Abstract
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis, the leading cause of death among all infectious diseases. There are 11 eukaryotic-like serine/threonine protein kinases (STPKs) in Mtb, which are thought to play pivotal roles in cell growth, signal transduction and pathogenesis. However, their underlying mechanisms of action remain largely uncharacterized. In this study, using a Mtb proteome microarray, we have globally identified the binding proteins in Mtb for all of the STPKs, and constructed the first STPK protein interaction (KPI) map that includes 492 binding proteins and 1,027 interactions. Bioinformatics analysis showed that the interacting proteins reflect diverse functions, including roles in two-component system, transcription, protein degradation, and cell wall integrity. Functional investigations confirmed that PknG regulates cell wall integrity through key components of peptidoglycan (PG) biosynthesis, e.g. MurC. The global STPK-KPIs network constructed here is expected to serve as a rich resource for understanding the key signaling pathways in Mtb, thus facilitating drug development and effective control of Mtb.
Tuberculosis (TB)1 is now the leading cause of death from infectious diseases, with more than 1.5 million deaths just in 2014 (1). With nearly 10 million newly identified cases in 2014, and the rapid emergence of multidrug resistant (MDR) and extensively drug resistant (XDR) strains of Mycobacterium tuberculosis (Mtb), a causative agent of tuberculosis, the threat of this disease to public health worldwide continues to grow substantially (2). There is thus an urgent need for effective vaccines and drugs to reduce the global burden of TB.
One of the most significant reasons for the slow progress in discovering and characterizing appropriate vaccine candidates and drug targets has been the general lack of knowledge about basic biological pathways in Mtb and how they may be regulated.
Phosphorylation is widely recognized to play a significant role in signal transduction in many pathways in both eukaryotes and prokaryotes (3). Serine/threonine protein kinases (STPKs) are one of the major groups of protein kinases in eukaryotes. In contrast to many of the widely studied bacterial pathogens and model organisms that have few or no STPKs but many two-component systems, the Mtb genome encodes 11 STPKs (4). According to their sequence similarity, these STPKs have been grouped into five clades, namely Clade I (pknA, pknB, pknL), Clade II (pknD, pknE, pknH), Clade III (pknF, pknI, pknJ), Clade IV (pknG) and Clade V (pknK) (5). It is known that these STPKs play critical roles in adaptation to various environmental conditions (6), cell wall synthesis (7), cell division (8) and pathogenicity (9). However, except for a few relatively well-studied STPKs, such as PknA, PknB (10, 11), and PknG (12), these STPKs are poorly characterized. Further, how these STPKs function on a systems-wide level is completely unknown.
To understand the roles of STPKs systematically, a highly effective first step is the characterization of the global protein-protein interactions (PPIs) in which the STPKs are involved. Although there are a handful of known STPK-protein interactions (KPIs) in Mtb, e.g. PknG and GarA (13), which regulates tricarboxylic acid cycle and glutamate synthesis, PknB and Wag31 (14), a component for cell division, PknB and a maltosyltransferase GlgE (15). These known KPIs are far too few to enable a systems-wide understanding of the roles of STPKs in Mtb.
Protein microarray is a powerful tool for proteome-wide study (16), including those that delineate global PPIs (17, 18). To accelerate our understanding of Mtb and tuberculosis, an Mtb proteome microarray, carrying most of the Mtb proteins, has recently been constructed particularly for the characterization of PPIs (19).
Herein, we applied the Mtb proteome microarray to identify the roles of the Mtb STPKs on a systems-wide level. Overall, we obtained a KPIs network of 492 binding proteins and 1027 interactions, which represents the first and the most comprehensive PPIs network of STPKs in Mtb. Bioinformatic analysis revealed that cell wall related activities were highly enriched in this network, whereas functional analysis confirmed this observation and demonstrated that PknG can regulate Mtb cell wall biogenesis through interaction with the Mur ligase MurC. We believe that the STPK-KPIs network presented here will become an invaluable resource in future biological and clinical studies of Mtb.
EXPERIMENTAL PROCEDURES
KPI Screening on the Mtb Proteome Microarray
M. tuberculosis protein microarrays containing about 4200 full-length GST fusion proteins spotted in duplicate were used in this study (19). Microarrays were blocked for 1 h at RT with shaking in blocking buffer (1× PBS at pH 7.4, 5% BSA, 0.1% Tween-20). Purified V5 tagged kinases probe was diluted in probing buffer (1× PBS, 0.1% Tween-20, 1% BSA). After blocking, the microarrays were incubated in 3 ml of the diluted kinase pool at a final concentration of 10–50 μg/ml at 4 °C overnight. The arrays were then washed 3 times, 10 min each in 25 ml of PBST (1× PBS at pH 7.4, 0.1% Tween-20) buffer at RT with shaking at 60 rpm. The microarrays were then probed with 1 ml of anti-V5 antibody (Sigma-Aldrich, Saint Louis, MI) at a concentration of 260 ng/ml in PBST buffer at RT for 1 h. Following antibody detection, the microarrays were probed with a cy5-conjugated antibody (Jackson Immuno Research, West Grove, PA) at a concentration of 260 ng/ml in PBST buffer at RT for 1 h. The microarrays were washed in 25 ml of PBST buffer for 3 times 10 min each and air dried. The microarray results were recorded using a 4200AL microarray scanner (Molecular Devices, Abingdon, UK). The image file was processed using Genepix software (Molecular Devices).
Protein Microarray Data Process Analysis
The process of data analysis includes four steps: (1) background correction, (2) normalization, (3) identification of positive hits, and (4) removal of nonspecific interactions. The first step, background correction, is critical to reduce background noise. Next, we quantified the signal intensity of each protein on the array through dividing the median foreground intensity by the median background. To identify positive hits on the microarrays, we estimated the standard deviation from the signal intensity distribution, as described in detail previously (20). A cutoff of four standard deviations above the mean was chosen in this study. Since each protein was printed in duplicate, a protein was scored as a positive hit only if both spots of the same protein showed signal intensity higher than a cutoff value determined for that microarray experiment.
Biolayer Interferometry Analysis
The binding kinetics of kinase and their interactors were measured using a ForteBio 70 Octet system. Affinity purified kinases PknB, PknG, PknD and PknH was biotinylated using an EZ-Link Sulfo-NHS-LC-Biotin protein biotinylation kit (Thermo Scientific, Bremen, Germany) according to the manufacturer's instructions. Biotinylated kinases were tethered on the tip surface of a streptavidin-coated sensor. The binding partner in S.D. buffer (1 × PBS, pH 7.4 with 0.02% Tween-20 and 0.1% BSA) was then exposed to tethered biotinylated kinases and binding was measured by coincident change in the interference pattern. SSA tips (Pall ForteBio, Menlo Park, NJ) were prewet in S.D. buffer, which served as the background buffer for the immobilization. This involved establishing a stable baseline (60 s), loading a 50 μg/ml kinase (300 s), balancing tips with S.D. buffer (60 s), association with 500 mm substrate (dissolved in S.D. buffer) (300 s) and then eluting nonspecific binding with S.D. buffer (300 s). Final immobilization levels were 1.0 ± 0.2 nm. Data was analyzed using the ForteBio Data Analysis Software 7. The data was fit to a 1:1 binding model to calculate an association and dissociation rate, and KD was calculated using the ratio Kd/Ka.
Yeast-two-hybrid (Y2H) Analysis
The PknG coding ORF sequence and PknB, PknD, PknH kinase domain sequences were cloned into the bait vector pGBKT7, and the interactors coding sequences into the prey vector pGADT7, which was modified into a Gateway system. 96 gene plasmids of interacting proteins with the BP clones were selected from our Mtb BP clone library, and straightly joined to the prey vector with one step of LR reaction. Y2H interaction assays were performed using the Matchmaker 2-hybrid system (Takara Bio, Tokyo, Japan) according to the manufacturer's instructions. The two recombinant plasmids were cotransformed to the Yeast strains (AH109) and grown on the medium of S.D./-Leu, -Trp for selecting the positive transformants. And then with the S.D./-Leu, -Trp, -His, -Ade medium to select the interactors.
Mycobacterial Protein Complementation Assay
The mycobacterial protein fragment complementation (MPFC) assay was performed as described (21). The genes of interest were PCR-amplified and cloned into pUAB300, PknB and PknD were cloned into pUAB400, whereas pUAB100 (expressing mDHFR fragment F1, 2) and pUAB200 (expressing mDHFR fragment F3) were set as positive controls. M. smegmatis (Msm) was cotransformed with both plasmids; the transformants were selected on 7H10 agar plates with 25 μg/ml kanamycin and 50 μg/ml hygromycin and tested for growth over 4 days on 7H10 kanamycin/hygromycin plates supplemented with 20 μg/ml trimethoprim (TMP).
Experimental Design and Statistical Rationale
Purification of STPKs With Its Interaction Proteins
The four kinases (PknB, PknD, PknG and PknH) were cloned to pET28a and expressed in E. coli BL21 (DE3). While the interaction proteins (15 substrates/kinase) were constructed with vector pDEST15 using the Gateway system and expressed in E. coli BL21 (DE3). The STPKs and the interacting proteins were purified with Ni2+ Sepharose beads and GST-Sepharose beads, respectively, according to the manufacturer's instruction. The proteins were quantified with silver staining (Beyotime, Nantong, China).
