Abstract
Human neutrophil elastase (HNE) is an important N-glycosylated serine protease in the innate immune system, but the structure and immune-modulating functions of HNE N-glycosylation remain undescribed. Herein, LC-MS/MS-based glycan, glycopeptide and glycoprotein profiling were utilized to first determine the heterogeneous N-glycosylation of HNE purified from neutrophil lysates and then from isolated neutrophil granules of healthy individuals. The spatiotemporal expression of HNE during neutrophil activation and the biological importance of its N-glycosylation were also investigated using immunoblotting, cell surface capture, native MS, receptor interaction, protease inhibition, and bacteria growth assays. Site-specific HNE glycoprofiling demonstrated that unusual paucimannosidic N-glycans, particularly Manα1,6Manβ1,4GlcNAcβ1,4(Fucα1,6)GlcNAcβ, predominantly occupied Asn124 and Asn173. The equally unusual core fucosylated monoantenna complex-type N-sialoglycans also decorated these two fully occupied sites. In contrast, the mostly unoccupied Asn88 carried nonfucosylated paucimannosidic N-glycans probably resulting from low glycosylation site solvent accessibility. Asn185 was not glycosylated. Subcellular- and site-specific glycoprofiling showed highly uniform N-glycosylation of HNE residing in distinct neutrophil compartments. Stimulation-induced cell surface mobilization demonstrated a spatiotemporal regulation, but not cell surface-specific glycosylation signatures, of HNE in activated human neutrophils. The three glycosylation sites of HNE were located distal to the active site indicating glycan functions other than interference with HNE enzyme activity. Functionally, the paucimannosidic HNE glycoforms displayed preferential binding to human mannose binding lectin compared with the HNE sialoglycoforms, suggesting a glycoform-dependent involvement of HNE in complement activation. The heavily N-glycosylated HNE protease inhibitor, α1-antitrypsin, displayed concentration-dependent complex formation and preferred glycoform-glycoform interactions with HNE. Finally, both enzymatically active HNE and isolated HNE N-glycans demonstrated low micromolar concentration-dependent growth inhibition of clinically-relevant Pseudomonas aeruginosa, suggesting some bacteriostatic activity is conferred by the HNE N-glycans. Taken together, these observations support that the unusual HNE N-glycosylation, here reported for the first time, is involved in modulating multiple immune functions central to inflammation and infection.
Neutrophils, the most abundant immune cell population in blood, are integral to the human innate immune system by forming a first line of defense against invading pathogens and by assisting in resolving inflammation. Chemically distinct granule compartments, which are characterized by their unique proteome content including an arsenal of antimicrobial proteins and peptides, are found within neutrophils, most notably, the primary (azurophilic), secondary (specific) and tertiary (gelatinase) granules (1). Other neutrophilic compartments including the secretory vesicles and the ficolin granules have been reported (2).
The large family of serine proteases, which includes human neutrophil elastase (HNE)1, proteinase 3, azurocidin, and neutrophil cathepsin G (nCG), are preferentially stored in the azurophilic granules, but also reside in the specific and gelatinase granules (3) and on the cell surfaces of activated neutrophils albeit at lower levels (4). These potent serine proteases are serving crucial immune functions central to the diapedesis, chemotaxis, and bactericidal activities of neutrophils including phagocytosis and degranulation (5). Protective functions of serine proteases were demonstrated in mice by a higher frequency of Mycobacterium tuberculosis infection upon inhibition of the serine protease activity (6). This was supported by the observation that serine protease deficient human neutrophils lack bactericidal activity, leaving affected individuals severely immune compromised (7). Among the serine proteases, HNE is particularly important in chronic inflammatory lung diseases e.g. cystic fibrosis (CF) (8) and in hematological disorders e.g. cyclic and severe congenital neutropenia (9–11).
The ELANE gene encoding the monomeric and heavily glycosylated HNE is highly expressed in the promyelocytic stage of cells committed to undergo myeloid lineage differentiation into neutrophils within the bone marrow (12). The directly encoded gene product is an inactive preproprotein precursor that is converted to the mature serine protease upon extensive proteolytic processing and post-translational modification (13). The 27 amino acid-long signal peptide of HNE is quickly removed by signal peptidases, followed by the addition and processing of N-glycosylation and the removal of the N- and C-terminal propeptides by other peptidases (14). It has been proposed that adaptor protein 3 (15) and serglycin (16) direct HNE and other proteins expressed in promyelocytes to the azurophilic granules by timing rather than by specific protein signals/motifs (17). Mature HNE is predicted to consist of 218 amino acid residues, and harbors four disulfide bonds and four putative N-glycosylation sites (Asn88, Asn124, Asn173, Asn185, numbering based on the preproprotein sequence) (supplemental Fig. S1), leaving the mature protein product with an apparent molecular mass of 25–27 kDa (18). The catalytic activity of HNE relies on its His-Asp-Ser triad, a conserved structural feature mirrored in other serine proteases, in which Ser202 confers the proteolytical activity (19). The potent HNE can be inhibited at the site of inflammation by endogenous serine protease inhibitors (serpins) including α1-antitrypsin (A1AT), an abundant plasma N-glycoprotein produced by hepatocytes (20).
HNE may be released from neutrophils upon stimuli-induced activation by lipopolysaccharide (LPS) or N-formylmethionine-leucyl-phenylalanine (fMLP) (21, 22). In the extracellular environment, HNE serves multiple protective and regulatory roles within the innate immune system. For example, HNE inhibits the growth of Gram-negative bacteria such as Pseudomonas aeruginosa in the airways (23) and facilitates the release of anti-inflammatory cortisol from corticosteroid binding globulin (CBG) at inflammatory sites (24). However, because of its high enzymatic activity, excessive secretion of HNE can damage the connective tissues of the lungs by causing proteolytic degradation of elastin, fibronectin and collagen, as commonly observed in chronic obstructive pulmonary disease and in chronic inflammation (25). Imbalanced A1AT-based inhibition may contribute to this uncontrolled proteolysis by HNE in such conditions (26). HNE is also implicated in the formation of neutrophil extracellular traps (NETs) (27, 28), but the role(s) of NETs in innate immune defense are not fully elucidated (29).
Despite the established biological significance of HNE, the site-specific structure and function of the HNE N-glycosylation remains undocumented. To the best of our knowledge, only studies using nuclear magnetic resonance and X-ray crystallography have provided qualitative and still incomplete structural insights into the N-glycosylation of HNE. Short mannose-terminating N-glycans were primarily reported on Asn124 and Asn173, whereas Asn88 was generally reported as being unoccupied on HNE (30, 31). These otherwise powerful analytical techniques for structural elucidation are, however, typically not suitable for accurate profiling of N-glycoproteins. The distribution of HNE glycoforms, glycosylation site occupancy and the presence of other post-translational modifications on HNE were also not described in these past studies.
Functionally, genetic mutations were induced near, but not directly at, the putative N-glycosylation sites of HNE in a myeloid cell line to assess how the local structures around the N-glycosylation sites affect HNE folding, stability and activity (32). Mutations near Asn88 and Asn124 altered the N-glycan processing, secretion and the proteolytic activity of HNE, but the N-glycosylation structure and function relationship of HNE was not investigated. In fact, given the importance of HNE in mediating a functional host immune response (33, 34), and the increasing appreciation that protein N-glycosylation significantly modulates the function of proteins (35, 36), the structure/function relationship of the N-glycosylation of this vital serine protease remains understudied.
Herein, we present the first detailed site-specific structural and functional characterization of HNE N-glycosylation from resting and activated human neutrophils from healthy individuals. Structural analyses revealed unusual N-glycans occupying three utilized HNE N-glycosylation sites i.e. Asn88, Asn124, and Asn173. The putative Asn185 site was not glycosylated. It was found that the N-glycosylation of HNE is more likely to contribute to immunomodulatory functions during innate immune processes than interfering directly with the HNE enzymatic activity. These observations are important to increase our understanding of the structure, functional importance and spatiotemporal regulation of central neutrophil proteins carrying unusual N-glycosylation.
EXPERIMENTAL PROCEDURES
Isolation of Blood-derived Human Neutrophils
Human resting neutrophils were isolated from the peripheral blood of a single healthy male donor after informed consent was obtained. The collection, handling and biomolecular analysis of healthy human neutrophils were approved by the Human Research Ethics Committee at Macquarie University, Sydney, Australia (Reference no. 5201500409). Neutrophils were isolated over multiple separate experiments from 40 ml freshly drawn blood collected in EDTA tubes (BD Biosciences, Australia). The donor and the collection volume, time and day were kept constant to reduce unnecessary donor variation. The neutrophils were isolated with polymorphprep density centrifugation (Axis Shield, Norway) followed by hypotonic lysis of the remaining erythrocytes with cold filtered high purity (MilliQ) water. Cell counts (> 1 × 107 cells/ml) and viabilities (>90%) of isolated neutrophils were determined using a Muse cell analyzer (Merck-Millipore, Australia). The purity of the neutrophil cell population (>95%) was estimated based on Wright-Giemsa stained cells prepared using a cytospin centrifuge (Thermo Scientific, Australia). Low abundance (∼2–3%) of other interfering cell types including erythrocytes, basophils and eosinophils were identified. The isolated neutrophils were used immediately for the structural and functional experiments.
