins are first digested with LysC overnight at room temperature, then with trypsin for 6 h at 37C. Digestion was quenched by the addition of TFA, and peptides were desalted with C18 solid-phase extraction columns as previously described (28). Peptides were dried in a speed vac, then re-suspended in 50% Acetonitrile/5% formic acid and quantified by BCA assay. A 50 μg aliquot was made for each sample for proteomic analysis, and a 2 mg aliquot was portioned for each sample for subsequent phospho-peptide enrichment and analysis. Peptide aliquots were lyophilized prior to further analysis.
Phospho-peptide Enrichment
Phospho-peptides were enriched by TiO2 as previously described (29, 30). Briefly, 8 mg of titanium dioxide beads were used per 2 mg of peptide starting material. Enriched peptides were desalted with solid-phase extraction columns as described above, then lyophilized and stored at −80°C until they were labeled for quantitation.
Tandem Mass Tag Labeling and Fractionation
In both cases, proteomic profiling and phosphorylation profiling, peptides were labeled with tandem mass tag (TMT) reagents (31, 32) (Fig. 1A). TMT reagents, at a concentration of 20 μg/μl in dry acetonitrile were used for labeling. Lyophilized peptides were re-suspended in 30% acetonitrile in 200 mm HEPES, pH 8.5 and 8 μl of the appropriate TMT reagent was added to each sample. The labeling reaction was conducted for 1 h at room temperature, and then quenched by the addition of 9 μl of 5% hydroxylamine, which was allowed to react for 15 min at room temperature. Labeled samples were then acidified by adding 50 μl of 1% trifluoroacetic acid. Differentially labeled samples were pooled into multiplex experiments and then desalted via solid-phase extraction as described above, lyophilized and stored at −80°C until fractionation.
Sample fractionation was accomplished by basic pH reverse-phase liquid chromatography (33). Lyophilized samples were re-suspended in 5% formic acid/5% acetonitrile and separated on a 4.6 mm x 250 mm C18 column on an Ultimate 3000 HPLC rigged with a fraction collector, degasser and variable wavelength detector. Separation was performed over a linear gradient of 8 to 28% acetonitrile in 10 mm ammonium bicarbonate in 60 min with a flow rate of 0.5 ml/minute. In total, 96 fractions were collected and combined in a concatenated manner as previously described (12 for proteomic analysis and 24 for phospho-proteomic analysis) (28). Combined fractions were lyophilized and re-suspended in 5% formic acid/5% acetonitrile for identification and quantification by LC-MS2/MS3 (Fig. 2A).
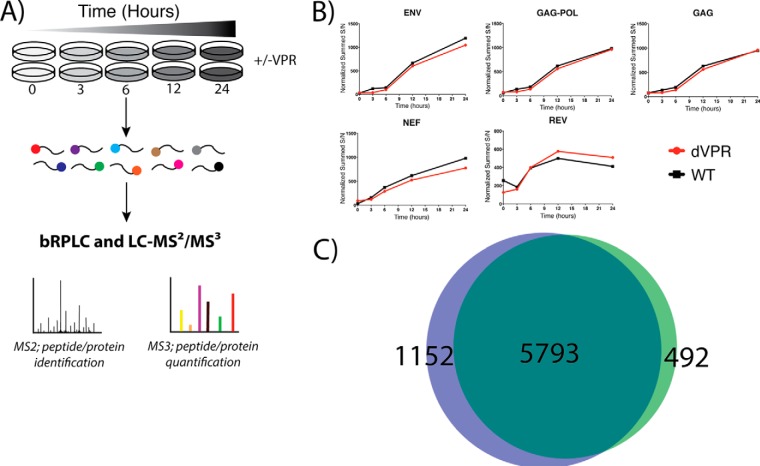
Fig. 2.
- Quantitative Temporal Viromics. A, Wild-type and vpr-deleted HIV expressing cell lines were analyzed in biological duplicate. Cells were lysed and digested prior to labeling with TMT reagents for quantitative proteomic and phospho-proteomic profiling. Proteins are identified at the MS2 level and quantified at the MS3 level. B, Temporal protein profiles show a time-dependent increase in detected viral proteins. This increase is reproducible between wild-type (WT) and vpr-deleted cell lines (ΔVPR). C, Venn Diagram showing overlap of proteins identified and quantified between WT (green) and ΔVPR cell lines (blue). The 5793 proteins in common were used for subsequent analyses.
LC-MS2/MS3 Analysis
All LC-MS2/MS3 experiments were performed on an Orbitrap Fusion mass spectrometer with an in-line Easy-nLC 1000 with chilled autosampler. Peptides were separated on a home pulled, home packed column (inner diameter, 100 μm; outer diameter, 360 μm). Columns were packed first with ∼0.5 cm of C4 resin (5 μm, 100 Å) followed by ∼0.5 cm of C18 resin (3 μm, 200 Å) and then to a final length of 30 cm with C18 (1.8 μm, 120 Å). Peptides were eluted with a linear gradient from 11 to 30% acetonitrile in 0.125% formic acid over 165 min at a flow rate of 300 nL/minute and heating the column to 60C. Electrospray ionization was achieved by applying 2000V through a stainless-steel T-junction at the inlet of the column.
The mass spectrometer was operated in a data-dependent mode, with a survey scan performed over a mass to charge (m/z) range of 500–1200 at a resolution of 120,000 in the Orbitrap. For the MS1 survey scan, automatic gain control (AGC) was set to 5 × 105 with a maximum injection time of 100ms and the s-lens set to an RF of 60. The most abundant ions observed in the survey scan were subjected to MS2 and MS3 analysis for identification and quantitation respectively. For this analysis, Top Speed mode was utilized, enabling the instrument to acquire a maximum number of spectra in a 5 s experimental cycle. Data collected at MS1, MS2, and MS3 levels were centroided.
For MS2 analysis, ions above an intensity threshold of 5 × 105 were selected for fragmentation. They were isolated in the quadrupole portion of the mass spectrometer with an isolation window of 0.5 m/z, and then fragmented with collision-induced dissociation with a normalized energy of 30%. Fragment ions were detected with rapid scan rate setting enabled in the ion trap, with an AGC setting of 1 × 104 and maximum injection time of 35 ms.
MS3 analysis was performed using the synchronous precursor selection setting to maximize sensitivity for quantification of TMT reporter ions (31). A maximum of 3 MS2 ions were simultaneously isolated and fragmented for MS3 analysis of phospho-peptides and 10 MS2 ions for normal proteomics experiments. An isolation window of 2 m/z was utilized to isolate MS2 fragments, and these were fragmented further with high energy collision dissociation at a normalized energy of 50%. Fragment ions were detected in the Orbitrap at a resolution of 60,000 with a low mass of 110 m/z for MS3 analysis. The MS3 AGC was set to 5 × 104 with a maximum ion injection time of 150 ms. MS2 ions in the range of 40m/z below and 15 m/z above the precursor (MS1) were excluded from selection for MS3.