In Vitro Kinase Phosphorylation
In vitro phosphorylation was performed as described (15). Briefly, the reaction is composed of 50 μg substrate in 200 μl of kinase buffer (50 mm Tris-HCl, pH 7.0, 1 mm DTT, 5 mm MgCl2, 2 mm MnCl2) with 5 mm ATP, and 10 μg kinase, and a control phosphorylation assay without ATP was included for each of the selected KPIs. As a sum, 60 substrates were carried out for kinase reaction and followed with 60 control reaction. The reactions were carried out for 2 h at 37 °C. The phosphorylation events were then read out by immunoblot with an anti-ser/thr phosphorylation antibody (Abcam, Cambridge, USA). The experiments were carried out twice.
In-solution Trypsin Digestion and Enrichment of Phosphopeptides with TiO2 Resin
After phosphorylation reactions, to obtain high efficiency for MS analysis, >10 samples were grouped as one. In total, 8 groups were prepared. The grouped samples were concentrated to a protein adsorption film (PALL, Port Washington, NY), washed with 50 mm ammonium bicarbonate for 3 times at 4 °C centrifuged at 10,000 × g for 20 min, and reduced with carboxyamidomethyl at 60 °C for 30 min. Proteins were digested with sequencing grade modified trypsin (Promega, Madison, WI) at 37 °C for 16 h - 24 h. The tryptic digested protein sample was eluted from the membrane with 50 mm ammonium bicarbonate for 2 times, and then dried by lyophilization. Phosphopeptides from digested peptides were enriched by using Phosphopeptide Enrichment TiO2 Mag Sepharose (GE healthcare life sciences, Parramatta, Australia) according to the manufacturer's instruction. Briefly, the dried peptides were redissolved in 200 μl TiO2 phospho binding buffer containing 1 m glycolic acid in 80% acetonitrile, 5% trifluoroacetic acid and then mixed with 50 μl TiO2 phospho binding Resin. After 1 h incubation, the supernatant was discarded, and TiO2 resin was washed 3 times with the wash buffer (80% acetonitrile, 1% trifluoroacetic acid). After that, the phosphopeptides were eluted by adding 50 μl elution buffer (5% ammonium hydroxide, pH ∼ 12). The eluted fractions were combined and dried by lyophilization and reconstituted in 20 μl of 0.1% formic acid (FA) for LC-MS/MS analysis (22).
Mass Spectrometry Analysis
The phosphorylation peptides were analyzed by online nanoflow liquid chromatography tandem mass spectrometry (LC-MS/MS) on a Q Exactive Plus quadrupole orbitrap mass spectrometer (Thermo fisher Scientific). Samples (1 μl) were loaded by an autosampler onto a 2 cm packed pre-column (75 μm ID × 360 μm OD) in 0.1% HCOOH/water (buffer A) at a flow rate of 1 μl/min for 5 min. Analytical separation was performed over a 15 cm packed column (75 μm ID x 360 μm OD) at 300 nl/min with a 60 min gradient of increasing CH3CN (buffer B, 0.1% HCOOH/CH3CN). Both precolumn (5 μm diameter, 200 Å pore size) and analytical column (3 μm diameter, 100 Å pore size) were packed with C18-reversed phase silica (DIKMA-inspireTM) using a pressure bomb. Following sample loading, buffer B was increased rapidly from 3% to 6% over 5 min and then shallowly to 22% over 36 min, and then to 35% over 9 min followed by a quick increase to 95% over 3 min, and hold at 95% for 7 min. The total acquisition duration lasted for 60 min. Survey full scan MS spectra (m/z 350–1800) were acquired in the Orbitrap with 70 000 resolution (m/z 200) after accumulation of ions to a 3 × 106 target value based on predictive AGC from the previous full scan. Dynamic exclusion was set to 60 s.
Mass Spectrometry Data Processing
The raw files were processed using the LC/MS software Data Analysis 4.0 (Bruker Compass software). For the improved identification of phosphopeptides, all MS/MS samples were analyzed using MASCOT 2.3 (23). MASCOT were set up to search a database of M.tuberculosis database from SwissProt (http://web.expasy.org/docs/swiss-prot_guideline.html) containing 2067 annotated protein sequences. The search criteria were as follows: trypsin digestion; carbamidomethylation (Cys) was set as fixed modification, whereas oxidation (M), phospho (ST), and phospho (Y) were considered as variable modifications; and two missed cleavages were allowed. Allowed maximum mass deviation was 0.4 Da (monoisotopic) for the precursor ion and fragment maximum mass deviation was 0.6 Da (monoisotopic). For results searching, the peptide-spectrum matches (PSMs) were filtered based on the score threshold of a 1% false discovery rate (FDR), according to the formula: FDR = 2[nDecoy/(nDecoy + nTarget)] (24). And also, all fragmentation spectra of the phosphorylated peptides assigned to the corresponding proteins were manually evaluated and identified according to the neutral loss of ion MH+-80. Finally, to pinpoint the actual phosphorylation site within the identified peptide, the localization probabilities for phosphorylation sites were calculated from the posttranslational modification score algorithm with PhosphoRS analysis using Protein Discovery software (Thermo). Phosphorylation sites that were occupied with a probability > 0.95 were identified as phosphorylation sites.
Coimmunoprecipitation
Bacteria harboring pTetInt-PknG, pGrna-MurC pGrna-MurC was grown at 37 °C with shaking at 200 rpm overnight to OD600 = 0.6. And then induced with anhydrote-tracycline (ATc) at concentrations of 20 ng/μl for 24 h to harvested. The supernatant was resuspended with PBS, and lyzed through high pressure cracker (Union-Biotech, Shanghai, China). Lysate was centrifuged to separate the soluble and insoluble fractions. Soluble fractions were used for coimmunoprecipitation. Briefly, mouse anti-flag antibody (Sigma-Aldrich, St. Louis, MO) was added to 2 mg of soluble lysate (1:100 dilution), and the mixture was incubated at 4 °C with shaking for overnight. Affi-Gel protein G agarose resin (Roche, Basel, Switzerland) was then added to the lysate and incubated at at 4 °C with shaking for 4 h to bind to the antibody-antigen complexes. The resin was then washed with PBST for 6 times, and SDS-PAGE loading buffer (Beyotime, Nantong, China) was added. The resulting samples were resolved on SDS-PAGE and transferred onto a nitrocellulose membrane for Western blot analysis with the rabbit anti-PknG antibodies.
MurC Enzyme Assay
The MurC reaction mixture (100 μl) contains 50 mm Bis-tris propane buffer (pH 7.0), 10 mm MgCl2, 10 mm KCl, 1 mm UDP-GlcNAc, 1 mm PEP, 1 mm NADPH, 2 mm l-Ala, 2 mm ATP and 200 μg cell lysate. The reaction was performed at 37 °C for 1 h and terminated by the addition of 500 μl methanol for mass spectrometry identification.
Measure the Activity of MurC With Mass Spectrometry
LC-HRMS was performed as described previously (Vogliardi et al., 2011) on a Waters ACQUITY UPLC system equipped with a binary solvent delivery manager and a sample manager, coupled with a Waters Micromass Q-TOF Premier Mass Spectrometer equipped with an electrospray interface (Waters Corporation, Milford, MA). Briefly, LC was performed on a Syncronis HILIC column (50 × 2.1 mm, 1.7 μm). The column was eluted with 200 mm ammonium formate aqueous solution and acetonitrile in gradient mode at a flow rate of 0.30 ml/min at 30 °C. MS was performed using negative polarity, 2.4KV capillary voltage, 30 V sampling cone, 4eV collision energy, source temperature of 110 °C, and a desolvation temperature of 350 °C. The flow rate for the desolvation gas was set at 600 L/h. Scan range was set to m/z 50∼1000, scan time to 0.3 s and interscan time to 0.02 s.
Acid-fast Staining
The Msm strains were grown to log phase and 100 μl of culture were spread onto a glass slide. The slides were heated at 100 °C for 2 min, dipped into 10% formalin for 30 min, dried and stained using the TB Fluorescent Stain Kit M (BD, Franklin Lakes, NJ) or the TB Stain Kit (BD) according to the manufacturer's instructions.
Biofilm Formation
For biofilm cultures grown on liquid medium, 10 ml of biofilm medium with a modified version of M63 in a 90 × 15 mm PVC Petri dish was inoculated with 10 μl of a saturated culture and incubated at 30 °C without disturbance (25).