Origin and Handling of Purified HNE
Enzymatically active human HNE (UniProtKB accession number: P08426) was purified from blood-isolated resting neutrophils from a pool of healthy individuals (Lee BioSolutions, MI, product number 342–40-1) using active site-based affinity and ion exchange liquid chromatography. Upon arrival, HNE was highly pure (>95%) and displayed full structural and functional integrity (i.e. showed high enzyme activity on various substrates) as assessed by SDS-PAGE (Invitrogen, Australia) and immunoblotting with rabbit anti-HNE (1:500, kind gift from Prof Niels Borregaard, University of Copenhagen, Denmark) and by its ability to rapidly cleave CBG (24). HNE was stored in a soluble form at 1.9 μg/μl in 50 mm sodium acetate and 600 mm sodium chloride buffer, pH 5.5 at 4 °C until use. Protein concentrations were determined at 280 nm absorbance with a percent extinction coefficient (ε1%) of 10 using a Nanodrop spectrophotometer (Thermo Scientific).
N-glycan Release and Clean-up
HNE (20 μg) was proteolytically inactivated with 1.5 mm PMSF, 90 min, 22 °C. Subsequently, the cysteine residues of HNE were reduced using 10 mm dithiothreitol (DTT) in 100 mm ammonium bicarbonate (pH 8.4), 45 min, 56 °C and alkylated using 25 mm iodoacetamide in 100 mm ammonium bicarbonate (pH 8.4) (both final concentrations), 30 min in the dark, 22 °C. The protein was then blotted on a primed 0.45 μm PVDF membrane (Merck-Millipore) and stained with Direct Blue (Sigma-Aldrich, Australia). The stained protein spots were excised, transferred to separate wells in a flat bottom polypropylene 96-well plate (Corning Life Sciences, Australia), blocked with 1% (w/v) polyvinylpyrrolidone in 50% (v/v) methanol and washed with MilliQ water. N-glycans were then released and handled as previously described (37). In brief, enzymatic release was performed using 3.5 U Flavobacterium meningosepticum N-glycosidase F (Roche, Australia) per 20 μg HNE in 10 μl water/well, 16 h, 37 °C. The unstable amino groups (-NH2) of the reducing end GlcNAc residues of N-glycosidase F-released N-glycans were allowed to spontaneously convert to hydroxyl groups (-OH) in weak acid using 100 mm ammonium acetate, pH 5, 1 h, 22 °C to facilitate for subsequent quantitative reduction to glycan alditols. Reduction was carried out using 1 m sodium borohydride in 50 mm potassium hydroxide, 3 h, 50 °C. The reaction was stopped by glacial acetic acid quenching. Dual desalting was then performed in micro-solid phase extraction (SPE) formats using strong cation exchange/C18 (where N-glycans are not retained) and porous graphitised carbon (PGC) (where N-glycans are retained) as stationary phases, respectively. The desalted N-glycans were eluted from the PGC-SPE columns using 40% (v/v) ACN containing 0.1% (v/v) aqueous TFA, dried, and dissolved in 10 μl MilliQ water for N-glycan analysis. Bovine fetuin carrying sialo-N-glycans (Sigma-Aldrich) and the paucimannose-rich nCG (38) were included as control glycoproteins to ensure efficient N-glycan release, clean-up and analysis.
Glycan standards were used to determine some structural aspects of HNE N-glycans. M2 N-glycan isoforms were generated from chicken ovalbumin (Sigma-Aldrich) by sequential exoglycosidase digestion using 1 U Streptococcus pneumoniae β-N-acetylhexosaminidase and 1 U Jack bean α1,3/6-linkage specific mannosidase (both from ProZyme, CA) using buffers and incubation times as recommended by the manufacturer. Synthetically generated and structurally validated M2 N-glycan (Product number MC0420) was obtained from Dextra Laboratories, Reading, UK. The N-glycan standards were reduced and desalted using PGC-SPE as described above prior to PGC-LC-MS/MS analysis.
N-glycan Analysis
Purified N-glycans were separated and detected by PGC-LC-MS/MS performed on an Agilent 1260 Infinity HPLC connected to an ESI ion trap mass spectrometer (LC/MSD Trap XCT Plus Series 1100, Agilent Technologies, Australia). The N-glycans (3 μl injection volume, ∼200 pmol) were loaded on a PGC HPLC capillary column (Hypercarb KAPPA, 5 μm particle size, 200 Å pore size, 180 μm inner diameter x 100 mm length, Thermo Scientific) and separated using a linear gradient of 0–45% (v/v) ACN/10 mm ammonium bicarbonate over 85 min at a 2 μl/min flow rate. The acquisition range was m/z 300–2200 and detection was performed in negative ionisation polarity mode with data-dependent acquisition. The top-three most abundant precursors in each full scan spectrum were selected for MS/MS using resonance activation (ion trap) CID performed with smart fragmentation (start/end amplitude 30–200%) at 1.0 V and an isolation window of 4 Th with a maximum accumulation time of 200 ms. Smart ion charge control was enabled with a smart parameter setting target of m/z 900. ESI was performed using a capillary voltage of +3.2 kV, a nitrogen drying gas flow of 6 liters/min at 325 °C, and a nitrogen-based nebuliser pressure of 12 psi. Dynamic exclusion was inactivated over the entire LC-MS/MS acquisition period. The mass spectrometer was calibrated using a tune mix (Agilent Technologies). The mass accuracy of the precursor and product ions was typically better than 0.5 Da. The N-glycan mixtures were analyzed in technical triplicates (n = 3). Mass spectra were viewed and inspected using DataAnalysis v4.0 (Bruker Daltonics, Australia). GlycoMod (http://web.expasy.org/glycomod/) and GlycoWorkbench v2.1 assisted in the annotation and visualization of the N-glycans (39, 40).
N-glycopeptide Generation and Enrichment
Approximately 10 μg HNE was reduced and alkylated as described above, but without PMSF-based inactivation to allow HNE to partially “self-digest” to generate appropriate (glyco)peptides for downstream LC-MS/MS analysis. Alkylation was quenched with 30 mm DTT (final concentration) before in-solution proteolytic digestion was carried out using porcine trypsin (sequencing grade, Promega, Australia) at a 1:20 enzyme/substrate ratio (w/w) in 50 mm ammonium bicarbonate, pH 8.4, 18 h, 37 °C. The majority (∼70%) of the resulting peptide mixture was enriched for glycopeptides as described previously, but with a minor modification (41). After first drying and redissolving this fraction in 10 μl loading solvent consisting of 80% ACN:1% TFA:19% H2O (v/v/v), the sample was then applied to custom-made hydrophilic interaction liquid chromatography (HILIC) SPE columns packed to an extended column height of 22 mm in GELoader tips (Eppendorf, Australia) with zwitterionic HILIC resin (ZIC-HILIC, 10 μm particle size, 200 Å pore size, kindly donated by Merck KGaA, Darmstadt, Germany). The relative long high-capacity SPE columns were utilized because we have experience that peptides displaying shorter (truncated) N-glycans may not be well retained using ZIC-HILIC (38). Columns were packed on top of a C8 Empore SPE disc for resin retention (Sigma-Aldrich) (42). Columns were then equilibrated in 50 μl loading solvent and the peptide samples were loaded and reloaded in successive rounds before the columns were washed twice with 50 μl loading solvent. The enriched HNE glycopeptides were eluted with 2 × 50 μl 1% (v/v) TFA and then with 50 μl loading solvent to ensure complete glycopeptide elution. The remaining 30% of the peptide mixture was used directly for LC-MS/MS analysis after desalting on C18 micro-SPE stage tips (Merck-Millipore). The Asn88 N-glycan site occupancy of HNE (the only site displaying macro-heterogeneity) was determined after de-N-glycosylation, which was achieved by incubating the nonenriched peptide mixture with 10 U N-glycosidase F (Roche) in 100 mm ammonium bicarbonate (pH 8.4), 16 h, 37 °C. This is an accurate method for estimating N-glycosylation site occupancy (43). The resulting peptide mixture was desalted with C18 micro-SPE tips. All desalted peptide fractions were dried and taken up in 15 μl 0.1% (v/v) formic acid (FA) before LC-MS/MS analysis.
N-glycopeptide Analysis
The HNE peptide mixtures were analyzed by ESI-LC-MS/MS in positive polarity mode using an HCT 3D ion trap (Bruker Daltonics) coupled to a Dionex Ultimate 3000 HPLC (Thermo Scientific). The samples were loaded directly onto a C18 column (Proteocol HQ303, 300 μm inner diameter x 10 cm length, 3 μm particle size, 300 Å pore size, SGE Analytical Science, Australia), which was heated to 50 °C. The column was equilibrated in 100% solvent A consisting of 0.1% (v/v) FA. After 8 min isocratic flow in solvent A, multisegmented linear gradients of solvent B increasing up to 30% over 60 min and then up to 60% over 7 min were applied before the column was washed in either 80% or 100% solvent B for 10 min and re-equilibrated back to the starting condition. Solvent B consisted of 0.1% (v/v) FA in ACN. A constant flow rate of 5 μl/min was used. Injections (5 μl/injection) were performed using the following two LC-MS/MS strategies that shared the same ESI conditions i.e. a capillary voltage of -4 kV, a nitrogen drying gas flow of 5 liters/min at 300 °C and a nitrogen nebuliser pressure of 15 psi: (1) MS full scans (m/z 300–2,200, scan speed of 8100 m/z/s) were followed by data-dependent fragmentation of the three most abundant signals in each full scan using resonance activation (ion trap) CID performed at 35% normalized collision energy, maximum accumulation time of 200 ms with smart ion charge control and smart fragmentation enabled (start/end amplitude of 30–200%), and (2) MS full scans (m/z 200–1800, scan speed of 8100 m/z/s) were followed by electron transfer dissociation (ETD) of the three most abundant signals present in each full scan. The ETD used an ion count control reactant target of 600,000 ions of fluoranthene, the maximum reactant accumulation time was 200 ms (actual accumulation time was 4–20 ms), and the reaction time was 150 ms. The ETD reaction used ultra-high purity methane (99.999% purity, BOC, Australia) as a carrier gas at 35–37 psi. Precursor ions were dynamically excluded for 1.5 min after three MS/MS events in both LC-MS/MS acquisition strategies. The mass spectrometer was calibrated using a tune mix (Agilent Technologies) prior to acquisition and the mass accuracy of the precursor and product ions were generally better than 0.5 Da. To determine the percentage relative abundance of each glycopeptide, the peptide mixture was analyzed with and without glycopeptide enrichment in technical triplicates. Two independent digestion experiments were carried out (two sets of technical triplicates in each digestion experiment, in total n = 6). To estimate the Asn88 macro-heterogeneity, the N-glycosidase F treated nonenriched HNE peptide mixtures were analyzed in technical triplicates (n = 3). Mass spectra were viewed and manually annotated using Data Analysis v4.0 (Bruker Daltonics) and GPMAW v10.0 (Lighthouse Data, Odense, Denmark) using the canonical protein sequence of HNE (UniProtKB, P08246).