Data Processing and Analysis
Data were processed using the ProteomeDiscoverer 2.1.0.81 software package. The built-in version of SequestHT (34) was utilized to assign identities to MS2 spectra searching against the Uniprot database of Human entries concatenated to the HIV-1 database (downloaded Feb. 27, 2016 with 69,961 total entries). The database was appended to include a decoy database comprised of all protein sequences in reversed order for downstream false discovery estimation (35–37). Search parameters include a 50ppm MS1 mass tolerance (29, 38) (supplemental Fig. S1), 0.6 Da fragment ion tolerance, fully-enzymatic trypsin with a maximum of two missed cleavages per peptide, static modifications of 10-plex TMT tags on lysines and peptide n-termini and carbamidomethylation of cysteines. Variable modifications included oxidation of methionines and phosphorylation of serine, threonine and tyrosine residues. Data were filtered to a peptide and protein false discovery rate of less than 1% using the target-decoy search strategy (35). Peptides matching to multiple proteins were assigned to the protein containing the largest number of matched redundant peptides following the law of parsimony (29).
TMT reporter ion intensities were extracted from MS3 spectra for quantitative analysis. Spectra used for quantitation had to meet the requirements of greater than 10 average signal-to-noise per label and isolation interference of less than 25% (39). Data were normalized in a multistep process, whereby they are first normalized to the average for each peptide, and then to the median of all averages for phospho-peptides, and a similar process at the protein level for proteomics analysis. To account for slight differences in amounts of protein labeled, these values are then normalized to the median of the entire data set and reported as final normalized signal-to-noise ratios per peptide per sample. Further, phospho-peptide abundances were normalized to protein levels. The normalization strategy employed here is analogous to the type of normalization done on an mRNA expression array (40). A step-by-step tutorial and R-script are included in the supplemental Materials detailing this strategy (supplemental Files S1 and S2). Both data sets and corresponding annotated spectra have been deposited to the ProteomeXchange through MassIVE (accessions PXD006230 and MSV000080877 respectively).
Proteins and phospho-peptides were grouped per temporal profile changes across the time course of viral induction via k-means clustering (41). The optimal number of clusters per data set was determined in the R environment via the elbow estimation method, and short time-course expression miner analysis (STEM) (42) software was used to visualize the clusters. Gene heat maps of resultant clusters and subsequent Gene ontology analysis (below) were visualized in GENE-E (https://software.broadinstitute.org/GENE-E/index.html).
Gene ontology (GO) analysis through the DAVID server (43, 44) was utilized to identify significant groups of proteins and peptides from each cluster. Output was filtered as previously described (45).
STRING-db (46) was utilized through the String App for Cytoscape (Version 3.5) to visualize connections between proteins and phospho-peptides determined to be dependent upon Vpr for depletion. Connections were limited to high confidence (0.7) with a maximum of 25 connections for the second shell. First shell of interactions was restricted to the query only.
RESULTS
Inducible HIV-1 CD4+ T-Cell Model Characterization
A well-known phenotype associated with HIV Vpr is delay of the cell cycle in G2/M phase (15). We hypothesized that a quantitative proteomic and phospho-proteomic analysis could be employed to elucidate this mechanism. Before engaging in proteomic analyses, we constructed and characterized a model system in which T cells of the continuous line Jurkat contain integrated HIV-1 genomes, either wild type (WT) or lacking vpr (ΔVPR), whose expression is tightly controllable by doxycycline. The doxycycline-inducible promoter driving viral expression allowed us to overcome challenges associated with the synchronicity and level of infection that are associated with viral inoculation models (17). Viral gene expression was assessed by measuring the induction of p24 (Gag) (Fig. 1A), which exhibited a time dependent increase in both the WT- and ΔVPR-expressing cells after treatment with doxycycline. We then probed for cell cycle progression (Fig. 1B). The expected accumulation of cells in G2/M phase in WT-expressing cells was observed in a time-dependent manner, whereas ΔVPR-expressing cells showed normal cell cycle progression.
Temporal Proteomics During Viral Induction
We sought to define global proteome dynamics through the course of HIV-1 induction in an unbiased, systems scale manner (Fig. 2A). Utilizing our model cell systems, we first assessed protein level dynamics, including those associated with depletion by Vpr.
We first assessed whether the quantitative proteomics platform could detect and quantify HIV-1 proteins. Indeed, we detected and accurately quantified five of the nine proteins of HIV-1 (Fig. 2B). During the time course of induction, a lag phase was observed, in which the proteins were at or below the detection limit (0 and 3 h), followed by a linear increase in viral protein levels (6, 12, and 24 h post induction). This pattern of expression was validated via Western blotting (Fig. 5) for Gag, with GAPDH as a loading control. Similar patterns of viral expression were recently reported in CEM-T4 T cells inoculated with HIV (17).
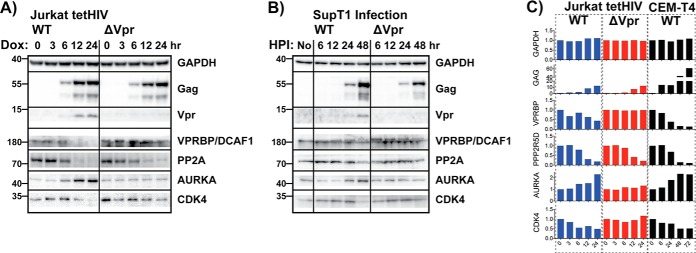
Fig. 5.
- Validation of quantitative mass spectrometry findings via Western blotting. A, Western blotting analysis of HIV and host proteins in WT and ΔVPR Jurkat tetHIV inducible cell lines. B, Validation of tetHIV model using a SupT1 infection model. Similar protein expression patterns are observed, but there is a lag time from the infection process which required a later time point (48 h). C, Average mass spectrometry derived values for blots in A and B. Vpr was not detected in our proteomics experiments or the CEM-T4 experiments performed by Greenwood et al. Values are shown relative to the zero-time point, which was normalized to 1.
For our analysis, we leveraged the full multiplexing capability of TMT 10-plex reagents, analyzing the time course in biological duplicate for WT and ΔVPR-expressing cells. Between two 10-plex experiments, we identified and quantified 7437 total proteins, with 5793 proteins in common between the two data sets (Fig. 2C, supplemental File S3). Subsequent proteomics analyses were derived from the intersection of these data sets (5793 proteins).
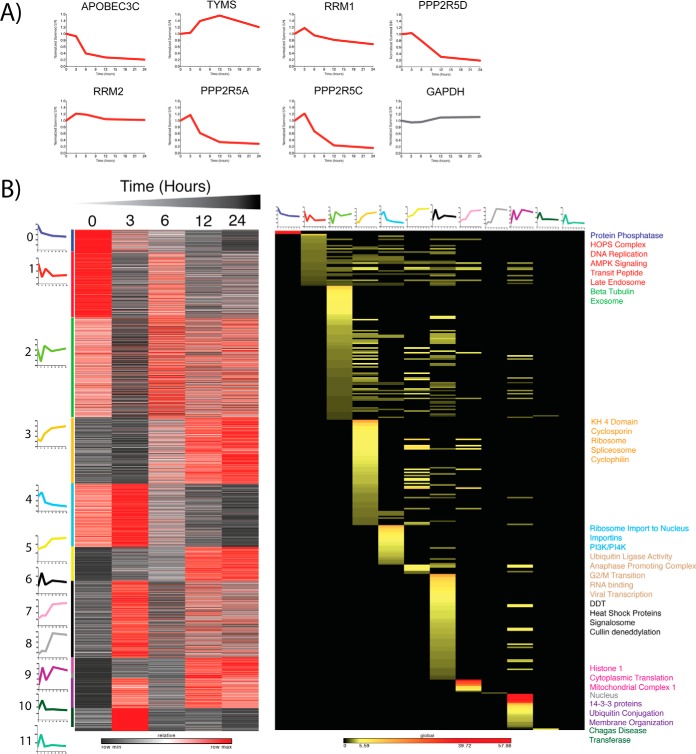
To further validate the model system, we analyzed the temporal profiles of several proteins that are known to be down-regulated by WT HIV-1. As reported (17), we detected a decrease in APOBEC3C, PPP2R5A, PPP2R5C, and PPP2R5D by 24 h after viral induction (Fig. 3A). The profiles of TYMS, RRM1, and RRM2, which were shown to decrease upon viral protein expression in CEM-T4 T cells (17), were also investigated. These proteins remained stable across the time course. One explanation for this observation is that our time course ended at 24 h, whereas the previously reported decreases were not noted until later time points.