RESULTS
Global Identification of KPIs Using Mtb Proteome Microarrays
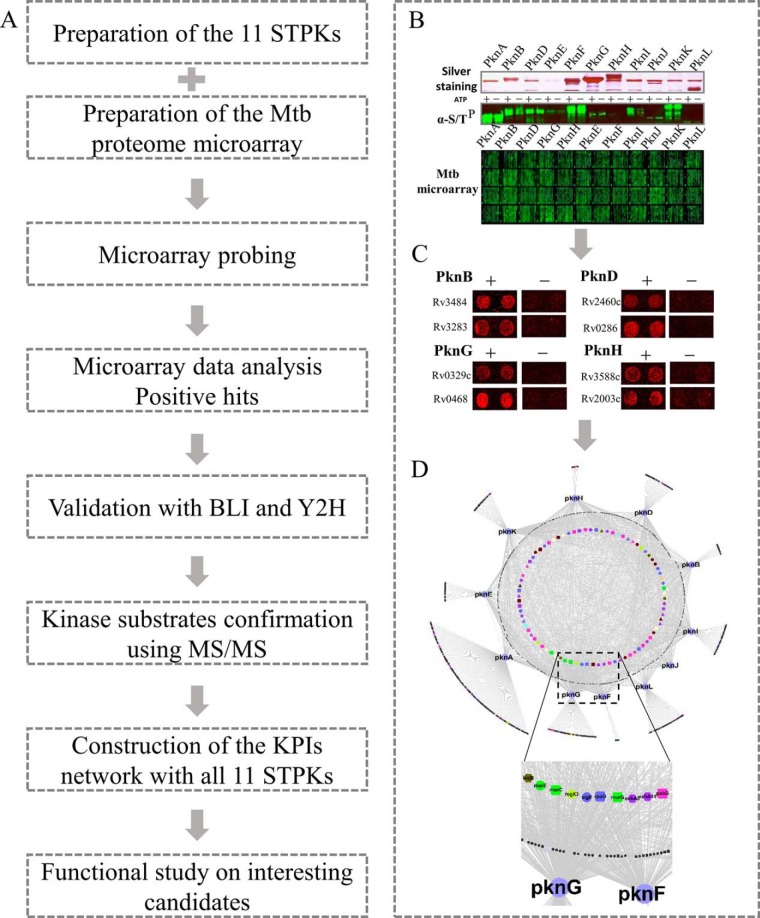
The overall design of this study is shown in Fig. 1A. Briefly, the study was composed of 4 parts: preparation of the Mtb proteome microarray and the functional kinases (Fig. 1B), screening KPIs on the microarray and validation (Fig. 1C), bioinformatics analysis and KPIs map construction (Fig. 1D), and functional studies of select, newly discovered KPIs.
Fig. 1.
The workflow and example of the STPK-KPIs network construction. A, The general procedure of the proteome microarray based global STPK-KPIs identification and network construction for all the 11 STPKs. B–D, An example of the KPIs study. Expression and purification of the 11 STPKs using yeast. The activities of the kinases were validated with an anti-ser/thr phosphorylation antibody after auto-phosporylation (B). Positive KPIs obtained from the Mtb proteome microarray after kinase probing (C). A representative network was generated using the KPIs (D).
To systematically identify STPK-KPIs, we used the newly constructed Mtb proteome microarray, which contains 4262 unique Mtb proteins (19). The proteome microarrays were freshly prepared to ensure their quality. To facilitate expression and purification, all 11 Mtb STPKs were cloned into the expression vector pEGH-A with an additional C-terminal V5 epitope. The kinases were then affinity purified through an N-terminal GST tag. Silver staining clearly demonstrated that all the 11 STPKs were of right size and successfully purified (Fig. 1B, supplemental Fig. S1A). To verify that the expressed kinases are active, we performed an auto-phosphorylation assay with ATP for each kinase (15), and a control without ATP was also included. The reactions were then readout using an anti-serine/threonine phosphorylation antibody (Fig. 1B, supplemental Fig. S1B). Significantly higher signals were observed for most of the kinases from the reactions with ATP, compared with that of the reactions without ATP, indicated that the purified kinases are of good activity. For kinases with lower activity, i.e. PknF, PknJ, and PknL, relatively higher concentration of these kinases were included for probing on the Mtb proteome microarray. For each kinase, we probed 2 microarrays, along with a negative control following the same probing procedure except without the kinase.
To identify the positive KPIs, the microarray results were extracted by Gene-Pix 6.1 and subjected to scoring by an algorithm designed to measure the relative signal intensity of each protein spot (supplemental Fig. S1C) (20). After normalization within each chip to eliminate spatial artifacts, a cutoff value of four standard deviations above the mean intensity was applied, as previously described (20). Finally, we obtained 1,027 KPIs involving 492 proteins for the Mtb STPKs (Fig. 1D, supplemental Table S1).
Systematic Analysis of STPKs-KPIs
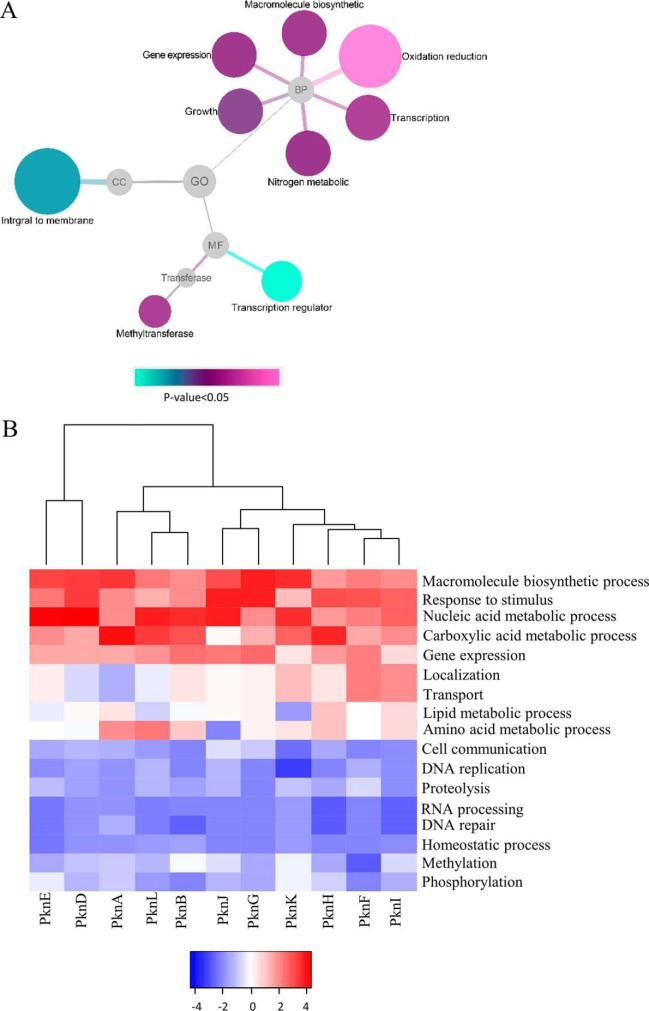
Initial analysis showed that the number of interacting proteins for each kinase ranged from 49 to 193 with an average of 93 proteins per kinase (supplemental Table S1). More specifically, more than 53% (262/492) of the proteins specifically interacted to only one kinase, 22% (109/492), 13% (64/492) and 12% (62/492) of the proteins interacted with 2, 3, and more than 3 kinases, respectively (supplemental Table S1). These results demonstrate that STPKs not only have their uniqueness, but also retain commonality for certain group of proteins, a phenomenon that has been reported previously (26). To our knowledge, this study is the most comprehensive KPIs study for the Mtb STPKs. To demonstrate the roles of these STPKs, the functional diversity of the STPKs binding proteins were illustrated in a network (Fig. 2A), where the interacting proteins were grouped based on the enriched Gene Ontology (molecular function, biological process, and cell compartment) terms related to signaling by DAVID (27). In terms of biological process, the most significant enriched group is oxidation reduction, whereas other enriched processes include gene expression, macromolecular biosynthetic and nitrogen compound metabolic process. GO analysis also showed that membrane-associated proteins are highly enriched (p value: 0.016), involving 84/492. Since 9 of the 11 STPKs are membrane proteins, it is not surprising that many of the interacting proteins are membrane associated. These results indicate that STPKs play important roles in regulating cell metabolism and membrane-associated functions.
Fig. 2.
The STPK binding proteins are functionally diversified. A, Gene ontology (GO) analysis of all the STPK binding proteins. The results were visualized by Cytoscape with p value <0.05. B, STPK classification based on functional diversity. The STPKs-KPIs database were analyzed with STRING through Biological Process, and the heatmap was drawn with R language.
Because we have the most comprehensive Mtb STPK-KPIs set, we next determined whether we can classify the 11 STPKs based on their GO Biological Processes [GO-BP] classifications. STPKs have important roles in regulating cell metabolic and development, it is not surprising that STPKs have functions in macromolecule biosynthetic process (13), nucleic acid metabolic process (19), and carboxylic acid metabolic process (24). Unexpectedly, our results also showed that STPKs may be involved in response to stimulus (19) and gene expression (13). Except their common functions in regulation, STPKs have diverse functions based on the hierarchical clustering, and they were grouped into four major classes (Fig. 2B). In the class of PknI, PknF, PknH, and PknK, they were enriched in the function of localization and transport. Meanwhile, the other group of PknB, PknL, and PknA play significant roles in regulating amino acid and lipid metabolism. Interestingly, the classification of STPKs by GO analysis has a high similarity with the known 5 clades phylogenetic tree of STPKs (5), except for PknH, and slight inconsistence for PknK and PknJ. For example, in the clade I (PknA, PknB, and PknL), the functions of these kinases are known to be relevant with cell division and growth, and our results are consistent with this knowledge.