Analysis of Intact HNE and HNE Protein Complexes
Intact HNE (∼1 μg) was analyzed by ESI-LC-MS in positive ion polarity mode using a high resolution/high mass accuracy Q-TOF 6538 mass spectrometer (Agilent Technologies) coupled to a capillary HPLC (Agilent 1260 Infinity). Purified HNE was loaded directly onto a C4 column (Proteocol C4Q, 3 μm particle size, 300 Å pore size, 300 μm inner diameter × 10 cm length, SGE Analytical Science) using 1 μl injections. The column was equilibrated and operated in identical mobile phases as for the glycopeptide analysis on the C18 column (described above). From a starting condition of 100% solvent A, a linear gradient up to 60% (v/v) over 30 min (2%/min slope) of solvent B was applied before the column was washed in 99% (v/v) solvent B for 10 min and re-equilibrated back to the starting condition. The flow rate was kept constant at 5 μl/min. A fragmentor potential of +200 V was used with the following MS settings in high resolution (4 GHz) mode: The MS full scan range was m/z 400–2500, the nitrogen drying gas flow rate was 8 liters/min at 300 °C, the nitrogen nebuliser pressure was 10 psi, the capillary potential was +4.3 kV, and the skimmer potential was +65 V. Beam-type CID-MS/MS fragmentation of the m/z 1146.3 signal, which was the most abundant ion of intact HNE, was performed at 55% normalized collision energy. HNE protein complexes (described below) were analyzed using the same conditions. Both intact (from here called “native”) HNE and HNE protein complexes were analyzed in technical triplicates (n = 3). The mass spectrometer was calibrated using a tune mix (Agilent Technologies) prior to acquisition and the mass accuracy of native HNE and the glycoprotein complexes were typically better than 20 ppm as assessed after spectral deconvolution. Mass spectra were viewed and analyzed with MassHunter work station vB.06 (Agilent Technologies).
Glycoprofiling of HNE N-glycans, N-glycopeptides and Intact Glycoprotein
HNE N-glycans were manually characterized in deep structural details using the PGC-LC-MS/MS data (44). In short, the characterization was based on accurate matches of the theoretical and experimental monoisotopic precursor masses, and substantial support of the candidate glycans by the observation of predicted glycan fragment ions in the corresponding CID-MS/MS spectra and predicted relative and absolute PGC-LC retention patterns that followed the retention rules for reduced but otherwise native N-glycans (38). The mass tolerance between theoretical and experimental N-glycan masses was 1 Da to accommodate the limited mass accuracy/resolution of the ion trap. The structural details of the glycan species such as the α1,3/6-mannose arm position of the single sialyl LacNAc or the β1,2-linked GlcNAc residue in the nonreducing end of the glycan were verified by the observation of the particularly useful diagnostic ion (D ion) in the CID-MS/MS spectra (45, 46). Using this information, monosaccharide compositions, glycan sequences, topologies, and some N-glycan glycosidic linkages could be assigned. Other structural details of the glycan species e.g. the existence of a β-linked mannosidic chitobiose in the reducing end of the glycan were inferred based on established knowledge of human N-glycosylation and the biosynthetic relatedness between the observed structures (47, 48). N-glycans are depicted throughout according to the established symbol nomenclature (49).
HNE N-glycopeptides were manually characterized using the reversed phase (RP)-LC-MS/MS data (38). In short, candidate glycopeptides were identified based on 1) matches (better than 1 Da) of the theoretical and experimental monoisotopic precursor masses, 2) the presence of relevant oxonium ions (e.g. m/z 366.1, 528.1, 657.1) in the CID fragment spectra, and 3) the retention time of the glycopeptides eluting in clusters within 1–3 min (50). The theoretical N-glycopeptide masses were generated using GPMAW v10.0 (Lighthouse Data), which assisted the glycopeptide identification. Verification of the identity of these glycopeptides was determined by the observation of the predicted peptide fragment ions i.e. b/y and c/z ion series and the glycan fragment ions e.g. the abundant peptide + GlcNAc (Y1 ion) with and without a dHex for core fucosylated glycopeptides in the corresponding CID- and ETD-MS/MS spectra.
Quantitative site-specific glycoprofiling of HNE was performed as follows: 1) To determine the percentage relative abundance of each glycopeptide form with a N-glycosylation site (micro-heterogeneity), the area-under-curve of extracted ion chromatograms (XICs) of the respective glycopeptides was quantified after a minimal degree of Gaussian smoothing of all observed charge states of the individual N-glycosylated peptides against all observed glycopeptides covering that particular N-glycosylation site within the nonenriched peptide mixture. In doing this, similar ionisation efficiencies of these related molecular species were assumed (43, 51), 2) The Asn88 site occupancy (macro-heterogeneity) was estimated by comparing the XIC areas of N-glycosidase F-generated de-N-glycosylated peptides (observed as deamidated i.e. Δm = +1 Da) and nonglycosylated peptides. A minor degree of spontaneous nonenzymatic deamidation was observed in a deamidation control without N-glycosidase F treatment performed in parallel under identical incubation conditions. The spontaneous deamidation was considered when assessing the site occupancy. This approach has the advantage of limiting potential ionisation differences between glycosylated and nonglycosylated peptides and simplifying the N-glycosylation site occupancy estimation (43). The Asn124 and Asn173 sites were fully occupied by N-glycosylation based on the lack of nonglycosylated and spontaneously deamidated peptides covering these two N-glycosylation sites in the unenriched HNE peptide mixture.
The glycoforms of native HNE were profiled by deconvoluting the multi-charged signals within the corresponding mass spectra using the maximum entropy algorithm in Bioconfirm within the MassHunter Work station vB.06 (Agilent Technologies). A mass range of 20–30 kDa, mass stepping of 1 and height filters set to a peak signal-to-noise ratio of 0.5 were used for spectral deconvolution. The HNE N-glycoforms were assigned based on accurate matches of the experimental average precursor masses (apex of deconvoluted signals) to the masses of theoretical glycoforms predicted to exist based on the prior site-specific N-glycopeptide and N-glycan profiling. The maximum mass difference allowed between the theoretical and experimental glycoprotein masses was 20 ppm. For comparative studies, the individual HNE glycoforms were quantitatively profiled using the relative height of the identified signals within the deconvoluted spectra.
Compartment-specific N-glycosylation Analysis of HNE from Human Neutrophil Granules
The distinct proteomes residing in the subcellular compartments of resting human neutrophils isolated from a healthy donor have been mapped (3). This proteomics data set was re-interrogated to perform compartment-specific HNE glycoprofiling. In short, resting human neutrophils were purified from freshly drawn peripheral blood using dextran sedimentation, followed by density centrifugation using Histopaque (GE Healthcare, Denmark). The remaining erythrocytes were lysed as described above (3). Proteins were extracted from the individual neutrophil granules isolated by Percoll density gradients after cellular cavitation using a Parr bomb pressurised with scientific grade nitrogen and then separated using SDS-PAGE. The purity of the isolated granules was validated using ELISA assays performed using antibodies with validated specificity toward granule-specific proteins. The granule-specific proteomics data was acquired from multiple SDS-PAGE gel fractions using high resolution/high mass accuracy LC-MS/MS on an LTQ Orbitrap XL mass spectrometer (Thermo Scientific) with linear ion trap CID fragmentation. The raw data is available at ProteomeXchange under the identifier PXD005559. Only the SDS-PAGE gel fractions containing HNE in the highest abundance (i.e. fraction 6) was used for the subcellular-specific glycoprofiling of HNE. The azurophilic, specific, gelatinase, and ficolin granules as well as the secretory vesicle and the plasma membrane fractions were investigated. The raw LC-MS/MS data files originating from two technical replicate experiments were used directly without any further processing for “first-pass” N-glycopeptide identification using Byonic v2.6 (Protein Metrics Inc, CA). Searches were performed using the entire human proteome (Homo sapiens, 20,198 reviewed entries in UniProtKB, released September 2015) and the common mammalian N-glycome (309 monosaccharide compositions, default N-glycome database in Byonic). The raw data files were first analyzed by Preview (Protein Metrics Inc). The following search parameters as recommended by Preview were used: Carbamidomethylation was a fixed Cys modification, whereas Met oxidation and Asn/Gln deamidation were variable modifications. The mass tolerance was set to 20 ppm and 0.6 Da for precursor and product ions, respectively. Decoys were included and a maximum protein false discovery rate of 1% was allowed for the search. Trypsin cleavage with two missed cleavages and full protease specificity was utilized. Precursor isotope selection was performed at “narrow” with the maximum precursor mass of 10,000 Da. The precursor and charge assignments were computed from the MS1 spectra. The CID-MS/MS spectra of the identified HNE glycopeptides with a Byonic score above 100 (52) that matched previously observed HNE glycopeptides from whole neutrophil lysates or were biosynthetically-related, were manually annotated and validated with Xcalibur (Thermo Scientific) using the same criteria as for the glycopeptide characterization of HNE from neutrophil lysates (see above). The confirmed HNE glycopeptides were quantified based on their relative XIC peak areas as described above. Granule-specific heat maps and regression profiles were generated using Microsoft Excel based on the relative abundance of the observed Asn88 and Asn173 N-glycopeptides. Low abundance N-glycoforms (< 0.5%) were excluded in the heat maps. Asn124 N-glycopeptides were not observed in the dataset. Hierarchical clustering was performed using Euclidean distance with GenePatterns 2.0 (53).