Fig. 3.
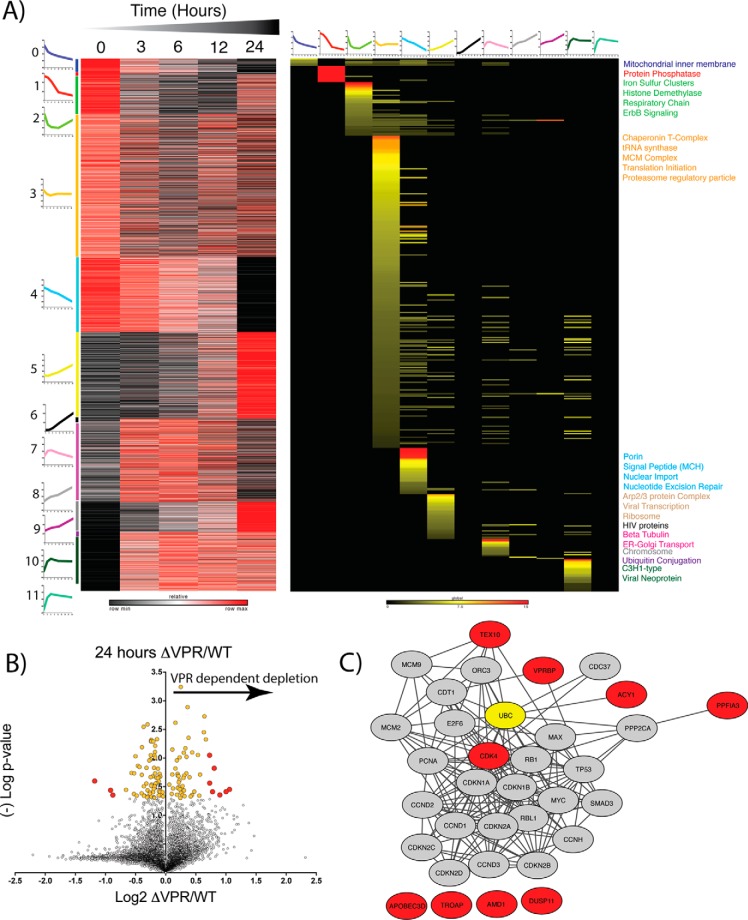
- Proteomic Profiling of Wild-Type HIV expressing cells reveals enriched clusters of protein expression. A, Temporal protein profiles recapitulate findings from other CD4+ T-cell models. B, K-means clustering of proteins allows Gene Ontology enrichment analysis of profiles of enriched clusters of protein functions. Average cluster profiles are depicted beside and on top of the gene and GO based heat maps respectively. Heat map of proteins is relative z-score per row with black representing minimum and red maximum. Heat map of Gene ontology analysis reflects significant enrichment factors and is on a global scale. Black is not present, yellow low enrichment and red high enrichment. Terms on the right of the plot are colored dependent upon the colored bar on the left of the protein heat map corresponding to the adjacent k-means cluster.
Next, k-means clustering was utilized to group proteins by their temporal profiles. Applying the elbow method, the optimal number of clusters was determined to be 12 for both the WT and ΔVPR sample sets. Proteins from each cluster were analyzed by Gene Ontology enrichment, with significantly enriched terms reported (Fig. 3B and Fig. 4A, supplemental Figs. S2 and S3, supplemental Files S4 and S5) for both WT and ΔVPR-expressing cells. Interestingly, in the cells expressing WT HIV, cluster 0, which demonstrates a time-dependent decrease in protein level, is enriched for protein phosphatases. This is consistent with our findings for PPP2R5A, PPP2R5C, and PPP2R5D (Fig. 3A). Other significantly enriched terms included ubiquitin ligase activity, ubiquitin conjugation, and viral transcription associated with clusters 5 and 9, which show time dependent increases in protein levels for cells expressing WT HIV. These results are not unexpected, as HIV accessory proteins are known to modify host protein ubiquitination (47, 48). Additionally, in cluster 5, we observed an enrichment for proteins associated with the G2/M transition. This is also expected in the cells expressing WT, as Vpr is known to arrest or delay infected cells in G2/M phase (49).
Fig. 4.
- Proteomic Profiling of ΔVPR cells reveals potential host targets of Vpr. A, K-means clustering of proteins allows Gene ontology enrichment analysis of profiles of enriched clusters of protein functions. Average cluster profiles are depicted beside and on top of the gene and GO based heat maps respectively. Heat map of proteins is relative z-score per row with black representing minimum and red maximum. Heat map of Gene ontology analysis reflects significant enrichment factors and is on a global scale. Black is not present, yellow low enrichment and red high enrichment. Terms on the right of the plot are colored dependent upon the colored bar on the left of the protein heat map corresponding to the adjacent k-means cluster. B, Volcano plot of vpr-deletion (ΔVPR) relative to wild-type (WT) protein level changes at 24 h post viral induction. X-axis shows log2 fold change and y axis (-)Log of Student's t test based p value. Yellow dots are significantly enriched proteins (p < 0.05) and red dots are significantly enriched and greater than two standard deviations from the median. C, STRING-db analysis of proteins significantly enriched (red dots) in ΔVPR relative to WT HIV. Red nodes correlate to red dots, and ubiquitin is highlighted in yellow.
For cells expressing the ΔVPR genome, protein phosphatases also demonstrate a time-dependent decrease (Cluster 1, Fig. 4A), indicating that they are not dependent on Vpr for their depletion. Intriguingly, Cluster 6 (Fig. 4A) shows no Gene Ontology enrichment, however this cluster contains many of the HIV proteins detected in this experiment. Absent from the ΔVPR sample is a cluster that shows enrichment for G2/M phase. This observation is expected, as removal of a functional vpr gene from the HIV genome should allow the cells to progress through the cell cycle (50).
To identify potential host targets of Vpr, we compared protein abundances for WT and ΔVPR cells at 24 h after viral induction. This 24-hour point was selected because in the protein measurements, the largest effects were observed now. This effect was likely because of a combination of the time required for degradation pathways to be fully engaged as well as the lag time between induction and viral protein expression. We hypothesized that proteins enriched in cells expressing ΔVPR compared with cells expressing WT required Vpr for their depletion. Additionally, we hypothesized that these proteins would be associated with cell cycle progression, given Vpr's influence on the cell cycle (50). Of the proteins significantly enriched in the case of ΔVPR relative to WT (Fig. 4B, supplemental File S6, red circles, p < 0.05, fold-change>2× standard deviations from median), four were connected to the cell cycle through STRING (46) analysis (Fig. 4C). Of these, VPRBP (also known as DCAF1), has been well characterized to interact with Vpr (51, 52). Additionally, CDK4, which is important for the progression from G1 phase into S phase (53, 54), is relatively enriched in the cells expressing ΔVPR. TEX10 and ACY1 were not explicitly connected to the cell cycle, however they were connected to ubiquitin, which is a well-documented PTM that mediates degradation of host proteins (47, 48).