Validation of the KPIs by Bio-Layer Interferometry (BLI) and Yeast-Two-Hybrid (Y2H) Assays
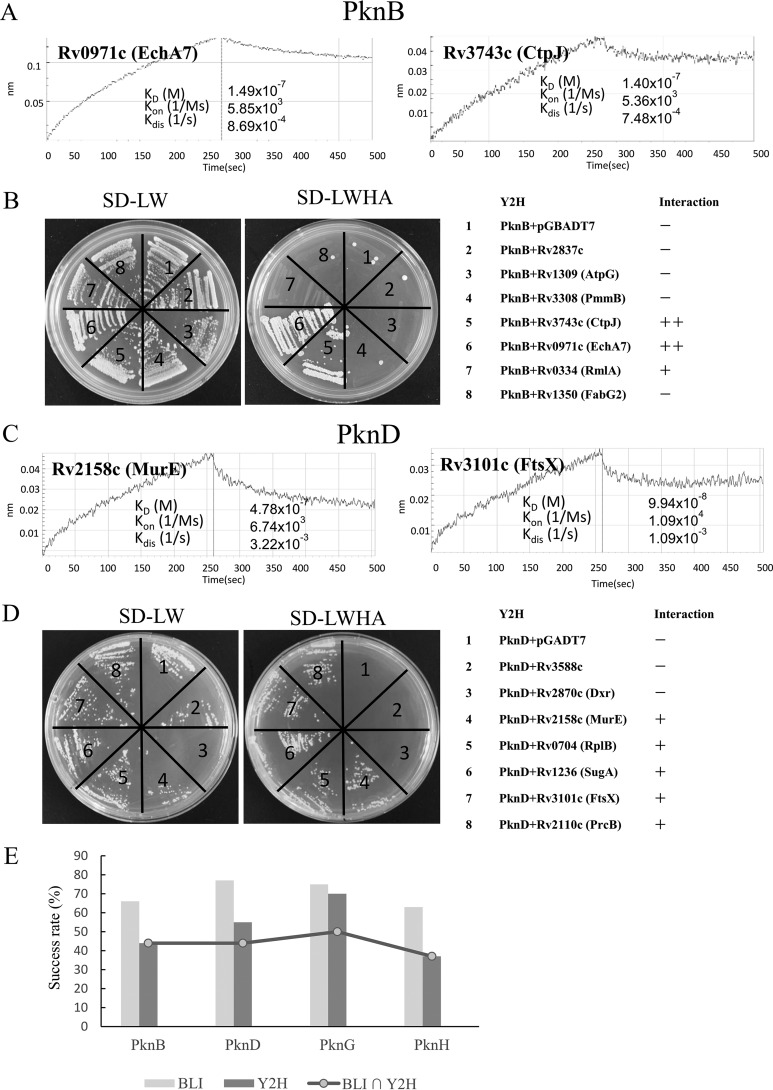
We randomly selected about 80 KPIs identified on the Mtb proteome microarray for validation. This set of KPIs is composed of 4 STPKs (PknB, PknD, PknG and PknH) and 75 interacting proteins (supplemental Table S2). All the 75 interacting proteins were cloned into an N-terminal GST tagged vector whereas the 4 kinases were cloned into a C-terminal 6×HIS tagged vector, and all proteins were expressed in E. coli and isolated by affinity purification. After several rounds of purification, all the interacting proteins were isolated solubly. The purified proteins and their corresponding interacting STPKs were examined using BLI. Clear BLI bindings were observed for 70% (53/75) of these proteins (Figs. 3A, 3C and 3E, supplemental Figs. S2 and S3, supplemental Table S2). The success rate for PknB, PknD, PknG, PknH was 66% (12/18), 77% (14/18), 75% (15/20) and 63% (12/19), respectively. Rv1827 (GarA) was included as a positive control because it is known to bind to PknB (28) whereas GST was included as a negative control (supplemental Fig. S2). We note that this methodology also identifies those interactions that are particularly strong (KD value of less than 10−7 m) (29). In this way, we found that many of the interactions between the proteins and STPKs were indeed strong, including PknB - EchA7, PknD - FtsX, PknG - PhoH2, and PknH - AccD6.
Fig. 3.
STPK-KPIs were validated by Bilayer Interferometry and Yeast-Two-Hybrid. A–B, Validation of KPIs of PknB by BLI (A) and Y2H (B). C–D, Validation of KPIs of PknD by BLI (C) and Y2H (D). For BLI, the binding was measured by coincident changes in the interference pattern. For Y2H, all the strains could grow on S.D.-LW media, whereas only positive strains could grow on S.D.-LWHA media. “+” indicates positive interaction, “++” indicates strong interaction. (E). The success rate of the validation for BLI and Y2H. In total, 75 KPIs were validated by BLI and Y2H for 4 representative STPKs, i.e. PknB, PknD, PknG and PknH.
To further validate the KPIs, we applied Y2H to the same set of KPIs (supplemental Table S2) as that of BLI validation. A total of 75 genes were successfully constructed into the pGADT7 vector that carries DNA activation domain as prey. Of the 4 STPKs, PknB, PknD, and PknH are membrane proteins whereas PknG is a not. Since it is known that membrane proteins are not applicable for traditional Y2H (30), only the kinase active domains of PknB, PknD, and PknH were cloned. The vector pGBKT7 with a DNA binding domain was used for cloning the 4 kinases as bait. The Y2H results showed that 52% (39/75) of the KPIs could be validated by Y2H (Figs. 3B, 3D, and 3E, supplemental Fig. S4, supplemental Table S2). Specifically, the success rates for PknB, PknD, PknG, and PknH were 44% (8/18), 55% (10/18), 70% (14/20), and 37% (7/19), respectively (Fig. 3E). As the membrane domains may be required to support the correct conformation of the kinase main. Thus, the lower Y2H success rate of the membrane kinases may be because of the lack of the supporting membrane domains as compare with that of PknG. The success rates of PknD and PknG are significantly higher than that of PknB and PknH, and interestingly, on the proteome microarray, the KPIs signals of PknD and PknG were in general higher than that of PknB and PknH. Thus, overall, the results from BLI and Y2H are highly consistent (Fig. 3E), and the KPIs of 33 proteins were successfully validated by both BLI and Y2H. To further validate the kinase-protein interactions, the M-PFC protein-protein assay, reassembly of the complementary fragments [F1, 2], and [F3] of mDHFR confers resistance to TMP was applied. GCN4-GCN4 was included as positive control. The results clearly showed that most the Y2H validated kinase-protein pairs could restore mycobacterial growth on 7H10 agar plates, further confirming their interaction in vivo (supplemental Fig. S5). Hence, these findings indicate that the KPIs discovered using the proteome microarray are reliable.
STPKs Can Phosphorylate Many of Their Interacting Proteins
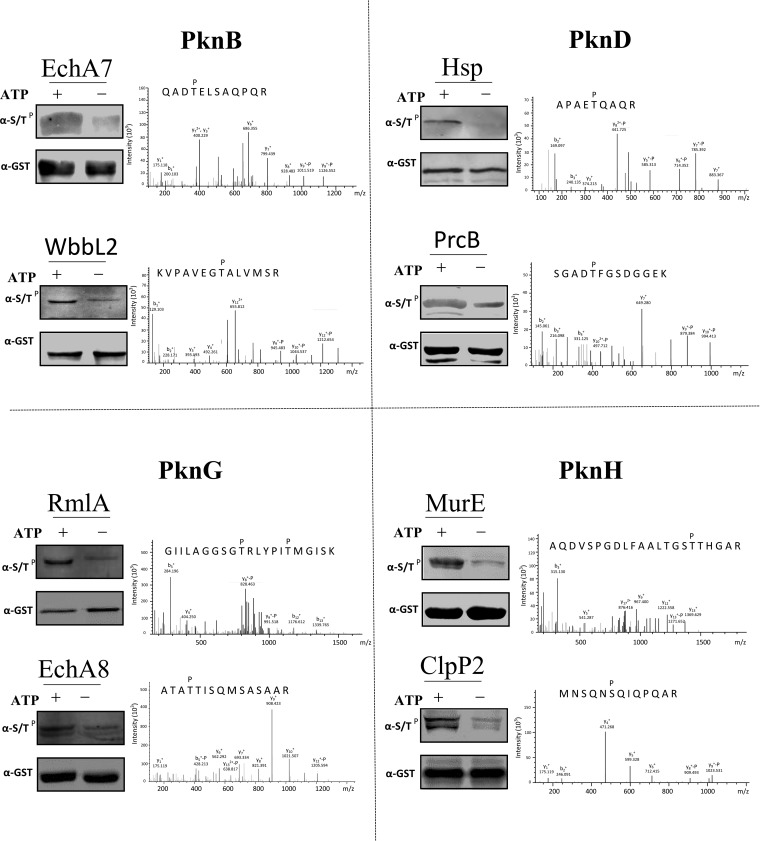
It is known that protein kinases usually phosphorylate their interacting proteins (17, 31). To test whether this is also the case for the STPK-KPIs discovered in this study, the kinase substrates relationships (KSRs) were tested for several kinases. We investigated the same set of 4 STPKs as that of validation (PknB, PknD, PknG, and PknH). A total of 60 KPIs were selected for phosphorylation site mapping by mass spectrometry. The selected proteins were incubated with their corresponding interacting STPKs and ATP for in vitro phosphorylation. The interacting proteins were expressed and affinity purified from E. coli with an N-terminal GST tag. As these proteins could also be endogenously phosphorylated, a control phosphorylation assay without ATP was included for each of the selected KPIs. By comparing the phosphorylated sites identified from the reactions with ATP and without ATP, we found that some of the interacting proteins are indeed the substrates of their corresponding STPKs and the ratios of the phosphorylated peptides/non-phosphorylated peptides range from 37%∼75%, these results indicate strong phosphorylation of these proteins by the serine/threonine kinases in vitro. Specifically, the success rates for PknB, PknD, PknG, and PknH were 33% (5/15), 20% (3/15), 20% (3/15), and 28% (4/15) (Fig. 4, supplemental Fig. S6, supplemental Table S3). Known phosphorylation sites were confirmed for several of the substrates, such as EchA8 and ClpB (32), though most of the sites and KSRs are novel.