Visualization and Solvent Accessibility of HNE
In total, nineteen three-dimensional (3D) protein structures of naturally occurring HNE obtained by X-ray crystallography were available from the protein data bank (PDB, http://www.rcsb.org/pdb). These structures were used to visualize and accurately determine the solvent accessibility of the HNE N-glycosylation sites. Visualization was performed using PyMOL Molecular Graphic System, v1.74 (Schrödinger, LLC, MA) using a representative high-resolution structure of HNE (PDB: 3Q77, 1.86 Å). The solvent accessibilities of the glycosylation sites of monomeric HNE were determined using NACCESS (54). Specifically, the atomic accessible areas (van der Waal's interactions) of the individual Asn residues forming the three occupied N-glycosylation sites were measured in arbitrary (unit-less) but comparable values by rolling a spherical probe with a radius of 5 Å on the protein surface of HNE. The most abundant N-glycans were modeled in silico on Asn124 and Asn173 of HNE (PDB: 3Q77) with the default torsion angles provided by the Glycoprotein builder functionality of GLYCAM web (http://glycam.org).
HNE Cell Surface Expression Assay
Human resting neutrophils isolated from blood were stimulated in three technical replicates (n = 3) using ∼1 × 107 neutrophils per replicate. As a vehicle control, 0.1% (v/v) DMSO in sterile filtered calcium containing Hanks' balanced salt solution (1.3 mm CaCl2,5.4 mm KCl, 0.3 mm Na2HPO4, 0.4 mm KH2PO4, 4.2 mm NaHCO3, 137 mm NaCl, 5.6 mm d-(+)glucose, pH 7.4) was used. The stimulation mixture consisted of 1 mm phorbol myristate acetate (PMA) in DMSO (Sigma-Aldrich), 5 μg/ml E. coli 0127:E8 LPS and 5 μg/ml Salmonella enterica serotype typhimurium LPS (Sigma-Aldrich), 1.25 mm latruncilin B in DMSO (Abcam, Australia), 2.5 μm ionomycin calcium salts in DMSO (Abcam) and 1 μm fMLP in DMSO (Sigma-Aldrich) (all final concentrations diluted in calcium containing HBSS). Cells were stimulated for 30 min at 37 °C under shaking condition. The activation of neutrophils was validated in technical (activation) triplicates (n = 3) using a myeloperoxidase (MPO) activity assay. Briefly, 100 μl of secreted and cell lysate protein fractions (cells lysed with 0.1% Triton X-100), were obtained from the same number of stimulated and nonstimulated neutrophils. After the addition of 150 μl 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich) reagent, all samples were incubated for 30 min, 22 °C. The reactions were stopped using 1 m sulfuric acid (Sigma-Aldrich) and the products of the reactions were measured as a proxy for MPO activity at an absorbance of 450 nm using a plate reader (BMD technologies, Australia). The cell viabilities were monitored using trypan blue exclusion. The degree of MPO release was calculated using the following formula: (OD450 nm, secreted protein fraction)/(OD450 nm, secreted protein fraction + OD450 nm, cell lysate protein fraction) × 100%.
Activated and resting neutrophils were washed with 1× phosphate buffered saline (PBS). Cell surface biotinylation was then carried out on pooled activated and resting neutrophils as previously described (55) using a trifunctional labeling agent provided in a cell surface capture kit (Pierce, Thermo Scientific). In short, cell surface proteins were labeled with N-hydroxysuccinimide ester (NHS)-biotinylation and captured on a Neutravidin stationary phase. The flow-through fraction was collected and these nonbiotinylated proteins were considered as intracellular proteins. The cell surface enriched (biotinylated) proteins were eluted with SDS-PAGE buffer containing 50 mm DTT and the proteins were precipitated overnight in cold acetone (−20 °C), resuspended in 2% (w/v) SDS in 25 mm ammonium bicarbonate and separated and visualized on a Coomassie Brilliant Blue (CMB)-stained 4–12% SDS-PAGE (Invitrogen) under reducing conditions. The gel separated proteins were transferred to nitrocellulose or PVDF membranes (Bio-Rad, Australia) for immunoblotting with Trans-Blot Turbo (Bio-Rad), blocked with 3% (w/v) bovine serum albumin in Tris buffered saline with 0.1% (v/v) Tween 20 (TBST) for 1 h at room temperature. The membranes were incubated with different primary antibodies in TBS overnight, washed in TBST and then incubated with matching secondary antibodies. Specifically, HNE was detected using rabbit anti-HNE (1:500, overnight, 4 °C) followed by an IRDye® fluorophore-conjugated goat anti-rabbit IgG (1:13,000, 30 min, 22 °C) (Licor Biosciences, NE). Paucimannosidic epitopes were determined using paucimannose-recognizing mouse IgM (called Mannitou, undiluted concentration, overnight, 4 °C, kind gift from Dr Simone Diestel, University of Bonn, Germany) and HRP-conjugated anti-mouse IgM (1:3000, 1 h, 22 °C) (Life Technologies, Australia). Mannitou has been previously validated to recognize paucimannosidic N-glycans (56). Blots were visualized on the Odyssey imager (Licor Biosciences) using the green fluorescence channel and the chemiluminescence channel on the ChemiDoc MP system (Bio-Rad) upon application of the HRP chemiluminescent substrates (Bio-Rad). Band intensities were quantified using Image studio (Licor Biosciences) and Image J (NIH) after normalization for cells and protein loading using the total protein intensity per lane as measured on matching CMB stained gels analyzed in parallel. This method yields a reasonable accurate quantitative estimate of the relative protein abundance (57). The sample preparation, LC-MS/MS-acquisition and annotation of peptides and glycopeptides from cell-surface expressed HNE were performed as described above.
Human MBL Binding Assay
Binding of HNE to human mannose binding lectin (MBL) was assessed by firstly diluting 100 μg HNE in 2 ml binding buffer (20 mm CaCl2, 1.25 m NaCl, 10 mm Tris-HCl, pH 7.4) and then applying this solution to 2 ml MBL-agarose slurry (product number 22212, Thermo Scientific) packed in a polystyrene tube (product number 29922, Thermo Scientific) to a final column volume of 1 ml. The column was washed with 6 ml binding buffer at 4 °C prior to sample loading. HNE was loaded onto the column using gravity and the flow-through was re-loaded back onto the same column. While the sample was applied the second time, the column exit was capped and the column was incubated overnight at 4 °C. The column was then washed with binding buffer at 4 °C until all non-specifically bound proteins were depleted from the column as measured by 280 nm absorbance using a biospectrometer (Eppendorf). This unbound protein fraction was collected for parallel analysis. The column and the elution buffer consisting of 10 mm EDTA, 1.25 m NaCl, 10 mm Tris-HCl, pH 7.4 were then equilibrated at room temperature for at least 1 h. Elution was carried out at room temperature by applying 1.2 ml elution buffer to the column using a gravity-based flow. The eluent was collected and re-loaded back to the column, which was then incubated with a capped column exit at room temperature for at least 1 h. The eluate was hereafter collected, which was pooled with additional eluting fractions using the same elution buffer. Complete elution of bound protein was ensured by measuring the protein absorbance of the eluting fractions at 280 nm. The eluates were concentrated, desalted and buffer exchanged into 0.1% (v/v) FA using 10 kDa molecular weight cut off (MWCO) filters (Merck-Millipore) prior to LC-MS analysis. HNE, which was not incubated with MBL, but otherwise treated like the rest of the samples (here called “native HNE”), and the MBL-unbound and MBL-bound HNE were analyzed in their intact form using high resolution LC-Q-TOF-MS. HNE glycoforms were quantitatively profiled using the relative height of the relevant signals within the deconvoluted spectra. Technical triplicate (n = 3) analyses of the binding experiments were carried out.
A1AT-based Inhibition Assay
The A1AT-based inhibition of HNE was assessed by incubating human A1AT purified from pooled plasma of healthy individuals (Sigma-Aldrich, A9024) with HNE in a 1:1 (19.6 μm:19.6 μm) and in a 3:1 molar ratio (58.8 μm:19.6 μm) in a physiological buffer consisting of 100 mm Tris-HCl, pH 7.6 for 30 min at room temperature without mixing. Technical (inhibition) triplicates (n = 3) were carried out at both ratios. Reactions were quenched by the addition of 1.5 mm PMSF (final concentration) after incubation. A fraction of the incubated samples containing covalently bound A1AT/HNE complexes and uncomplexed A1AT and HNE were separated and visualized on a CMB stained 4–12% SDS-PAGE gel (Invitrogen) under nonreducing conditions. Matching gels were used for immunoblotting by utilizing rabbit anti-human HNE (1:500) after transferring the proteins to a nitrocellulose membrane (Bio-Rad) exactly as described above. Visualization was performed with an IRDye® fluorophore-conjugated goat anti-rabbit IgG (1:13,000, Licor Biosciences) using the Odyssey imager (Licor Biosciences) with the green fluorescence channel. The remainder of the protein mixtures was concentrated, desalted and buffer exchanged into 0.1% (v/v) FA using 10 kDa MWCO filters (Merck-Millipore) prior to native LC-MS analysis (see above). Uncomplexed HNE, which eluted separately in the LC-MS analysis, was identified and quantified after deconvolution using the relative height of the apex of the relevant MS signals while the glycoforms of coeluting uncomplexed A1AT and the HNE:A1AT complexes were identified and quantified with the assistance of existing N-glycoprofiles of human A1AT (58, 59) after spectral deconvolution using a mass stepping of 1 and height filters set to a peak signal-to-noise ratio of 0.5.