As an initial means of validating the proteomics data, a reanalysis of the raw data from Greenwood et al. (PDX004187) (17) was performed using the same computational platform utilized herein (supplemental Fig. S4, supplemental File S7). The Greenwood model is an infection model in a different T cell line, CEM-T4 cells. In total, 4760 proteins were common between our model of inducible HIV in Jurkat cells and the infection model in CEM-T4 cells (supplemental Fig. S4A). Pearson correlation revealed a positive correlation, albeit low, between the 24-hour time point in the inducible WT HIV model and the 48-hour time point in the CEM-T4 cells when both data sets were compared with their 0-hour time points in log 2 space (supplemental Fig. S4B). We suspect that this low correlation was because of several factors, including the different lag times before viral gene expression during infection and induction from pre-integrated proviruses (our model), which was visible in abundance profiles (Fig. 5 and supplemental Figs. S5–S8), and to the differences in the cell lines. To the latter point, Jurkat cells are of male origin (55) whereas CEM-T4 cells are of female origin (56). This is exemplified by the presence of numerous Y-chromosome proteins (USPY, EIFIAY, DDX3Y, ZFY, RPS4Y1, TBL1Y) found in the Jurkat cells that were absent from the CEM-T4 cells. When we restricted our analysis to a subset of 38 proteins that included the HIV proteins, APOBECs and a variety of cell cycle related proteins, the correlation increased to 0.828 (supplemental Fig. S4C). The cell cycle-related proteins were of interest because of Vpr's phenotype of cell cycle arrest and our above findings from Gene Ontology analysis.
When analyzing the relative levels of proteins in these experiments, we found it useful to split the proteins into four groups: 1) cellular proteins that behaved the same across all three samples (ΔVPR and WT inducible Jurkat cells as well as CEM-T4 cells infected with the WT), that is, proteins not modulated by Vpr, 2) HIV proteins, 3) cellular proteins that were increased in the presence of Vpr in WT inducible Jurkats and in WT infected CEM T4 cells, and 4) cellular proteins that were decreased in the presence of Vpr in both systems and are thus potential targets of Vpr-dependent degradation (supplemental Figs. S5–S8).
Proteins from group 1 and group 2 were less interesting, as we suspected that Vpr did not have a role in their dynamics based on lack of positive or negative regulation. Proteins in the third group, which were up-regulated in the presence of Vpr, were largely centromere and cell division-associated proteins (supplemental File S8). This observation seems consistent with Vpr's role in arresting cells in G2/M phase. The increased abundance of these proteins could be secondary to the G2/M arrest of these cells or because of Vpr competitively usurping the degradation machinery that normally targets these proteins for destruction.
Among those proteins in the fourth group, which were decreased by Vpr, was VPRBP/DCAF1, a well-known interactor of Vpr. We hypothesized that the additional proteins in this group were potential targets for degradation directed by Vpr. Among these proteins was the cyclin-dependent kinase CDK4, which emerged as a candidate from initial screening (Fig. 4B and 4C).
Proteomic findings regarding proteins from the various groups were validated via Western blotting utilizing not only the Jurkat induction model but also an additional CD4+ T cell infection model using SupT1 cells (Fig. 5). SupT1 cells were chosen for their greater susceptibility to HIV-1 infection compared with Jurkat cells (because of their higher levels of CD4 expression), allowing for high levels of infection without needing to pseudotype the virus with a non-HIV envelope. A greater lag time to viral protein expression was noted with infection of SupT1 cells compared with induction from integrated proviruses (Jurkat model), so a later time point (48 h after infection) was added for Western blotting validation of the infected cells. Very importantly, these Western blotting data confirmed the protein modulations indicated by quantitative proteomics: the HIV-associated depletion of PP2A, Vpr-associated depletion of VPRBP and CDK4, and the Vpr-associated increase in Aurora kinase A were all observed by Western blotting.
Temporal Phospho-Proteomics During Viral Induction
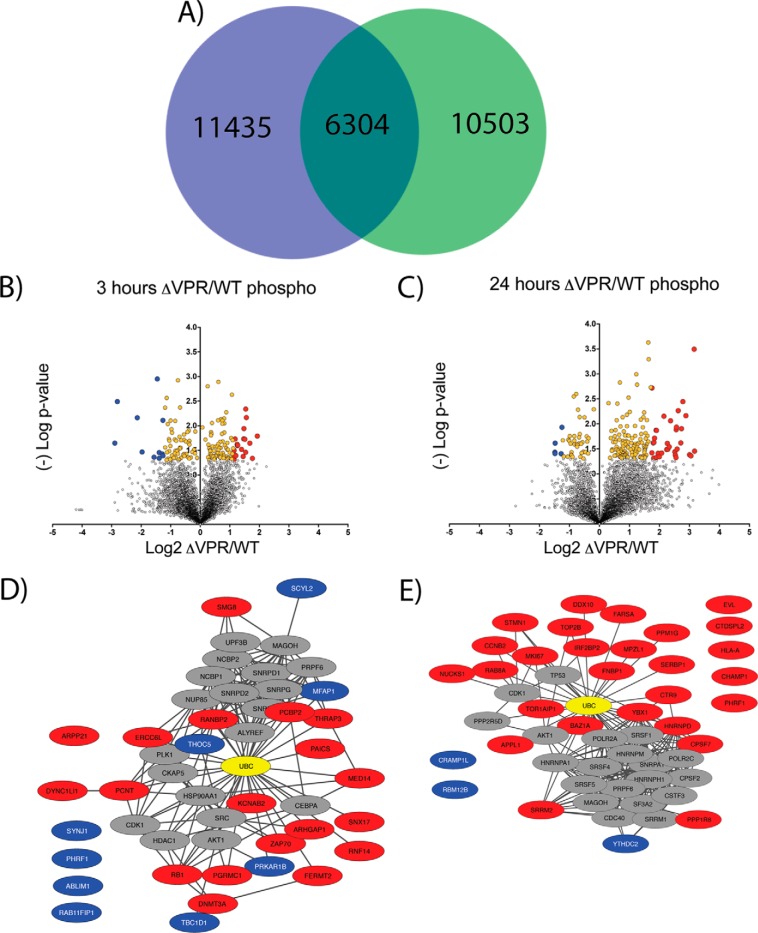
As a second means of interrogation, the global phospho-proteome dynamics during HIV-1 viral induction were defined (Fig. 6, supplemental File S11). Utilizing the Jurkat inducible HIV model system we assessed phospho-peptide level dynamics associated with HIV expression either with or without Vpr. In total, over 28,000 nonredundant phospho-peptides were identified and quantified, with 6304 in common (intersection) between WT and ΔVPR -expressing cells (Fig. 6A). The common intersection set of phospho-peptides was used for subsequent analyses.
Fig. 6.