Fig. 4.
The STPK interacting proteins were also the substrates of the corresponding kinase. Phosphorylation reactions were carried out by adding both the STPKs and the purified interacting proteins. Four representative STPKs, i.e. PknB, PknD, PknG and PknH were included, the phosphorylation reactions were performed with 2 randomly selected interacting proteins for each of the STPK. Reactions without ATP were also included as negative controls. The proteins were then subjected for mass spectrometry analysis to determine the phosphorylated serine/threonine sites.
Cross-talking is very common among kinases (3, 32). We also found crosstalk among the four STPKs. For example, RmlA could be phosphorylated by both PknB and PknG at the same sites (supplemental Table S3). Our data showed that threonine phosphorylation is more abundant than that of serine, which is consistent with earlier work (32), though in eukaryotes, where serine phosphorylation may account for 80–90% of total phosphorylation sites (33). Tyrosine phosphorylation was also identified, such as that in WbbL2 by PknB (supplemental Table S3), which is also consistent with previous work (34).
Key Biological Functions Were Enriched in the STPK-KPIs Map
To systematically understand the protein interactions of the 11 STPKs, all KPIs identified in this study were subjected to further bioinformatics analysis. The relationships between all STPKs and their interacting proteins were displayed in a map determined with CYTOSCAPE (supplemental Fig. S7). Most of the functional proteins were in the inner regions of the map according to STRING analysis (35), whereas the colors represent different biological processes. From this analysis, we found that the STPKs play significant roles in metabolic processes and regulate many proteins of unknown function, which could be a good start for discovering the functions of these proteins.
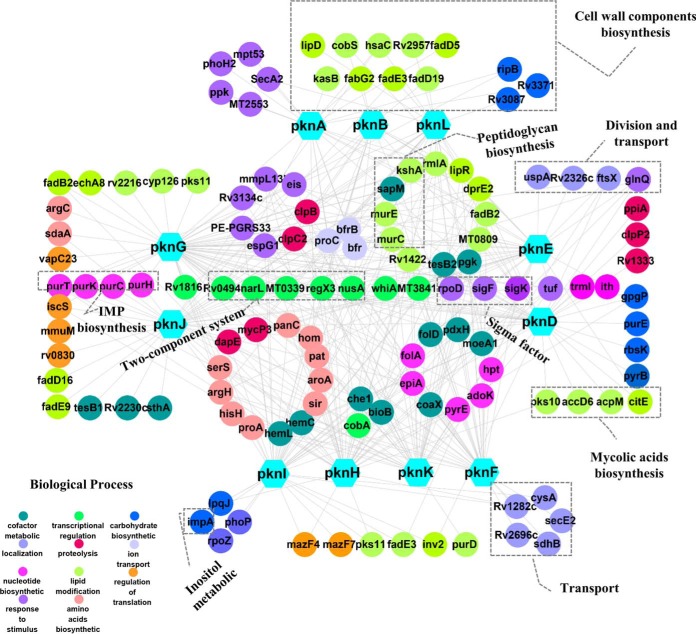
The biological roles of many Mtb proteins are still poorly understood according to the latest version of TBDB (36). To understand the KPIs map in a more biologically meaningful way, the interacting proteins with clear functional annotations were selected for further analysis. The corresponding STPK-KPIs of just these proteins were subjected for network analysis with STRING and CYTOSCAPE and a compact STPK-KPIs network was generated (Fig. 5). According to this network, many proteins are involved in cell wall lipid biosynthetic, such as RmlA for peptidoglycan (PG), LipR and FadB2 for mycolic acids formation, amino acids metabolic enzymes, such as SerS, ArgH, HisH, and ProA, nucleotide biosynthetic process including AdoK, PryE, EpiA, and Hpt, which is consistent to a notion that STPKs play key roles in regulating cell growth and division (37, 38). Meanwhile, some proteins like MmpL13b, EspG1, and PE-PGRS33, that respond to chemicals were also enriched in these networks. Further, this analysis identified more than 25 transcription-related proteins, including three sigma factors (SigK, SigF, and SigA), and four two-component system proteins (TCSs) (NarL, RegX3, NusA, and Rv1816). These proteins are involved in sensing various signals, and adapting the metabolism to environmental cues, and maintaining the integrity of the cell envelope during stress (39–41). It is possible that STPKs may regulate cell growth and response to host by altering protein transcriptional processes. Surprisingly, proteins involved in proteolysis processes were also enriched, namely ClpB and ClpP. Previous work has shown that the full Clp complexes family are indispensable for M. tuberculosis growth (42, 43). These results strongly suggest that the activity of Clp protease complex and the closely related biological processes may be tightly regulated by STPKs.
Fig. 5.
A variety of biological functions were enriched according to GO analysis based on the STPK-KPIs map. One hundred and twenty-eight STPKs interacting proteins with clear functional annotation were selected for GO and network analysis. The results of the network analysis were then further analyzed and visualized using Cytoscape.
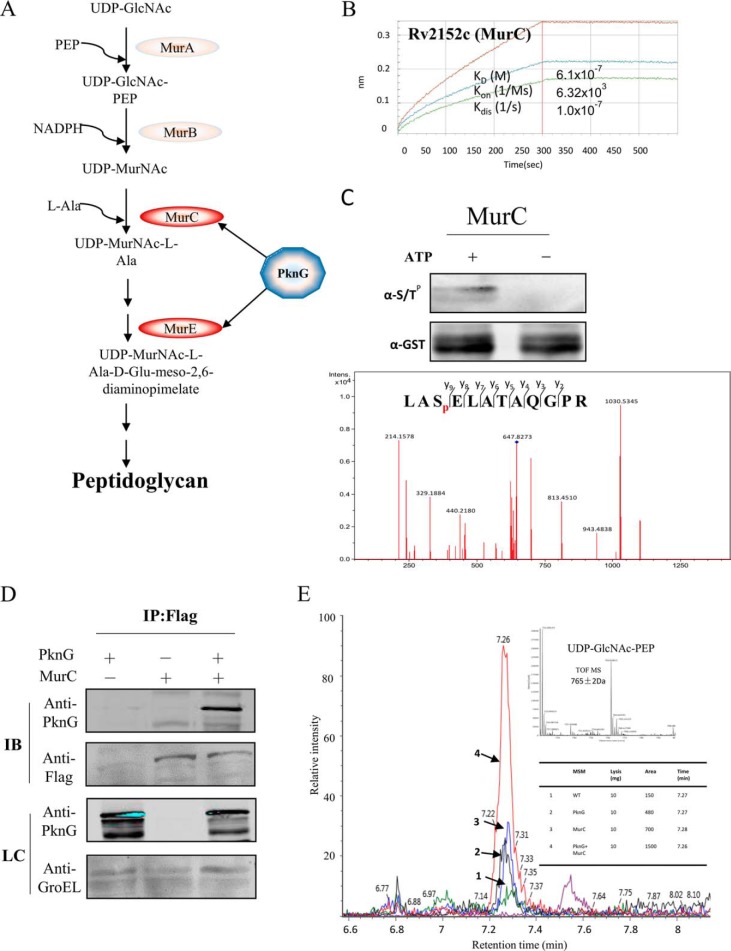
ATP-dependent Mur ligases are key enzymes of the peptidoglycan (PG, a key component of cell wall synthesis) biosynthetic pathway. Our results include 2 of the Mur ligases (namely MurC and MurE), suggesting that the STPKs may regulate cell wall synthesis through the interaction with Mur ligases.
In summary, constructing an interaction map for M. tuberculosis STPKs permits the ready identification of highly connected interactions among the STPK-KPIs. This network highlighted the significant role of STPKs in regulating cell metabolism, growth, division, and proteolysis, thus extending our understanding of range of critical cellular functions that involve STPKs.