Bacteriostatic Assay
The clinically relevant Pseudomonas aeruginosa PASS1 strain was isolated and cultured to mid-log growth phase as previously described (60, 61). PASS1 was then washed twice in 1× PBS and diluted to an OD600 nm of 0.1 in Luria Beria broth before use. N-glycans were released from HNE by N-glycosidase F and desalted using PGC-SPE as described above, but the N-glycans were used in their free nonreduced form in the bacteriostatic assay. Enzymatically active HNE (1.8 μm and 3.6 μm), the released N-glycans from the same concentrations of HNE and appropriate controls (containing media only) were separately added to PASS1 cultures in total volumes of 100 μl Luria Beria broth per well in a 96-well flat bottom microtiter plate kept at 37 °C. The growth of PASS1 was then monitored over a period of 12 h using absorbance (OD600 nm), which was measured automatically with 30 min intervals using a microplate reader (BMG technologies). Technical (growth inhibition) triplicates (n = 3) of all culture experiments were carried out.
Statistics
Data points and error are presented as mean ± standard deviation (S.D.) or standard error of mean (S.E.) as indicated for the individual experiments. Statistical regression analyses and significance testing were carried out using Microsoft Excel. The significance of the individual experiments was assessed by paired and unpaired two-tailed type 2 Student's t-tests as indicated where p < 0.05 was chosen as the minimum acceptable level of confidence to support a rejection of the proposed null hypothesis, e.g. difference of two means. The nature of the replicates (biological or technical) and the sample numbers (n) used for the individual experiments are described above.
Experimental Design and Statistical Rationale
The experimental design, number of replicates and statistical rationale have been described above for the individual experiments performed in this study.
RESULTS AND DISCUSSION
Site-specific N-glycoprofiling Reveals Unusual Glycosylation Signatures of HNE
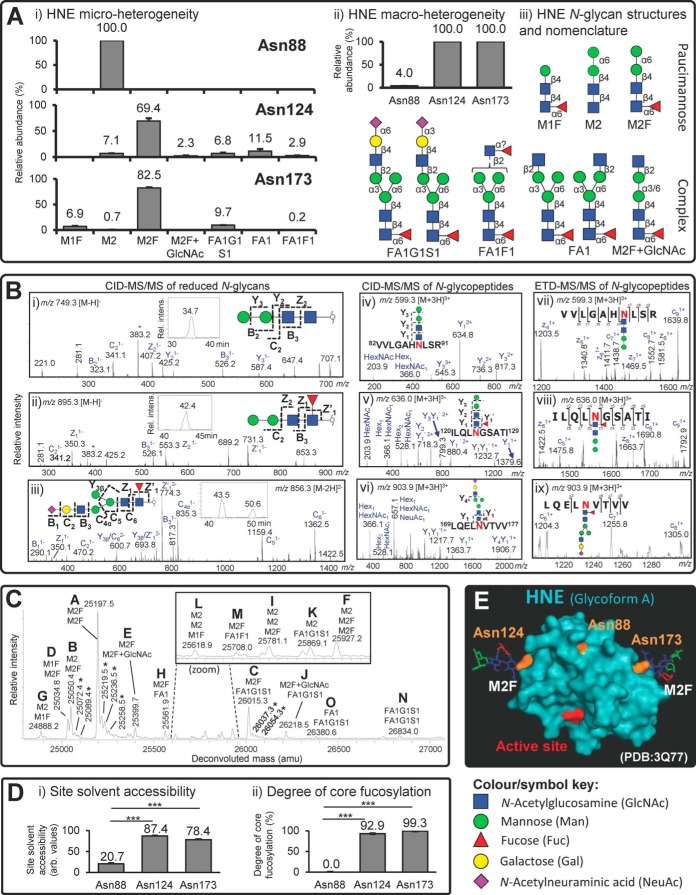
N-glycopeptide, N-glycan, and intact N-glycoprotein analyses were used for the site-specific characterization of the N-glycosylation of naturally occurring and enzymatically active HNE derived from resting neutrophils isolated from healthy individuals. LC-MS/MS-based site-specific glycoprofiling of the enriched and nonenriched intact N-glycopeptides facilitated complete coverage of all four putative N-glycosylation sites of HNE i.e. Asn88, Asn124, Asn173, and Asn185 (supplemental Fig. S2, supplemental Fig. S4B, and supplemental Table S1). Asn185 was not utilized as a glycosylation site. The three occupied N-glycosylation sites (Asn88, Asn124, and Asn173) displayed relatively limited micro-heterogeneity (Fig. 1A i), and showed site-specific differences in their occupancy (macro-heterogeneity) (Fig. 1A ii). Asn124 and Asn173 were both fully occupied (100% site occupancy, n = 3) by five or six monosaccharide compositions at each site based on the lack of both nonglycosylated and spontaneously deamidated peptides covering these two N-glycosylation sites in the unenriched HNE (glyco)peptide mixture. In contrast, a single monosaccharide composition only infrequently occupied the Asn88 site (4.0% ± 1.1% site occupancy, n = 3). The observed monosaccharide compositions corresponded to unconventional paucimannosidic and complex-type N-glycans. PGC-LC-MS/MS of reduced, but otherwise native, HNE N-glycans revealed their exact structures including their topology (sequence and branching patterns) and glycosidic linkages (supplemental Fig. S3A). In total, only nine N-glycan isomers were observed at both the N-glycan and N-glycopeptide level (Fig. 1A iii and supplemental Table S2). All reported structures were validated using CID-MS/MS for the HNE N-glycans (Fig. 1B i–iii and supplemental Fig. S3B) whereas complementary CID- and ETD-MS/MS dissociation methods were applied to the HNE N-glycopeptides (Fig. 1B iv-ix and supplemental Fig. S4A). These analyses demonstrated that the single Asn88-glycan was Manα1,6Manβ1,4GlcNAcβ1,4GlcNAcβ (hereafter called M2). Because of their biosynthetic relatedness, the abundant N-glycan of Asn124 and Asn173 was suggested to be the matching core fucosylated species also only observed as a single isomer i.e. Manα1,6Manβ1,4GlcNAcβ1,4(Fucα1,6)GlcNAcβ (hereafter called M2F) (relative abundance: Asn124: 69.4% ± 5.2% and Asn173: 82.5% ± 0.8%). Interestingly, both bimannosylchitobiose structures appeared only as the α1,6-Man-terminating isomer (see inserts in Fig. 1B i–ii showing single eluting isomers), in agreement with past observations reported in other neutrophil N-glycome studies (38, 60, 62, 63). The presence of the α1,6-Man-terminating isomer was supported by its PGC-LC retention time that agreed with the PGC retention time of a synthetic α1,6-Man-terminating M2 reference compound and the α1,6-Man-terminating isomer of artificial M2 generated from chicken ovalbumin (see insert in supplemental Fig. S3B, Glycan #2). The α1,3-linked Man isomer was absent for both M2 and M2F. This suggests that the α1,3-linked Man of the conserved trimannosyl-chitobiose core and any monosaccharide residues decorating the N-glycan core including β1,2-GlcNAc residues, are efficiently and selectively removed by glycosidases during or after HNE biosynthesis. Given that other neutrophil proteins including nCG, azurocidin and proteinase 3 have been reported to be highly paucimannosylated (38, 60, 63, 64), it indicates that paucimannosylation is a commonly utilized N-glycoprotein feature of human neutrophils. The complex N-glycans decorating Asn124 and Asn173 appeared as two highly unusual isobaric sialyl-linkage isomers (see insert in Fig. 1B iii for the PGC-LC separated isomers) i.e. NeuAc2,3/6Galβ1,4GlcNAcβ1,2Manα1,6(Manα1,3)Manβ1,4GlcNAcβ1,4(Fucα1,6) GlcNAcβ (hereafter collectively called FA1G1S1) (relative abundance: Asn124: 6.8% ± 1.8% and Asn173: 9.7% ± 0.4%) (Fig. 1A i). Interestingly, the fully processed and mature monoantenna of these two isomeric sialylated structures was exclusively positioned on the 3′-Man arm based on the absence of D ions in the MS/MS spectra, thereby yielding protection from degrading α1,3-mannosidases that appear active in neutrophils as judged by the prevalent M2 and M2F. Other unconventional biosynthetically-related N-glycans of the monoantennary complex type (FA1F1 and FA1) and the paucimannosidic type (M1F) were also observed to be conjugated to Asn124 and Asn173 (Fig. 1A i). Although rarely reported, we have recently characterized similar monoantennary sialoglycans on nCG (38) and these structures were also seen in relative high abundance in pathogen-infected sputum and in isolated resting neutrophils (60). Interestingly, these structures have been reported as free N-glycans in stomach cancer-derived cell lines (65) and mouse embryonic fibroblasts (66), where the monoantenna was also proposed to be positioned on the 3′-Man arm. In contrast to nCG, HNE did not display the highly truncated chitobiose type N-glycans i.e. (±GlcNAcβ1,4) (±Fucα1,6)GlcNAcβ (38). Taken together, these observations support that human neutrophils can generate highly unusual N-glycosylation including paucimannosylation and monoantennary sialylation in a site-specific manner on HNE.