- Phospho-proteomic profiling of ΔVPR cells reveals potential host targets of Vpr. A, Venn Diagram showing overlap of phospho-peptides identified and quantified between wild-type (green) and ΔVPR cell lines (blue). The 6,304 phospho-peptides in common were used for subsequent analyses. B, Volcano plot of ΔVPR relative to wild-type (WT) phospho-peptide level changes at 3 h post viral induction. X-axis shows log2 fold change and y axis (-)Log of Student's t test based p value. Yellow dots are significantly enriched proteins (p < 0.05) and blue and red dots are significantly enriched and greater than two standard deviations from the median up-(blue) or down-regulated (red) relative to ΔVPR. C, Volcano plot of ΔVPR relative to WT phospho-peptide level changes at 24 h post viral induction. D, STRING-db analysis of phospho-proteins significantly enriched (red and blue dots) in ΔVPR relative to WT HIV at 3 h post induction. E, STRING-db analysis of phospho-proteins significantly enriched (red and blue dots) in ΔVPR relative to WT HIV at 24 h post induction. Ubiquitin is highlighted in yellow in STRING plots.
Given Vpr's role in modulating the cell cycle, phosphorylation dynamics were investigated for both early (3 h) and late (24 h) times post-viral induction (Fig. 6B–6E). At three-hours post-induction, a significant enrichment of 20 phospho-peptides in ΔVPR cells relative to WT was observed (Fig. 6B, supplemental File S9, red dots p < 0.05, fold-change>2X standard deviation, right side of plot). For these phospho-peptides, Vpr was apparently necessary for their relative depletion in cells expressing WT HIV. Phospho-proteomic analysis indicated a greater than 2-fold, significant increase in RB1 phosphorylation in the ΔVPR cells by three hours after viral induction (Fig. 6D). RB1 phosphorylations are known to be inactivating, and have a role in cell cycle progression (57). Because RB1 is a substrate of CDK4, the relative decrease in the phosphorylation of RB1 in the presence of Vpr is consistent with the decrease in the amount of CDK4, although the timing of the changes in phosphorylation seem earlier than expected if because of Vpr. Other significantly altered phosphorylation events closely tie to proteins that regulate the cell cycle and to ubiquitin ligases at the three-hour time point.
To determine the relationships between the proteins from which these phospho-peptides originated, STRING-db was utilized (46). For STRING analysis, phosphorylations that were higher in WT expressing cells were included as well. Our rationale for this was that cell cycle regulation is highly dependent on a complex network of kinases and phosphatases. For example, phosphorylation of the splicing factor MFAP1 was increased in WT versus ΔVPR, suggesting a role for Vpr in its phosphorylation through modulation of the responsible MFAP1 phosphatase. Intriguingly, depletion of the MFAP1 homolog in Drosophila causes cells to arrest in G2/M, providing a possible connection between this and the G2/M arrest caused by Vpr (58, 59). THRAP3 is another pre-mRNA splicing-associated protein whose phosphorylation appears to be regulated by Vpr, except in the opposite direction; THRAP3 phospho-peptides were significantly decreased in WT compared with ΔVPR. Interestingly, THRAP3 is part of a complex that stabilizes cyclin-D1 mRNA (59); the observation of a significant increase in THRAP3 phosphorylation suggests a role for Vpr in regulating cyclin-D1 levels, which in turn regulate the activity of CDK4 and CDK6 during the G1/S phase transition.
Examining phosphorylation differences at 24-hours after induction revealed 33 significantly enriched phospho-peptides in the absence of Vpr (Fig. 6C, supplemental File S10). Interestingly, by this late time point, an obvious shift occurs in differentially phosphorylated peptides: proteins associated with DNA repair, vesicle trafficking, and phosphatases were more highly phosphorylated in the absence of Vpr (Fig. 6E). This, again, suggests a role for Vpr in the depletion of these phosphorylated peptides in cells expressing WT virus. E3 ligases also contain significantly higher levels of phosphorylation in the absence of Vpr at 24-hours post-induction. These findings, along with the phosphatase and cell cycle related proteins, are consistent with the whole cell quantitative proteomics-based findings.
STRING-db was also used to evaluate the significantly regulated phospho-peptides at the 24-hour time point. Again, ubiquitin was a central connecting node. As noted above, most of the significantly regulated phospho-proteins were involved in cell cycle regulation. Of note, several helicases had significantly higher levels of phosphorylation in cells expressing the WT virus relative to those expressing ΔVPR.
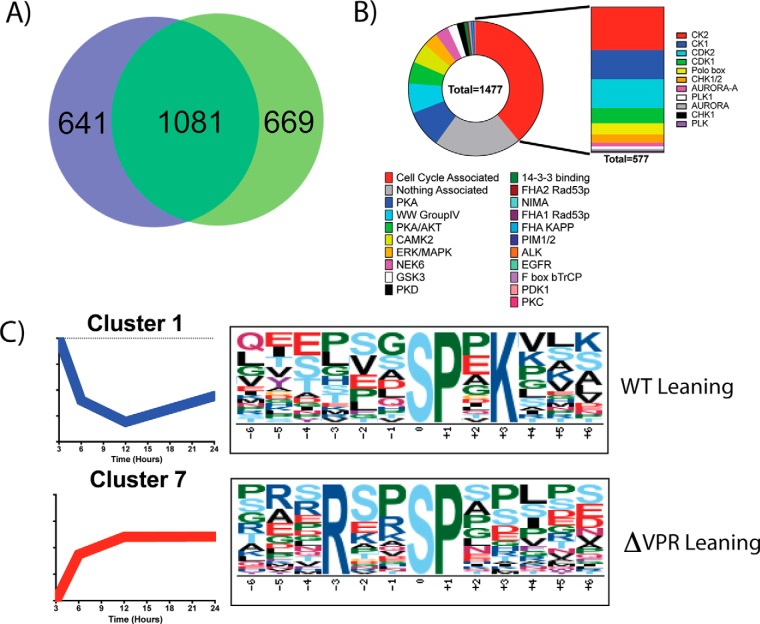
To assess the validity of the phospho-sites identified, the results were compared with the work of Wojcechowskyj et al. (13) (Fig. 7, supplemental File S12). In total, 1081 phospho-proteins overlapped between data sets (Fig. 7A). From these proteins, 1477 specific phospho-sites were in common. Wojecechowskyj et al. annotated motifs associated with each of these sites, allowing for the cross correlation of motifs (Fig. 7B). Of the 1477 sites defined in both studies, 577 (39%) were related to cell cycle regulation, which supports the observed phenotype of cell cycle arrest and the presented early and late time point analysis (Fig. 6).
Fig. 7.
- Phospho-proteomics reveals Vpr-modulated kinase motifs. A, Comparison of Inducible HIV with data from the Wojcechowskyj et al. data set shows a 1081 protein overlap for proteins with identified phosphorylation sites. B, Within the 1081 proteins, 1477 phospho-sites were identified as being the same between the two data sets. Previously annotated motifs show an enrichment for cell cycle associated domains. C, Motif analysis of phospho-peptides shows differential WT-leaning motifs (Higher in WT than ΔVPR) and ΔVPR-Leaning motifs (Higher in ΔVPR than WT). Full motif analysis can be found in supplemental Figs. S9–S14 and supplemental File S13.