PknG Regulates Cell Wall Biogenesis Through the Interaction with Mur Ligases
PknG has been found to exert its activity by directly interfering with host signaling cascades (12). In addition, PknG has been implicated in the regulation of glutamate metabolic processes that might indirectly influence virulence (13). Because of its potential functional significance, we next selected PknG for a more detailed functional examination. Among PknG interacting proteins, cell wall biogenesis-related proteins are highly enriched, including SigA, SigF, and enzymes for the synthesis of cell wall components, MurC, MurE, EchA, and FabG (Fig. 5). The physical interaction of PknG with Mur ligases are novel. PG, as a unique component to bacteria cell wall, provides a rigid support that gives the cell its shape and maintains its integrity (44). In M. tuberculosis, Mur ligases are central enzymes in the formation of PG (Fig. 6A). The interaction of PknG and MurC was successfully validated by BLI in vitro (Fig. 6B). And aslo the phosphorylation of MurC by PknG have been determined in vitro (Fig. 6C). To further validate the interaction of PknG with the MurC in vivo, PknG and MurC were coexpressed in Msm and MurC was fused with a C-terminal Flag tag. There is a clear band with anti-PknG antibody in the coexpressed strain, whereas no signal was found from the control strains by immuno precipitation using anti-Flag conjugated magnetic beads (Fig. 6D). To further explore the effects of the interaction between MurC and PknG, the activity of MurC upon the overexpression of PknG, MurC or coexpression of PknG and MurC were monitored by mass spectrometry. MurC catalyze l-alanine and UDP-MurNAc to UDP-MurNAc-l-alanine, we then measured the substrate l-alanine and the product UDP-MurNAc-l-alanine by mass spectrometry. There was a remarkable increase with the production of UDP-MurNAc-l-alanine in the strains with PknG overexpression and cooverexpression of PknG and MurC as compared with that of the wild type strain (Fig. 6E), in addition the substrate l-Alanine was largely consumed (supplemental Fig. S8). These results confirmed that PknG promotes the activity of MurC.
Fig. 6.
PknG regulates MurC enzyme activity through interaction with MurC in vivo. A, Schematic of the PG formation with the Mur enzymes. B, In vitro validation of PknG and MurC interaction by BLI. C, In vitro phosphorylation of MurC by PknG. The purified of MurC and PknG were mix with 2:1 in the kinase buffer (50 mm Tris.HCl, 5 mm MgCl2, 2 mm MnCl2, 1 mm DTT, 5 mm ATP) and incubated in 37 °C for 2 h. D, In vivo validation of PknG and MurC interaction. Flag tagged MurC and PknG were expressed in Msm with ATc induction, PknG interacted with MurC were then immuno-precipitated using an anti-Flag antibody. Anti-PknG and GroEL antibody were loading control. E, MurC activity was enhanced by PknG. MurC activity assay was performed using UDP-GlcNAc, PEP and l-Ala as substrates. The major product of this reaction, UDP-MurNAc-l-Ala, was measured by ultra-performance liquid chromatography coupled with mass spectrometry.
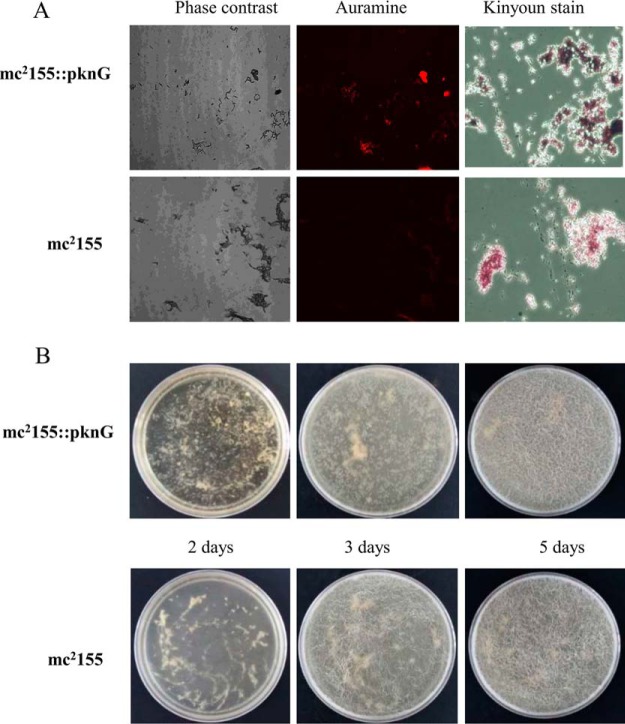
PG plays an important role in stabilizing cell wall structure, which is a linker for the mycolic acids in the outside of cell wall. We sought to determine whether PknG changes cell wall biogenesis through the interaction with MurC. To check the phenotypic influence of PknG overexpression, the Msm strain with PknG overexpression was cultured and subjected for acid-fast staining and biofilm formation analysis, using the Msm strain with the empty vector as a control. The strains were first analyzed by acid-fast staining using aurmine and kinyoun dyes, and we found that with PknG overexpression, the bacteria were more easily stained by both dyes as compared with wild type strain under the same conditions (Fig. 7A). This indicates that PknG may promote the thickness of cell wall. Since biofilm formation is closely related to the change of the cell wall components (25, 45), we next tested the effects of PknG overexpression on biofilm formation. After inoculation of the two Msm strains with 105 CFUs, the results were recorded every 24 h. At day 2, Msm cells grew to form a uniform thin film. At day 3, surface texure, including ridges and troughs appeared. At day 5, small structures reminiscent of microcolonies appeared. Compared with that of the control, PknG overexpressed cells formed larger patches, and from day 3, they also grew much faster (Fig. 7B). These results strongly suggest that PknG may increase the content of cell wall components through the activation of MurC.
Fig. 7.
PknG regulates cell wall formation. A, Acid-fast staining was performed on Msm with PknG overexpression, the wild-type Msm were included as negative control. After the cultures were fixed on glass slides, acid-fast staining was performed on the fixed smears using either the BD TB Auramine Kit or the Kinyoun stain kit. B, Biofilm formation. Time course of biofilm development at a liquid-air surface. Dishes containing liquid growth medium were inoculated with either Msm mc2155 or Msm mc2155-PknG, and the biofilms were allowed to develop for 5 days at 30 °C.
DISCUSSION
By using our unique Mtb proteome microarray, we have constructed the most comprehensive KPIs map for the complete repertoire of STPKs in Mtb. The network is composed of 492 kinase-interacting proteins and 1027 binary interactions. The interacting proteins are involved in a wide range of important cellular functions, including lipid biosynthesis and cell wall division. More detailed functional studies revealed that PknG regulates Mtb cell wall composition by interacting with MurC.
As a powerful tool, proteome microarrays have already been widely applied for the discovery of novel protein-protein interactions (17, 18). It possesses several advantages over other techniques. First, once the target protein and microarray are available, only a few hours are needed for proteome-wide PPI identification. In this study, the discovery stage took less than 1 week for all the 11 STPKs. Second, because all the proteins on the proteome microarray are individually purified, the PPIs identified are direct interactions. Binary interactions also facilitate follow-up validations, as we have demonstrated in this study.
Nonetheless, there are features of this methodology that could be improved. One disadvantage of a proteome microarray-based PPIs investigation lies in the fact that it is an in vitro screening and so the binding may be false-positive. To rule out this possibility, we performed BLI-based kinetic analysis of the interactions for four kinases (PknB, PknD, PknG, and PknH) with 75 interacting proteins, and also for Y2H validations. We found that the majority were successfully validated by these two methods: 70% (53/75) and 52% (39/75) (supplemental Table S2), respectively. Compared with other techniques that are used to validate the reliability of interactions, such as mass spectrometry, these values are lower than what is observed for much more stable PPIs such as spliceosomal interactions (46). However, it compares well to the validation rates reported for other representative sets of PPIs with co-IP assays (47, 48).
Prior to this study, there was only one systematic study concerning Mtb STPKs with their substrates (32), and few known KPIs, such as PknB and InhA (7), PknD and MmpL (49), PknH and EmbR (50). In this study, we identified 492 STPKs binding proteins, including 21% (6/28) of the known KPIs, such as KasB, MurC, SahH, MmpL, DosR, and Rv1747. Thus, most of these KPIs are novel. Based on these interactions, we generated the most comprehensive, and to our knowledge, the first Mtb STPK-KPIs map. This network could serve as a rich resource for future Mtb functional studies. Mtb is presently a very poorly studied pathogen, with 1957 out of 4346 of its predicted protein coding genes annotated as “function unknown,” “hypothetical,” or “possible” according to the latest version of TBDB (http://tuberculist.epfl.ch/). In the case of the STPKs, only a few are relatively well studied, including PknB, PknD, and PknG, whereas the functions of the others are not clear. Using our STPK-KPIs network, we can assign new functions to the well-studied STPKs, as well as identify potential experimental-based functions for the poorly studied STPKs for the first time.