Fig. 1.
Site-specific characterization of HNE N-glycosylation. A, i) Limited structural micro-heterogeneity (relative glycan distribution, %, mean ± S.E., n = 6). ii) Differential macro-heterogeneity (site occupancy, %, mean ± S.E., n = 3) across the three utilized HNE N-sites i.e. Asn88, Asn124 and Asn173. iii) Overview of the characterized HNE N-glycan structures and their short-hand nomenclature (see bottom for key and symbol nomenclature) (49). B, Comprehensive glycan and glycopeptide MS/MS spectral data supporting the structural characterization. The spectra were annotated and the corresponding structures were assigned based on the identified glycan and peptide fragments (as well as molecular mass and retention time) following the established fragmentation rules and nomenclature. Resulting fragments have only been partially annotated for the sake of clarity; See supplemental Fig. S3B and supplemental Fig. S4A for all supporting fully annotated spectra. * denotes cross-ring fragment ion. Left: Annotated ion trap CID-MS/MS spectra of reduced N-glycans as exemplified by the three main HNE N-glycans i.e. i) M2, ii) M2F and iii) FA1G1S1 (α2,6 sialyl-linkage), see inserts for XIC-based PGC-LC elution pattern. Middle/Right: Annotated ion trap CID- and ETD-MS/MS spectra, respectively, of the major HNE N-glycopeptides, iv and vii) Asn88-M2, v and viii) Asn124-M2F, and vi and ix) Asn173-FA1G1S1, respectively. C, Glycoprofiling of intact HNE. The deconvoluted spectrum shows the fifteen identified HNE glycoforms (labeled A-O, in the order of high to low relative abundance) identified by high resolution LC-ESI-Q-TOF mass spectrometry. The proposed glycans conjugated to the intact mature HNE polypeptide chain (Ile30-Arg248) are indicated in a site-unspecific manner with the short-hand nomenclature by considering the previously determined site-specific glycopeptide and glycan data. * denotes adduct formation of HNE, see supplemental Fig. S5A for more. D, Relationship between the N-glycosylation and tertiary structure around the N-glycosylation sites of HNE as shown by the clear correlation between i) the solvent accessibility of the three HNE N-glycosylation sites assessed using nineteen PDB structures of neutrophil-derived HNE (mean ± S.E. of arbitrary (unit-less) but comparable values, n = 19 3D structures, *** denotes p < 0.001 using paired two-tailed type 2 Student's t-tests), see supplemental Table S3 for more details and ii) the degree of N-glycan core fucosylation (in %) of the individual HNE N-glycosylation sites based on N-glycopeptide analysis (mean ± S.E., n = 6 technical replicates; *** denotes p < 0.001 using unpaired two-tailed type 2 Student's t-tests). E, Spatial location of the active site relative to the three utilized N-glycosylation sites on a representative 3D structure (PDB: 3Q77) of human neutrophil-derived monomeric HNE. M2F was modeled in silico on Asn124 and Asn173 by utilizing their default torsion angles provided by GLYCAM whereas Asn88 was left unoccupied (i.e. Glycoform A was visualized). Protein color scheme: The active site (His39, Asp117, Ser217 based on preproprotein amino acid sequence) is in red and the three N-glycosylation sites are in orange. The monosaccharide colors follow the symbol nomenclature used throughout (see key below) (49).
Supported by an extensive sequence coverage (>75%) (supplemental Fig. S4B), the HNE peptide mapping showed no evidence of other PTMs. The peptide analysis also indicated that lle30 forms the N-terminal of the mature, fully processed, HNE polypeptide chain in accordance with the predicted amino acid sequence of HNE based on previous cDNA sequencing (14) (supplemental Fig. S1). In contrast to the reported Gln247 (67), we observed that Arg248 instead forms the C-terminal of mature HNE similarly to an Arg-terminating proteoform of nCG (38). No polypeptide sequence heterogeneity of HNE with respect to C- and N-terminal truncation forms was observed in our peptide analysis. The C-terminal arginine residues of HNE and nCG may provide these already highly cationic serine proteases with even higher isoelectric points, but the biological importance of their basic nature remains insufficiently understood (16).
Reflecting the limited micro- and macro-heterogeneity observed here and elsewhere (68), high mass accuracy LC-Q-TOF-MS detected only fifteen compositional glycoforms of intact HNE, which all agreed very well with the prior HNE N-glycopeptide and N-glycan analyses (Fig. 1C and supplemental Fig. S5A). Beam-type CID-MS/MS fragmentation of intact HNE was used to confirm that Arg248 forms the true C-terminal of the glycoprotein (supplemental Fig. S5B). Glycoform A (molecular mass, M = 25197.5 Da) was the most abundant species, most likely corresponding to a monomeric HNE glycoform displaying unoccupied Asn88 and M2F conjugated to both Asn124 and Asn173. Also in agreement with the site-specific glycoprofiles, glycoform B (M = 25050.4 Da) carrying most likely M2 and M2F on Asn124 and Asn173 and displaying an unoccupied Asn88 was also abundant, as was glycoform C (M = 26015.3 Da) carrying monosaccharide compositions potentially corresponding to M2F and FA1G1S1 moieties. Several HNE glycoforms of lower abundance were detected to be carrying M2 as a third N-glycan presumably on Asn88 (glycoform F, I, and L). Although the HNE glycoforms are strongly supported by the glycopeptide data, top down electron transfer/capture dissociation experiments are required to confirm the site-specific distribution of these multi-glycosylated forms of HNE (69). Taken together, the accurate mass analysis of native HNE confirmed the site-specific glycopeptide profiles, validated the N- (Ile30) and C- (Arg248) termini of mature HNE and simultaneously demonstrated that unusual paucimannosidic N-glycans were carried by enzymatically active intact HNE, proved the absence of other PTMs, and confirmed that the molecular mass of the mature HNE proteoforms is 25–27 kDa in agreement with previous, less accurate, estimates based on gel electrophoresis (30, 70).
Solvent Accessibility and Spatial Positioning of N-glycosylation Sites on mature HNE
Asn88 has a unique glycosylation signatures (low site occupancy and lacking both core fucosylation and complex-type structures) relative to the two more processed glycosylated sites, Asn124 and Asn173. The possible protein structural differences around the glycosylation sites causing these site-specific glycosylation differences were investigated using nineteen 3D structures of neutrophil-derived HNE solved using X-ray crystallography available from the Protein Data Bank (supplemental Table S3). Significantly lower solvent accessibility of Asn88 was observed (NACCESS score: 20.7 ± 1.4) compared with the accessibilities of Asn124 (87.4 ± 0.7, p < 0.001) and Asn173 (78.4 ± 1.0, p < 0.001, paired two-tailed type 2 Student's t-tests) (all as mean ± S.E., n = 19) (Fig. 1D i). The differential site accessibilities agreed well with the observation that the relatively hidden Asn88 only carried N-glycans completely lacking core fucosylation, whereas the surface exposed Asn124 and Asn173 sites were highly core fucosylated (92.9% ± 0.3% and 99.3% ± 0.1%, respectively, both p < 0.001) (mean ± S.E., n = 6) (Fig. 1D ii). We previously showed that the extent of core fucosylation is heavily influenced by the local structure and the solvent accessibility of the N-glycosylation site being modified (71). The Asn124- and Asn173-glycans also displayed other features associated with a high degree of glycan processing including antenna extension and NeuAc capping; however, it remains unclear if such glycan structural elements further away from the protein surface are similarly affected by solvent exposure. The influence of the solvent accessibility on any glycan truncation by hydrolases (e.g. β-hexosaminidase and α-/β-mannosidases) after protein folding also remains unclear. Dissecting out such structural-biosynthetic mechanistic relationship(s) would advance our understanding of the biosynthetic generation of paucimannosidic proteins and the even shorter chitobiose type structures.
The transfer of the initial N-glycan precursor, which effectively determines the degree of site occupancy, takes place on unfolded or only partially folded glycoproteins early in the endoplasmic reticulum (48). Hence, the 3D structures of maturely folded HNE are not suitable to probe the mechanisms driving the site-specific occupancy differences. Although the Asn88 sequon (NLS) is followed immediately by an arginine residue (Arg91), which reportedly favors the N-glycan transfer rate because of its beneficial positive charge (72), Asn88 still displayed very low glycan site occupancy. Thus, it is likely that other yet-to-be-determined primary and secondary structural features around the glycosylation sites are responsible for generating these site occupancy differences. Finally, as demonstrated on a representative 3D structure of neutrophil-derived HNE (PDB: 3Q77), it is interesting to note that the three utilized N-glycosylation sites of HNE are positioned distal to the active site of the protease (Fig. 1E). This implies that the relative simple HNE N-glycans of short length and limited volume are more likely involved in other functions than in direct modulation of HNE proteolytic activity.