To further characterize host processes that are specifically targeted by Vpr, the Motif-x algorithm was used to identify phosphorylation motifs that were enriched across the time course of viral gene expression (Fig. 7C, supplemental File S13) (60). For this analysis, the fold change of ΔVPR/WT quantification values was calculated for each phospho-peptide at each time point relative to the zero-time point. Performing k means clustering on the resulting data identified 9 distinct temporal profiles (supplemental Figs. S9–S14). Using this information, phosphorylation-motifs were identified as enriched in either the WT or ΔVPR expressing cells across the entire time course. For example, the Cdk2 phosphorylation motif “SPxK” was the most enriched motif in Cluster 1, a WT-leaning cluster (Fig. 7C) (61). Cyclin A/Cdk2 is active during the transition from S to M phase (62), which is consistent with the cell cycle arrest phenotype described above. Furthermore, cyclin A and Cdk2 total protein levels are increased in the cells expressing WT compared with cells expressing ΔVPR (supplemental File S3).
Another motif, “RxxSP,” which is the highest enriched in cluster 7 (Fig. 7C), a ΔVPR-leaning cluster, is a recognition motif for SRPKs (63). SRPK activity is required for proper splicing of mRNA transcripts and has previously been shown to be downregulated by HIV expression (64). However, SRPK activity appears increased in the ΔVPR-expressing cells, which suggests that Vpr inhibits their function during the expression of WT HIV. Additional phosphorylation motifs identified include predicted motifs for casein kinase 2 (CK2), JNK, ERK, Fam20C, and PKA among others (supplemental Figs. S9–S14). The motifs predicted to be recognized by CK2 (SxxE, SxxD, SxE, SxD, SS) are of interest as CK2 is a known kinase for the HIV proteins Vpu and Rev (65, 66). These motifs are present in both WT and ΔVPR leaning clusters. Thus, although Vpr does not seem to degrade CK2 or inhibit it completely, it may modulate its specificity. Interestingly, of the putative CK2 phosphorylation sites identified, 144 are unique to WT leaning clusters and 324 are unique to ΔVPR leaning clusters.
DISCUSSION
Through this work, we sought to comprehensively, and in an unbiased manner, characterize proteome and phospho-proteome dynamics associated with HIV-1 in a new, inducible CD4+ T-cell based model system, with special emphasis on the role of the accessory protein Vpr The experimental model presented herein represents a complementary tool to traditional HIV infection models (17), and it has advantages in regards to the synchronicity and uniformity of viral gene expression. The use of a self-limiting virus (because of a self-inactivating deletion in the 3′ LTR) that is stably integrated into the genome of Jurkat cells provides the additional advantage of reducing hazards associated with HIV while still studying its effects in a biologically relevant cell type. Placing viral gene expression under the control of a tetracycline-inducible promoter provides the advantages of fine tuning and scaling, rendering the model ideal for proteomic studies. Induction of HIV gene expression with doxycycline does come with certain caveats (67), such as tetracycline-induced perturbations of mitochondria. However, such potential confounding effects were mitigated here by comparing two conditions—cells expressing the WT genome and cells expressing a vpr-negative genome—both of which were treated with doxycycline, so any effects of doxycycline on the cells were likely normalized away.
Proteomic analysis using this inducible HIV model identified and quantified over 7000 proteins, with nearly 5800 of these in common between the WT and ΔVPR expressing cells. From this set of 5793 identified and quantified proteins, a time dependent increase in the five HIV proteins detected in the experiment was observed. These findings were observed reproducibly across both cell lines (Fig. 2B), and corroborated by Western blotting (Fig. 5). Upon comparing the data generated here to a recently published study (17), this model was further validated through the observation of similar shifts of proteins in WT expressing cells (Fig. 3A, supplemental Figs. S5–S8). These included time-dependent decreases in APOBEC3C as well as PP2A subunits.
Though statistical significance was not reached, Vpr-dependent decreases for the known degradation targets UNG (9), HLTF (10, 11) and MUS81 (12) were noted (supplemental Fig. S10). Another target of Vpr, PLK1, is known to become increasingly phosphorylated on threonine 592 in response to Vpr (12). Although phosphorylation of PLK1 was not detected here, a time dependent increase in PLK1 protein levels was observed in WT HIV but not ΔVPR expressing cells (supplemental Fig. S15). Interestingly, PLK1 is a connecting node in Fig. 6D and PLK motifs are implicated in Fig. 7B, suggesting an important role for this kinase; however, a clear motif for PLK did not emerge from the Motif-x analysis (supplemental Figs. S9–S14).
Data obtained from both the proteomics and phospho-proteomics portions of this study support numerous previous observations of Vpr's role in cell cycle arrest during G2/M phase. For example, the proteomics data indicated enrichment of CDK4 in cells expressing the ΔVPR genome relative to cells expressing the WT. These data suggest that CDK4 might be targeted by Vpr for degradation. Because CDK4 is a known regulator of G1/S progression (53, 54), this observation seems reasonable. Additionally, the up-regulation of the phosphorylation of RB1 in the absence of Vpr is potentially consistent with the effects of Vpr on CDK4, which phosphorylates RB1. The presence of the well characterized Vpr-interacting protein VPRBP/DCAF1 (51, 52) in this enriched protein data set further supports the hypothesis that Vpr was necessary for the depletion of proteins found to be of significantly higher abundance in the cells expressing ΔVPR than in cells expressing the WT. By identifying proteins that followed similar abundance dynamics to VPRBP, we identified new candidate targets for Vpr-mediated degradation (Fig. 5, supplemental Fig. S8); CDK4 was among these candidates.
Unexpectedly, the phospho-proteomic data in this study suggest a potential role for Vpr in the modulation of splicing via an influence on SRPK activity. Although WT HIV has been reported previously to affect SRPK activity, this effect has not been associated with Vpr until now. SRPKs are known to upregulate HIV protein expression (64). Follow-up studies of the mRNA transcriptome now have the potential to reveal an impact of Vpr on the splicing of viral and cellular transcripts.
STRING based analysis of proteomic and phospho-proteomic data revealed the ties of enriched proteins and phospho-proteins to the cell cycle and centromere regulation, as well as to ubiquitin-based pathways. A common protein in each of these pathway analyses (Figs. 4C, 6D–6E) was ubiquitin as a major hub. Other clusters of proteins with known relationships to HIV became obvious through STRING analysis, including the spliceosome and proteins involved in the repair of DNA damage.
An important caveat to interpreting our study is the difficulty in sorting out which modulations are directly because of Vpr and which are secondary to Vpr's effects on the cell cycle. Direct effects of Vpr would presumably be because of its ability to degrade targets by recruiting the DCAF1-containing cullin 4-based E3 ubiquitin ligase; those potential effects are reflected in the quantitative whole cell proteomics herein. When these targets are kinases and phosphatases, the consequent impact of Vpr should be reflected in our phospho-peptide data. The potential relationships suggested herein provide a basis for cell biological and biochemical validation aimed at dissecting these complex interrelationships.
In summary, through a multiplexed proteomics approach, we defined protein dynamics for over 7000 proteins and 28,000 phospho-peptides in response to the expression of HIV-1 Vpr, representing an advance in the field of HIV systems level approaches. This study largely serves as a characterization of a novel model system to study HIV in the context of events surrounding viral protein expression. Our model was validated through proteomic and traditional means and compared against other HIV models for reproducibility of known effects. This study presents a new and valuable resource in our proteomic data set and a new model system to the field of HIV-1 research. Through the development of these novel tools and characterizations, we can gain insight into how HIV overcomes host defenses and coopts cellular processes, and so inform efforts to devise new approaches to treat and cure HIV infection.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited into MassIVE (http://massive.ucsd.edu) and submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with the data set identifiers MSV000080877 and PXD006230 respectively.