It is often found that kinase-binding proteins are also substrates of the kinase (31, 51). This was also found in the present work using phospho-peptide enrichment and mass spectrometry to directly identify phosphorylated threonine or serine on the kinase-interacting protein after incubation with the cognate kinases. Among these, there were 36 phosphorylated proteins in our interactors that were previously catalogued in the mass spectrometry phosphorylation database (32). Yet we also uncovered several novel phosphorylated proteins such as that for PknB, e.g. EchA7 and WbbL2. Obtaining reliable kinase-substrate information represents a major challenge in the development of novel regulatory pathway models to elucidate cellular regulation in strain growth and immunity. Our data strongly suggest that STPKs may regulate the function of the interacting proteins through phosphorylation.
These “eukaryotic-like” STPKs are thought to play essential roles in growth, virulence, persistence and reactivation, generally by signaling within bacterial cells. In our interactors list, most of the proteins are involved in cell growth. According to published work, only a few substrates are related with growth, such as the PknB-substrates, KasA and KasB (52), the PknA substrates InhA (7) and FtsZ (53, 54), and PknG substrate OdhI (54). Our results point to a significant role that PknG directly plays in regulating cell growth through its interaction with cell wall-related proteins.
The cell wall is a potential drug target, because it plays a significant role in the ability of the bacteria to acquire or obtain essential nutrients, persistence, transcription regulation, and energy metabolism (55). The Mtb cell wall is composed of three main components, namely PG, AG and MAs. In addition to the cell wall related proteins that interact with PknG, we have also found within our KPIs many other proteins that are involved in cell wall formation. UDP-MurNAc plays a role in the second step of PG biosynthesis under the UDP-GlcNAc. Many enzymes involved in this process have been identified, including MurA-B and MurC-F. Among these enzymes, only MurC was known to be phosphorylated by PknA (56). According to our results, we have discovered 3 enzymes (MurC, MurE, and MurF) that interact with kinases, and also identified the phosphorylated sites on MurC and MurE. e In summary, the importance of protein phosphorylation and interaction with STPKs in regulating key processes for Mtb cell wall biosynthesis, together with the extensive expertise in design and development of small molecule inhibitors of these enzymes, make them highly attractive targets for the development of new anti-tuberculosis agents.
Taken together, taking advantage of our newly constructed Mtb proteome microarray (19), we have constructed the first and the most comprehensive KPIs network for all Mtb STPKs, representing a wide range of biological functions. Novel annotations could be assigned to several of the poorly studied STPKs, such as the involvement of PknE in regulating cell division and transport processes, PknI in carbohydrate biosynthesis, and PknJ in nucleotide biosynthetic processes. We also added to the functional roles of the well-studied STPKs, such as PknB in the cell wall formation, PknD in mycolic acids synthesis, and PknG with nucleotide biosynthesis, amino acids synthesis and lipid biosynthesis. Functional studies demonstrated that one of the KPIs, PknG, and MurC, plays key role in Mtb cell wall biosynthesis. The STPK-KPIs network generated here is expected to serve as a valuable resource for studies designed to delineate regulation and functional activities of pathways in Mtb, yet also holds the great potential for Mtb specific drug development by providing promising novel drug targets.
DATA AVALIABILITY
The mass spectrometry proteomics data have been deposited to the PRIDE Archive (http://www.ebi.ac.uk/pride/archive/) via the PRIDE partner repository (57) with the data set identifier PXD006389 and 10.6019/PXD006389.
Supplementary Material
Acknowledgments
We thank Dr Kaixia Mi, Dr Jianping Xie, Dr Peifu Zhou, and Dr Adrie J. C. Steyn for providing the Mycobacterial protein fragment complementation system.
Footnotes
Author contributions: S.C.T. conceived and designed the project. F.L.W. and Y.L. performed most of the experiments and Y.Z.L. and Z.X. carried out most of the computational analyses. H.W.J., H.N.Z., X.H., and Z.W.X. contributed reagents or provided laboratory assistance. J.L.H. performed the mass spectrometry analysis. F.L.W. and S.C.T. interpreted results and wrote the manuscript. D.C., L.J.B., J.Y.D., and X.E.Z. commented on the manuscript.
* This study was supported in part by The National Key Research and Development Program of China Grant 2016YFA0500600, National Natural Science Foundation of China Grants 31670831, 31370813 and 31370750. This study was also supported by the National Natural Science Foundation of China Grant 31471232, Science and Technology Planning Projects of Guangdong Province Grant 2014B030301040 and Joint Research Fund for Overseas Natural Science of China Grant 3030901001222 to ZX. The authors declare that they have no conflict of interest.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- TB
- tuberculosis
- Mtb
- Mycobacterium tuberculosis
- STPK
- serine/threonine protein kinase
- KPI
- STPK protein interaction
- BLI
- bio-layer interferometry
- Y2H
- yeast-two-hybrid
- KSRs
- kinase substrates relationships
- PG
- peptidoglycan
- Msm
- Mycobacterium smegmatis
- ESI-MS
- electrospray ionization mass spectrometry.
REFERENCES
- 1. WHO. (2015) Global tuberculosis report 2015
- 2. Horsburgh C. R. Jr, Barry C. E. 3rd, and Lange C. (2015) Treatment of Tuberculosis. N. Engl. J. Med. 373, 2149–2160 [DOI] [PubMed] [Google Scholar]
- 3. Newman R. H., Hu J., Rho H. S., Xie Z., Woodard C., Neiswinger J., Cooper C., Shirley M., Clark H. M., Hu S., Hwang W., Jeong J. S., Wu G., Lin J., Gao X., Ni Q., Goel R., Xia S., Ji H., Dalby K. N., Birnbaum M. J., Cole P. A., Knapp S., Ryazanov A. G., Zack D. J., Blackshaw S., Pawson T., Gingras A. C., Desiderio S., Pandey A., Turk B. E., Zhang J., Zhu H., and Qian J. (2013) Construction of human activity-based phosphorylation networks. Mol. Syst. Biol. 9, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dworkin J. (2015) Ser/Thr phosphorylation as a regulatory mechanism in bacteria. Curr. Opin. Microbiol. 24, 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Narayan A., Sachdeva P., Sharma K., Saini A. K., Tyagi A. K., and Singh Y. (2007) Serine threonine protein kinases of mycobacterial genus: phylogeny to function. Physiol. Gen. 29, 66–75 [DOI] [PubMed] [Google Scholar]
- 6. Ortega C., Liao R., Anderson L. N., Rustad T., Ollodart A. R., Wright A. T., Sherman D. R., and Grundner C. (2014) Mycobacterium tuberculosis Ser/Thr protein kinase B mediates an oxygen-dependent replication switch. PLoS Biol. 12, e1001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khan S., Nagarajan S. N., Parikh A., Samantaray S., Singh A., Kumar D., Roy R. P., Bhatt A., and Nandicoori V. K. (2010) Phosphorylation of enoyl-acyl carrier protein reductase InhA impacts mycobacterial growth and survival. J. Biol. Chem. 285, 37860–37871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jani C., Eoh H., Lee J. J., Hamasha K., Sahana M. B., Han J.-S., Nyayapathy S., Lee J.-Y., Suh J.-W., Lee S. H., Rehse S. J., Crick D. C., and Kang C.-M. (2010) Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol. 10, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wolff K. A., de la Pena A. H., Nguyen H. T., Pham T. H., Amzel L. M., Gabelli S. B., and Nguyen L. (2015) A redox regulatory system critical for mycobacterial survival in macrophages and biofilm development. PLoS Pathogens 11, e1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gee C. L., Papavinasasundaram K. G., Blair S. R., Baer C. E., Falick A. M., King D. S., Griffin J. E., Venghatakrishnan H., Zukauskas A., Wei J. R., Dhiman R. K., Crick D. C., Rubin E. J., Sassetti C. M., and Alber T. (2012) A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci. Signal. 5, ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kang C. M., Nyayapathy S., Lee J. Y., Suh J. W., and Husson R. N. (2008) Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology-Sgm 154, 725–735 [DOI] [PubMed] [Google Scholar]
- 12. Walburger A., Koul A., Ferrari G., Nguyen L., Prescianotto-Baschong C., Huygen K., Klebl B., Thompson C., Bacher G., and Pieters J. (2004) Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304, 1800–1804 [DOI] [PubMed] [Google Scholar]
- 13. O'Hare H. M., Duran R., Cervenansky C., Bellinzoni M., Wehenkel A. M., Pritsch O., Obal G., Baumgartner J., Vialaret J., Johnsson K., and Alzari P. M. (2008) Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol. Microbiol. 70, 1408–1423 [DOI] [PubMed] [Google Scholar]
- 14. Hamasha K., Sahana M. B., Jani C., Nyayapathy S., Kang C. M., and Rehse S. J. (2010) The effect of Wag31 phosphorylation on the cells and the cell envelope fraction of wild-type and conditional mutants of Mycobacterium smegmatis studied by visible-wavelength Raman spectroscopy. Biochem. Biophys. Res. Commun. 391, 664–668 [DOI] [PubMed] [Google Scholar]