Uniform N-glycosylation of HNE Residing in Various Neutrophil Compartments
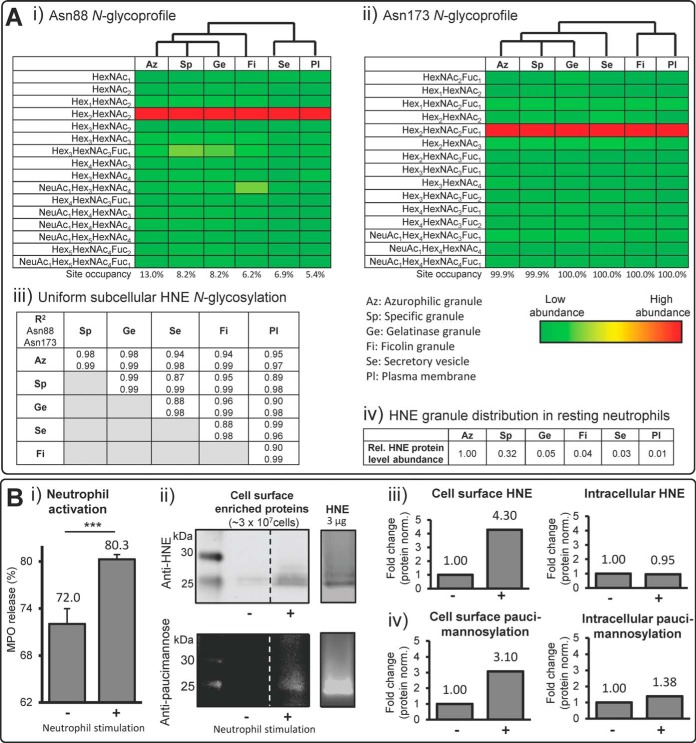
We have recently used subcellular proteomics to show that the individual granule compartments of human neutrophils harbor distinct subproteomes (3). This LC-MS/MS dataset was here manually reinterrogated with the purpose of performing a detailed subcellular- and site-specific N-glycoprofiling of HNE. Glycosylated and nonglycosylated tryptic peptides covering Asn88 and Asn173 (but not Asn124 because of tryptic peptides that were too large for efficient MS detection) were confidently identified (supplemental Table S4), facilitating a granule-specific assessment of both the micro- and macro-heterogeneity of these two HNE glycosylation sites. This highly sensitive, compartment-specific data provided identification of many relatively low-abundant monosaccharide compositions at each site, but importantly, the resulting glycoprofiles matched reasonably well our site-specific glycoprofiles of HNE extracted from whole resting neutrophils (see Fig. 1A i). Across all compartment lysates of Asn88 was largely unoccupied (∼87–95%), but harbored a significant proportion of M2 (∼3–7%), whereas Asn173 was almost fully occupied by M2F (∼55–75%) and complex-type (∼30%) sialylated and neutral N-glycans. Interestingly, these studies showed surprisingly little variation in the N-glycosylation of HNE residing in the various neutrophil compartments as illustrated by highly similar heat maps representing their site-specific glycan (i.e. monosaccharide composition) distribution (Fig. 2A i–ii). With unsupervised hierarchical clustering, the Asn88- and Asn173-glycosylation of HNE residing in the three major granules i.e. azurophilic, specific and gelatinase granules proved to be slightly more similar than the glycosylation signatures of HNE residing in other compartments and on the plasma membrane. Minor granule-specific differences were also observed with respect to the glycan occupancies of Asn88 and Asn173. However, irrespective of compartment origin, the site-specific N-glycosylation was surprisingly uniform across the granule compartments as demonstrated by very high correlation coefficients of both N-sites (R2 = 0.87–0.99) (Fig. 2A iii). The strong HNE glycoform similarities across the neutrophil granules are somewhat unexpected for a number of reasons i.e. 1) subcellular-specific N-glycosylation is a feature of several human cell types (73), 2) we have previously suggested that paucimannosylation is an N-glycan feature that appears to be preferentially displayed by proteins residing in the azurophilic compartment in human neutrophils (60), 3) the various neutrophil compartments, where the final glycoprotein processing presumably takes place, are thought to be formed at different times during myelopoiesis (17) and, as a result of temporal hydrolase expression during myelopoiesis, harbor different granular levels of various glycan processing enzymes (60) and, finally, 4) limited protein exchange reportedly occur between the neutrophil granules after their formation (3, 74). The compartment-specific proteome data also showed that HNE, under resting conditions, primarily resides in the azurophilic (relative level set to 100%) and specific (32%) granules, whereas less HNE protein is found in the gelatinase (5%) and ficolin (4%) granules or in the secretory vesicles (3%) and on the plasma membrane (1%) (Fig. 2A iv). Thus, it appears that human neutrophils can synthesize the same set of biosynthetically related HNE glycoforms and in similar ratios during the maturation stages, but that HNEs end up being unevenly distributed across the various intracellular compartments and on the cell surface of neutrophils. The latter observation is supported by the “targeting by timing” hypothesis proposed by Borregaard and coworkers suggesting that the various neutrophil granules are filled with different proteins reflecting the different protein expression profiles during time-separated granule formation in developing myelocytes. Hence, neutrophil proteins that are synthesized simultaneously in maturing neutrophils will end up in the same granule compartment (75).
Fig. 2.
Uniform but site-specific N-glycosylation of HNE residing in various neutrophil compartments. A, Subcellular- and site-specific N-glycosylation analysis of HNE isolated from azurophilic (Az), specific (Sp), gelatinase (Ge), and ficolin (Fi) granules and from the secretory vesicles (Se) and the plasma membrane (Pl) of human resting neutrophils. Data based on retrospective manual interrogation of compartment-specific proteomics datasets of human neutrophils (3). The i) Asn88 and ii) Asn173 N-glycosylation profiles in their respective compartments are represented using heat maps to the resolution of their monosaccharide compositions (see sliding tricoloured bar for color coding). The profiles largely matched the site-specific glycoprofiles of HNE from neutrophil lysates (see Fig. 1). The predictably very large tryptic Asn124 glycopeptides were not identified in these datasets. Unsupervised clustering showed that the Asn88- and Asn173-glycosylation of HNE residing in Az, Sp and Ge clustered stronger together than the N-glycosylation of HNE from the other compartments. iii) The N-glycosylation of HNE is highly uniform throughout the granule compartments as shown by the high regression correlation coefficients of the corresponding subcellular glycoprofiles. iv) HNE preferentially resides in Az proteomics data of human neutrophils (3). B, i) The activity of the neutrophil activation marker, MPO, was increased in secretions from human resting neutrophils as assessed by reinterrogation of the granule-specific stimulated (+) relative to resting (-) neutrophils while the cell viabilities remained unchanged, indicating active degranulation (mean ± S.D., n = 3 technical replicates, *** denotes p < 0.001 using unpaired two-tailed type 2 Student's t-tests). ii) HNE- (top gel) and paucimannose-specific (bottom gel) immunoblotting of stimulated (+) and resting (-) human neutrophils showed spatiotemporal regulation of paucimannosylated HNE on the cell surface of activated neutrophils. Cell surface proteins were extracted by NHS-biotinylation and captured on a Neutravidin stationary phase from ∼3 × 107 cells in both conditions (n = 3 technical replicates). Broken lines denote nonneighboring lanes from immuno blots on the same gel (same exposure and contrast). Purified HNE (3 μg) is shown in a separate lane to illustrate the reactivity to anti-HNE and anti-paucimannosidic antibodies and visualize the molecular mass of HNE, see Fig. S6 for more. iii) Quantitation of cell surface (left) and intracellular (right) HNE in the immunoblotted HNE gel region (∼25–27 kDa) of stimulated relative to resting human neutrophils. Signal intensities were quantified using densitometry after normalization for total protein loading and presented as fold difference of stimulated versus resting neutrophils, n = 3 pooled technical replicates. iv) Quantitation of the degree of cell surface (left) and intracellular (right) paucimannosylation in the immunoblotted HNE gel region (∼25–27 kDa) of stimulated relative to resting human neutrophils. Signal intensities were quantified using densitometry after normalization for total protein loading and presented as fold difference of stimulated versus resting neutrophils, n = 3 pooled technical replicates.
Interestingly, the peculiar FA1F1 structure carried by HNE on Asn124 and Asn173 was found across all granules, but present at too low relative abundance (< 3%) to assess whether significant differences in its granule distribution occurred. The unusual GlcNAc-Fuc epitope at the nonreducing end of this structure does not conform to the established N-glycan biosynthetic pathway, but indicates that this structural feature arises from the trimming of a more elongated antenna by the action of β-galactosidases (and possible also α-sialidases) present in the neutrophil granules (3, 76). The exact enzymatic pathway that allows the biosynthesis of this structure as well as many other spatial and temporal aspects of the glycosylation machinery of developing neutrophils remain to be determined.
Stimuli-induced Expression of Paucimannosidic-rich HNE on the Neutrophil Cell Surface
Antimicrobial glycoproteins and peptides secreted from neutrophil compartments via degranulation mechanisms are critical in the pathogen defense system and in other immune functions of the host (1). Although HNE has been reported to associate with the neutrophil cell surface upon activation (77, 78), the molecular details underpinning the mobilization and the retention of soluble HNE at the plasma membrane as well as the N-glycosylation of cell surface HNE of activated human neutrophils are yet to be explored. These research questions are particularly interesting considering that neutrophils are known to express an A1AT-resistant form of HNE on their cell surfaces (4, 22) and the fact that paucimannosidic N-glycoproteins are increasingly being associated with important extracellular functions in inflammation and infection (79, 80).