Supplementary Material
Footnotes
Author contributions: J.L., J.G., M.L., and D.G. conceived and designed the experiments. J.L. and M.L. performed the experiments. J.L., J.G., M.L., J.W., and D.G. participated in the discussions and analysis of the data. J.L., J.G., M.L., J.W., and D.G. wrote the manuscript.
* This work is supported by the UCSD Center for AIDS Research (5 P30 AI036214) (D.G.), Ray Thomas Edwards Foundation (D.G.), M.K.L. was supported by NIH K08 AI112394, and a Developmental Grant from the University of California, San Diego, Center for AIDS Research (CFAR), an NIH-funded program (P30 AI036214). J.G. was supported by NIH R37 AI081668. D.G. is supported by the University of California Office of the President. J.L. was supported by NIH K12 GM06852. J.W. was supported by Graduate Training in Cellular and Molecular Pharmacology Training Grant NIH T32 GM007752. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HIV
- human immunodeficiency virus
- AGC
- automatic gain control
- AIDS
- acquired immuno-deficiency syndrome
- BCA
- bicinchoninic acid
- CD4
- cluster of differentiation 4
- DAVID
- database for annotation, visualization and integrated discovery
- DNA
- deoxyribonucleic acid
- DTT
- dithoithreitol
- ΔVPR
- Vpr deletion
- EDTA
- ethylenediaminetetraacetic acid
- HEPES
- 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HPLC
- high performance liquid chromatography
- LC
- liquid chromatography
- MOI
- multiplicity of infection
- mRNA
- messenger ribonucleic acid
- MS2
- tandem mass spectrometry
- MS3
- three stage mass spectrometry
- NaCl
- sodium chloride
- NaF
- sodium fluoride
- PAGE
- polyacrylamide gel electrophoresis
- PBS
- phosphate buffered saline
- PMSF
- phenylmethylsulfonyl fluoride
- PTM
- post-translational modification
- PVDF
- polyvinylidene fluoride
- RF
- radio frequency
- RNA
- ribonucleic acid
- RPMI
- Roswell Park Memorial Institute medium
- STEM
- short time series expression miner
- STRING
- search tool for recurring instances of neighboring genes
- SDS
- sodium dodecyl sulfate
- TET
- tetracycline
- TFA
- trifluoroacetic acid
- TMT
- tandem mass tags
- Tris-HCl
- Tris(hydroxymethyl)aminomethane hydrochloride
- UCSD
- University of California, San Diego
- WT
- wild-type.
REFERENCES
- 1. UNAIDS. (2016) UNAIDS Fact Sheet 2016. 2016 Ed., Unaids.org
- 2. Gisslen M., Svedhem V., Lindborg L., Flamholc L., Norrgren H., Wendahl S., Axelsson M., and Sonnerborg A. (2016) Sweden, the first country to achieve the Joint United Nations Programme on HIV/AIDS (UNAIDS)/World Health Organization (WHO) 90–90-90 continuum of HIV care targets. HIV Med. 18, 305–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li M. (2015) Proteomics in the investigation of HIV-1 interactions with host proteins. Proteomics Clin. Appl. 9, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garcia F., Plana M., Vidal C., Cruceta A., O'Brien W. A., Pantaleo G., Pumarola T., Gallart T., Miro J. M., and Gatell J. M. (1999) Dynamics of viral load rebound and immunological changes after stopping effective antiretroviral therapy. AIDS 13, F79–F86 [DOI] [PubMed] [Google Scholar]
- 5. Frankel A. D., and Young J. A. (1998) HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67, 1–25 [DOI] [PubMed] [Google Scholar]
- 6. Malim M. H., and Emerman M. (2008) HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398 [DOI] [PubMed] [Google Scholar]
- 7. Margottin F., Bour S. P., Durand H., Selig L., Benichou S., Richard V., Thomas D., Strebel K., and Benarous R. (1998) A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1, 565–574 [DOI] [PubMed] [Google Scholar]
- 8. Ako-Adjei D., Fu W., Wallin C., Katz K. S., Song G., Darji D., Brister J. R., Ptak R. G., and Pruitt K. D. (2015) HIV-1, human interaction database: current status and new features. Nucleic Acids Res. 43, D566–D570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schrofelbauer B., Yu Q., Zeitlin S. G., and Landau N. R. (2005) Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 79, 10978–10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hrecka K., Hao C., Shun M. C., Kaur S., Swanson S. K., Florens L., Washburn M. P., and Skowronski J. (2016) HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E3921–E3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lahouassa H., Blondot M. L., Chauveau L., Chougui G., Morel M., Leduc M., Guillonneau F., Ramirez B. C., Schwartz O., and Margottin-Goguet F. (2016) HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages. Proc. Natl. Acad. Sci. U.S.A. 113, 5311–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laguette N., Bregnard C., Hue P., Basbous J., Yatim A., Larroque M., Kirchhoff F., Constantinou A., Sobhian B., and Benkirane M. (2014) Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156, 134–145 [DOI] [PubMed] [Google Scholar]
- 13. Wojcechowskyj J. A., Didigu C. A., Lee J. Y., Parrish N. F., Sinha R., Hahn B. H., Bushman F. D., Jensen S. T., Seeholzer S. H., and Doms R. W. (2013) Quantitative phosphoproteomics reveals extensive cellular reprogramming during HIV-1 entry. Cell Host Microbe 13, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Binette J., Dube M., Mercier J., Halawani D., Latterich M., and Cohen E. A. (2007) Requirements for the selective degradation of CD4 receptor molecules by the human immunodeficiency virus type 1 Vpu protein in the endoplasmic reticulum. Retrovirology 4, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romani B., Shaykh Baygloo N., Aghasadeghi M. R., and Allahbakhshi E. (2015) HIV-1 Vpr Protein Enhances Proteasomal Degradation of MCM10 DNA Replication Factor through the Cul4-DDB1[VprBP] E3 Ubiquitin Ligase to Induce G2/M Cell Cycle Arrest. J. Biol. Chem. 290, 17380–17389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinberg R. A. (1995) The retinoblastoma protein and cell cycle control. Cell 81, 323–330 [DOI] [PubMed] [Google Scholar]
- 17. Greenwood E. J., Matheson N. J., Wals K., van den Boomen D. J., Antrobus R., Williamson J. C., and Lehner P. J. (2016) Temporal proteomic analysis of HIV infection reveals remodelling of the host phosphoproteome by lentiviral Vif variants. Elife 5, e18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chesebro B., Wehrly K., Nishio J., and Perryman S. (1992) Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66, 6547–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Planelles V., Jowett J. B., Li Q. X., Xie Y., Hahn B., and Chen I. S. (1996) Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 70, 2516–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., Di Marzio P., Marmon S., Sutton R. E., Hill C. M., Davis C. B., Peiper S. C., Schall T. J., Littman D. R., and Landau N. R. (1996) Identification of a major co-receptor for primary isolates of HIV-1. Nature 381, 661–666 [DOI] [PubMed] [Google Scholar]
- 21. Morgenstern J. P., and Land H. (1990) Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18, 3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Charneau P., Mirambeau G., Roux P., Paulous S., Buc H., and Clavel F. (1994) HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241, 651–662 [DOI] [PubMed] [Google Scholar]
- 23. Day J. R., Martinez L. E., Sasik R., Hitchin D. L., Dueck M. E., Richman D. D., and Guatelli J. C. (2006) A computer-based, image-analysis method to quantify HIV-1 infection in a single-cycle infectious center assay. J. Virol. Methods 137, 125–133 [DOI] [PubMed] [Google Scholar]