- 15. Leiba J., Syson K., Baronian G., Zanella-Cleon I., Kalscheuer R., Kremer L., Bornemann S., and Molle V. (2013) Mycobacterium tuberculosis maltosyltransferase GlgE, a genetically validated antituberculosis target, is negatively regulated by Ser/Thr phosphorylation. J. Biol. Chem. 288, 16546–16556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., and Snyder M. (2001) Global analysis of protein activities using proteome chips. Science 293, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 17. Fasolo J., Sboner A., Sun M. G., Yu H., Chen R., Sharon D., Kim P. M., Gerstein M., and Snyder M. (2011) Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Develop. 25, 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Y., Yang L. N., Cheng L., Tu S., Guo S. J., Le H. Y., Xiong Q., Mo R., Li C. Y., Jeong J.-S., Jiang L., Blackshaw S., Bi L. J., Zhu H., Tao S. C., and Ge F. (2013) Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity. Mol. Cell. Proteomics 12, 2804–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng J., Bi L., Zhou L., Guo S. J., Fleming J., Jiang H. W., Zhou Y., Gu J., Zhong Q., Wang Z. X., Liu Z., Deng R. P., Gao J., Chen T., Li W., Wang J. F., Wang X., Li H., Ge F., Zhu G., Zhang H. N., Gu J., Wu F. L., Zhang Z., Wang D., Hang H., Li Y., Cheng L., He X., Tao S. C., and Zhang X. E. (2014) Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Reports 9, 2317–2329 [DOI] [PubMed] [Google Scholar]
- 20. Hu S., Xie Z., Onishi A., Yu X., Jiang L., Lin J., Rho H. S., Woodard C., Wang H., Jeong J. S., Long S., He X., Wade H., Blackshaw S., Qian J., and Zhu H. (2009) Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139, 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh A., Mai D., Kumar A., and Steyn A. J. (2006) Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. U.S.A. 103, 11346–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang M. K., Qiao Z. X., Zhang W. Y., Xiong Q., Zhang J., Li T., Ge F., and Zhao J. D. (2013) Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res. 12, 1909–1923 [DOI] [PubMed] [Google Scholar]
- 23. Fu Y., Yang Q., Sun R., Li D., Zeng R., Ling C. X., and Gao W. (2004) Exploiting the kernel trick to correlate fragment ions for peptide identification via tandem mass spectrometry. Bioinformatics 20, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 24. Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., and Mann M. (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707 [DOI] [PubMed] [Google Scholar]
- 25. Ojha A., Anand M., Bhatt A., Kremer L., Jacobs W. R. Jr, and Hatfull G. F. (2005) GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123, 861–873 [DOI] [PubMed] [Google Scholar]
- 26. Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R. R., Schmidt M. C., Rachidi N., Lee S. J., Mah A. S., Meng L., Stark M. J., Stern D. F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P. F., and Snyder M. (2005) Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 [DOI] [PubMed] [Google Scholar]
- 27. Huang, da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 28. Villarino A., Duran R., Wehenkel A., Fernandez P., England P., Brodin P., Cole S. T., Zimny-Arndt U., Jungblut P. R., Cervenansky C., and Alzari P. M. (2005) Proteomic identification of M. tuberculosis protein kinase substrates: PknB recruits GarA, a FHA domain-containing protein, through activation loop-mediated interactions. J. Mol. Biol. 350, 953–963 [DOI] [PubMed] [Google Scholar]
- 29. Gowthaman R., Miller S. A., Rogers S., Khowsathit J., Lan L., Bai N., Johnson D. K., Liu C., Xu L., Anbanandam A., Aube J., Roy A., and Karanicolas J. (2016) DARC: mapping surface topography by ray-casting for effective virtual screening at protein interaction sites. J. Med. Chem. 59, 4152–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., and Rothberg J. M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 31. Varjosalo M., Keskitalo S., Van Drogen A., Nurkkala H., Vichalkovski A., Aebersold R., and Gstaiger M. (2013) The Protein Interaction Landscape of the Human CMGC Kinase Group. Cell Reports 3, 1306–1320 [DOI] [PubMed] [Google Scholar]
- 32. Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C. M., Bemis G., Church G. M., Steen H., and Husson R. N. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U.S.A. 107, 7521–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 34. Kusebauch U., Ortega C., Ollodart A., Rogers R. S., Sherman D. R., Moritz R. L., and Grundner C. (2014) Mycobacterium tuberculosis supports protein tyrosine phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 111, 9265–9270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lew J. M., Kapopoulou A., Jones L. M., and Cole S. T. (2011) TubercuList–10 years after. Tuberculosis 91, 1–7 [DOI] [PubMed] [Google Scholar]
- 37. Parikh A., Verma S. K., Khan S., Prakash B., and Nandicoori V. K. (2009) PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J. Mol. Biol. 386, 451–464 [DOI] [PubMed] [Google Scholar]
- 38. Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., and Sassetti C. M. (2011) High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathogens 7, e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barik S., Sureka K., Mukherjee P., Basu J., and Kundu M. (2010) RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol. Microbiol. 75, 592–606 [DOI] [PubMed] [Google Scholar]
- 40. Hatzios S. K., Baer C. E., Rustad T. R., Siegrist M. S., Pang J. M., Ortega C., Alber T., Grundner C., Sherman D. R., and Bertozzi C. R. (2013) Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc. Natl. Acad. Sci. U.S.A. 110, E5069–E5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park S. J., Kim J. H., Ha T. S., and Shin J. I. (2013) Is there a link between Escherichia coli septicemia and the onset of systemic lupus erythematosus? Comment on: overlapping juvenile idiopathic arthritis and systemic lupus erythematosus: a case report (Rheumatol Int. 2011 May; 31(5):695–698). Rheumatol. Int. 33, 269–270 [DOI] [PubMed] [Google Scholar]
- 42. Vaubourgeix J., Lin G., Dhar N., Chenouard N., Jiang X., Botella H., Lupoli T., Mariani O., Yang G., Ouerfelli O., Unser M., Schnappinger D., McKinney J., and Nathan C. (2015) Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microbe 17, 178–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raju R. M., Jedrychowski M. P., Wei J. R., Pinkham J. T., Park A. S., O'Brien K., Rehren G., Schnappinger D., Gygi S. P., and Rubin E. J. (2014) Post-translational regulation via Clp protease is critical for survival of Mycobacterium tuberculosis. PLoS Pathogens 10, e1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kieser K. J., Baranowski C., Chao M. C., Long J. E., Sassetti C. M., Waldor M. K., Sacchettini J. C., Ioerger T. R., and Rubin E. J. (2015) Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc. Natl. Acad. Sci. U.S.A. 112, 13087–13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Katsivela E., Bonse D., Kruger A., Strompl C., Livingston A., and Wittich R. M. (1999) An extractive membrane biofilm reactor for degradation of 1,3-dichloropropene in industrial waste water. Appl. Microbiol. Biotechnol. 52, 853–862 [DOI] [PubMed] [Google Scholar]
- 46. Hegele A., Kamburov A., Grossmann A., Sourlis C., Wowro S., Weimann M., Will C. L., Pena V., Luehrmann R., and Stelzl U. (2012) Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell 45, 567–580 [DOI] [PubMed] [Google Scholar]
- 47. Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H., Sahalie J. M., Murray R. R., Roncari L., de Smet A. S., Venkatesan K., Rual J. F., Vandenhaute J., Cusick M. E., Pawson T., Hill D. E., Tavernier J., Wrana J. L., Roth F. P., and Vidal M. (2009) An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods 6, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weimann M., Grossmann A., Woodsmith J., Oezkan Z., Birth P., Meierhofer D., Benlasfer N., Valovka T., Timmermann B., Wanker E. E., Sauer S., and Stelzl U. (2013) A Y2H-seq approach defines the human protein methyltransferase interactome. Nat. Methods 10, 339–342 [DOI] [PubMed] [Google Scholar]
- 49. Perez J., Garcia R., Bach H., de Waard J. H., Jacobs W. R. Jr, Av-Gay Y., Bubis J., and Takiff H. E. (2006) Mycobacterium tuberculosis transporter MmpL7 is a potential substrate for kinase PknD. Biochem. Biophys. Res. Commun. 348, 6–12 [DOI] [PubMed] [Google Scholar]
- 50. Alderwick L. J., Molle V., Kremer L., Cozzone A. J., Dafforn T. R., Besra G. S., and Futterer K. (2006) Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 2558–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., Qin Z. S., Pawson T., Gingras A. C., Nesvizhskii A. I., and Tyers M. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Molle V., Brown A. K., Besra G. S., Cozzone A. J., and Kremer L. (2006) The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J. Biol. Chem. 281, 30094–30103 [DOI] [PubMed] [Google Scholar]
- 53. Thakur M., and Chakraborti P. K. (2006) GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J. Biol. Chem. 281, 40107–40113 [DOI] [PubMed] [Google Scholar]
- 54. Schultz C., Niebisch A., Schwaiger A., Viets U., Metzger S., Bramkamp M., and Bott M. (2009) Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Mol. Microbiol. 74, 724–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sharma K., Chopra P., and Singh Y. (2004) Recent advances towards identification of new drug targets for Mycobacterium tuberculosis. Expert Opin. Therap. Targets 8, 79–93 [DOI] [PubMed] [Google Scholar]
- 56. Fiuza M., Canova M. J., Patin D., Letek M., Zanella-Cleon I., Becchi M., Mateos L. M., Mengin-Lecreulx D., Molle V., and Gil J. A. (2008) The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum. J. Biol. Chem. 283, 36553–36563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.