- 24. Means R. E., Matthews T., Hoxie J. A., Malim M. H., Kodama T., and Desrosiers R. C. (2001) Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75, 3903–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Villen J., and Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haas W., Faherty B. K., Gerber S. A., Elias J. E., Beausoleil S. A., Bakalarski C. E., Li X., Villen J., and Gygi S. P. (2006) Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics 5, 1326–1337 [DOI] [PubMed] [Google Scholar]
- 27. Wessel D., and Flugge U. I. (1984) A Method for the Quantitative Recovery of Protein in Dilute-Solution in the Presence of Detergents and Lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 28. Tolonen A. C., and Haas W. (2014) Quantitative proteomics using reductive dimethylation for stable isotope labeling. Jove-J Vis Exp. 89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villen J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jedrychowski M. P., Huttlin E. L., Haas W., Sowa M. E., Rad R., and Gygi S. P. (2011) Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Mol. Cell. Proteomics 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McAlister G. C., Nusinow D. P., Jedrychowski M. P., Wuhr M., Huttlin E. L., Erickson B. K., Rad R., Haas W., and Gygi S. P. (2014) MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thompson A., Schafer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., and Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 33. Wang Y., Yang F., Gritsenko M. A., Wang Y., Clauss T., Liu T., Shen Y., Monroe M. E., Lopez-Ferrer D., Reno T., Moore R. J., Klemke R. L., Camp D. G. 2nd, and Smith R. D. (2011) Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11, 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eng J. K., Mccormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectr. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 35. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 36. Elias J. E., Haas W., Faherty B. K., and Gygi S. P. (2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat.Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 37. Peng J., Elias J. E., Thoreen C. C., Licklider L. J., and Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 38. Beausoleil S. A., Villen J., Gerber S. A., Rush J., and Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 39. Ting L., Rad R., Gygi S. P., and Haas W. (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quackenbush J. (2002) Microarray data normalization and transformation. Nat. Genet. 32, 496–501 [DOI] [PubMed] [Google Scholar]
- 41. Wu J. (2012) Advances in k-means clustering, Springer, New York [Google Scholar]
- 42. Ernst J., and Bar-Joseph Z. (2006) STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang, da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 44. Huang, da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nicolay B. N., Danielian P. S., Kottakis F., Lapek J. D. Jr, Sanidas I., Miles W. O., Dehnad M., Tschop K., Gierut J. J., Manning A. L., Morris R., Haigis K., Bardeesy N., Lees J. A., Haas W., and Dyson N. J. (2015) Proteomic analysis of pRb loss highlights a signature of decreased mitochondrial oxidative phosphorylation. Genes Dev. 29, 1875–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simon V., Bloch N., and Landau N. R. (2015) Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 16, 546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sugden S. M., Bego M. G., Pham T. N., and Cohen E. A. (2016) Remodeling of the host cell plasma membrane by HIV-1 Nef and Vpu: a strategy to ensure viral fitness and persistence. Viruses 8, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Andersen J. L., Le Rouzic E., and Planelles V. (2008) HIV-1 Vpr: mechanisms of G2 arrest and apoptosis. Exp. Mol. Pathol. 85, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jowett J. B., Planelles V., Poon B., Shah N. P., Chen M. L., and Chen I. S. (1995) The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 69, 6304–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Belzile J. P., Abrahamyan L. G., Gerard F. C., Rougeau N., and Cohen E. A. (2010) Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog. 6, e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hrecka K., Gierszewska M., Srivastava S., Kozaczkiewicz L., Swanson S. K., Florens L., Washburn M. P., and Skowronski J. (2007) Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Serrano M., Gomez-Lahoz E., DePinho R. A., Beach D., and Bar-Sagi D. (1995) Inhibition of ras-induced proliferation and cellular transformation by p16INK4. Science 267, 249–252 [DOI] [PubMed] [Google Scholar]
- 54. Sicinski P., Donaher J. L., Parker S. B., Li T., Fazeli A., Gardner H., Haslam S. Z., Bronson R. T., Elledge S. J., and Weinberg R. A. (1995) Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82, 621–630 [DOI] [PubMed] [Google Scholar]
- 55. Schneider U., Schwenk H. U., and Bornkamm G. (1977) Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer 19, 621–626 [DOI] [PubMed] [Google Scholar]
- 56. Foley G. E., Lazarus H., Farber S., Uzman B. G., Boone B. A., and McCarthy R. E. (1965) Continuous culture of human lymphoblasts from peripheral blood of a child with acute Leukemia. Cancer 18, 522–529 [DOI] [PubMed] [Google Scholar]
- 57. Narasimha A. M., Kaulich M., Shapiro G. S., Choi Y. J., Sicinski P., and Dowdy S. F. (2014) Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Andersen D. S., and Tapon N. (2008) Drosophila MFAP1 is required for pre-mRNA processing and G2/M progression. J. Biol. Chem. 283, 31256–31267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bracken C. P., Wall S. J., Barre B., Panov K. I., Ajuh P. M., and Perkins N. D. (2008) Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 68, 7621–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chou M. F., and Schwartz D. (2011) Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics Chapter 13, Unit 13 15–24, John Wiley and Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 61. Kitagawa M., Higashi H., Jung H. K., Suzuki-Takahashi I., Ikeda M., Tamai K., Kato J., Segawa K., Yoshida E., Nishimura S., and Taya Y. (1996) The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15, 7060–7069 [PMC free article] [PubMed] [Google Scholar]
- 62. Chung J. H., and Bunz F. (2010) Cdk2 is required for p53-independent G2/M checkpoint control. Plos Genet. 6, e1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lipp J. J., Marvin M. C., Shokat K. M., and Guthrie C. (2015) SR protein kinases promote splicing of nonconsensus introns. Nat. Struct. Mol. Biol. 22, 611–617 [DOI] [PubMed] [Google Scholar]
- 64. Fukuhara T., Hosoya T., Shimizu S., Sumi K., Oshiro T., Yoshinaka Y., Suzuki M., Yamamoto N., Herzenberg L. A., Herzenberg L. A., and Hagiwara M. (2006) Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. U.S.A. 103, 11329–11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schubert U., Schneider T., Henklein P., Hoffmann K., Berthold E., Hauser H., Pauli G., and Porstmann T. (1992) Human-immunodeficiency-virus-type-1-encoded Vpu protein is phosphorylated by casein kinase II. Eur. J. Biochem. 204, 875–883 [DOI] [PubMed] [Google Scholar]
- 66. Meggio F., D'Agostino D. M., Ciminale V., Chieco-Bianchi L., and Pinna L. A. (1996) Phosphorylation of HIV-1 Rev protein: implication of protein kinase CK2 and pro-directed kinases. Biochem. Biophys. Res. Commun. 226, 547–554 [DOI] [PubMed] [Google Scholar]
- 67. Moullan N., Mouchiroud L., Wang X., Ryu D., Williams E. G., Mottis A., Jovaisaite V., Frochaux M. V., Quiros P. M., Deplancke B., Houtkooper R. H., and Auwerx J. (2015) Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited into MassIVE (http://massive.ucsd.edu) and submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with the data set identifiers MSV000080877 and PXD006230 respectively.