In agreement with literature (81), human resting neutrophils were activated ex vivo upon chemical stimulation with a mixture of PMA, LPS, and chemo-attractant peptides as evaluated by the increased secretion of MPO (Fig. 2B i), which is a well-established marker of neutrophil activation. Importantly, the resting and activated neutrophils maintained similar viabilities (∼55–70%) as assessed by trypan blue exclusion over the 30 min stimulation experiment, ruling out significantly higher contributions of intracellular glycoproteins from potential cell death and leakage because of plasma membrane deterioration in activated neutrophils. By utilizing a cell surface protein capture approach, immunoblotting and mass spectrometry (Fig. 2B ii–iii and supplemental Fig. S6), we showed that activation enhanced HNE expression on the neutrophil cell surface relative to its constitutive expression on resting neutrophils and only caused a negligible reduction in the amount of intracellular HNE as evaluated by reactivity to anti-HNE antibodies. Complementary (opposite) changes of the intra- and extracellular levels of HNE upon neutrophil stimulation were expected because the de novo protein synthesis and degradation were considered to be negligible over the 30 min stimulation experiment. However, the negligible reduction in the intracellular levels of HNE upon neutrophil stimulation may be explained by the relatively weak HNE cell surface expression (1%) (see Fig. 2A iv). LC-MS/MS based peptide identification confirmed that the 25–27 kDa gel region contained HNE (supplemental Fig. S7). Thus, the experiments showed that the majority of HNE remained in an intracellular location upon stimulation in agreement with the observations that granular HNE can undergo both intra- and extracellular degranulation by granule fusion with intracellular phagolysosomes (82) and the plasma membrane (83), respectively. The lower propensity of the HNE-rich azurophilic granules to fuse with the plasma membrane compared with the specific and gelatinase granules upon stimulation and their ability to translocate to the nucleus of neutrophils support these observations (2, 28, 84).
To probe the N-glycosylation of cell surface-bound HNE of neutrophils undergoing activation, immunoblotting with a paucimanose-reactive antibody, Mannitou (56), was performed and the signals in the 25–27 kDa gel region were assessed. Stimulated neutrophils showed an increased level of cell surface paucimannosylation relative to the constitutive level under resting condition (Fig. 2B ii, iv and supplemental Fig. S6). This increase was comparable to the activation-induced increase in cell surface HNE (Fig. 2B iii). Mass spectrometry confirmed the presence of HNE paucimannosylation on the surface of activated neutrophils by the identification of Asn88-M2 and Asn124-M2F glycopeptides derived from cell surface captured HNE (supplemental Fig. S8), but the signal strength proved too weak to perform an accurate qualitative and quantitative site-specific analysis using LC-MS/MS as performed for plasma membrane HNE of resting neutrophils (Fig. 2A). The level of intracellular paucimannosylation remained relatively constant upon stimulation, further supporting that the bulk of paucimannosylated proteins including HNE reside in nonmobile granules or within granules that undergo intracellular fusion during neutrophil activation (28). Taken together, our analyses indicate that paucimannosidic-rich HNE glycoforms are expressed on the surface of neutrophils during resting conditions and that similarly paucimannosylated intracellular HNE can be mobilized to the cell surface upon neutrophil activation.
Although the exact molecular details of the mechanism(s) facilitating plasma membrane association remain unknown, the positively charged arginine-rich HNE was suggested to be retained on the surface of activated neutrophils by forming electrostatic interactions with membrane-tethered/associated chondroitin sulfate- and heparin sulfate-containing proteoglycans (77). Given the highly hydrophilic and flexible nature of N-glycans (85), the partially charged N-glycans on HNE may directly modulate such interactions and/or more indirectly mask protein surface charges as mechanistically shown for sialoglycans of other N-glycoproteins (24). Computational analysis suggests that the three occupied glycosylation sites of membrane-retained HNE display N-glycans that face and extend into the extracellular milieu (86). Thus, the paucimannose-rich N-glycosylation of HNE may likely mediate cell-cell communication with immune cells including dendritic cells and macrophages that are known to express mannose recognizing C-type lectin receptors (mrCLRs) with affinity to mannosylated glycoepitopes (87, 88). The interplay between neutrophils and other immune cells at inflammatory sites has received considerable attention recently; not least in cancer where tumor associated neutrophils have been reported (89, 90). In addition, the unusual HNE sialoglycans may also contribute to the extravasation process by interacting with selectins expressed by the endothelium (91), if these sialoglycoforms of HNE are presented and correctly oriented on the neutrophil surface at the appropriate time. The potential of paucimannosylated HNE to interact with mrCLRs from the cell surface of activated neutrophils was investigated to provide further insights into neutrophil-mediated immune communication.
Glycoform-dependent Binding of Mannosylated HNE to Human MBL
Given the highly unusual and mannose-rich N-glycosylation of the spatiotemporally-regulated HNE, we probed the binding potential and affinity preferences of glycosylated HNE to a central mrCLR in inflammation. This is pertinent because of (1) the accessible nature of the HNE N-glycans localized distal to the active site (see above), (2) the fact that HNE is an important glycoprotein involved in many innate immunological processes (92), and (3) the family of mrCLRs including the soluble MBL (93) and the mannosidic glycoprotein ligands are increasingly recognized as key players in innate immunity (79, 94). In fact, MBL is, similarly to HNE, enriched in inflamed/infected airways (95) where it was found to be resistant to HNE degradation (96). We therefore investigated if HNE and human MBL are interaction partners and, if so, whether paucimannosylated forms of HNE show binding preference to MBL over other HNE glycoforms.
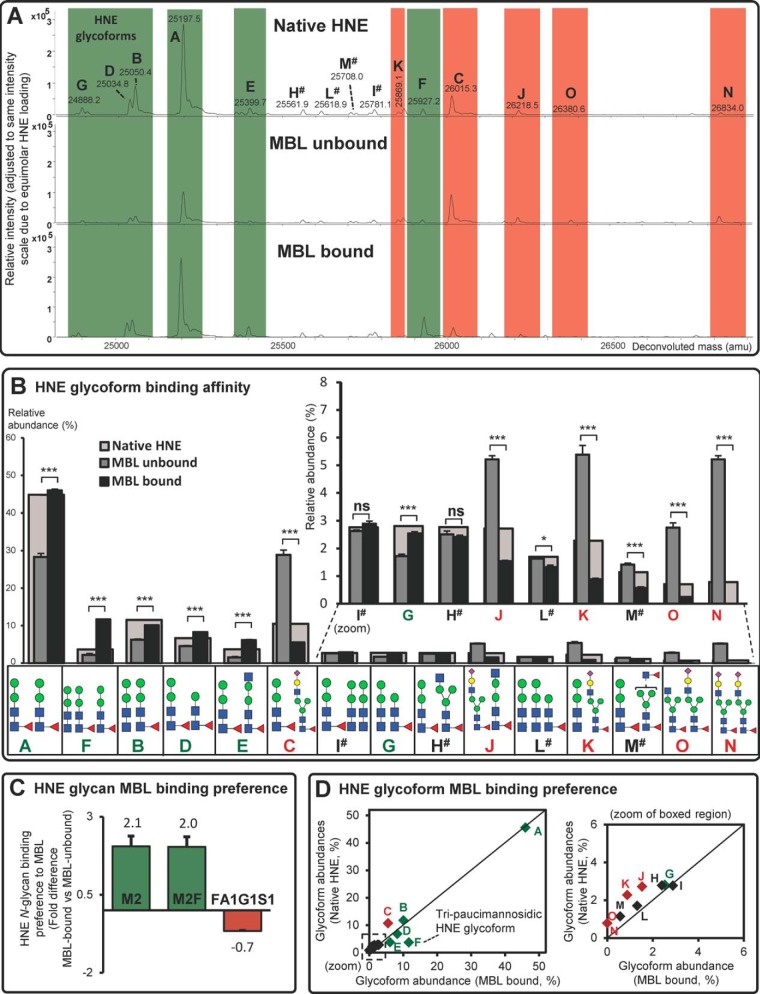
Immobilized human MBL affinity chromatography with LC-MS-based analysis of the native and MBL-bound/-unbound glycoforms of HNE revealed that some paucimannosidic N-glycoforms carrying M2F and M2 (e.g. glycoforms A, B, D, E, F, and G) appeared to be overrepresented in the MBL-bound relative to the MBL-unbound fractions (Fig. 3A, green shading). In total, the binding preference of six HNE glycoforms to human MBL was confirmed by quantitatively comparing the relative distribution of the MBL-bound and unbound HNE glycoforms based on the relative height of their respective deconvoluted signals (0.0001 < p < 0.001 for all six glycoforms) (Fig. 3B and supplemental Table S5). Further assessment of the individual N-glycans validated that both the mannosylated M2 and M2F clearly displayed preferential affinity to human MBL (2.1 ± 0.3 and 2.0 ± 0.3-fold binding preference, respectively) (all as mean ± S.E., n = 3) (Fig. 3C). In contrast, clear binding discrimination was shown against the FA1G1S1 sialoglycans (0.7 ± 0.01-fold discrimination), which could be appreciated by relative signal suppression of all five HNE sialoglycoforms (glycoforms C, J, K, N, and O in Fig. 3A–3B, red shading). By comparing the distribution of MBL-bound HNE glycoforms to the native HNE glycoform distribution, it became clear that the HNE glycoform F carrying three paucimannosidic N-glycans (two M2F N-glycans and a single M2 N-glycan), was a highly preferred binding partner compared with the other HNE glycoforms as shown by its deviation from the perfect regression line (identity line) in the scatter plot (Fig. 3D). This preferential binding to MBL could relate to the higher valency of the N-linked mannoglycans forming the glycoform F relative to glycoform A (87). Despite also carrying three paucimannosidic N-glycans, HNE glycoform I and L surprisingly did not display enhanced MBL binding. M2F and GlcNAc (± Fuc)-capped trimannosyl-chitobiose structures carried by the HNE glycoforms H and M also did not display enhanced MBL binding. The lack of MBL binding preference of these glycoforms may relate to their decoration by only a single high-affinity M2F and/or be because of a spatial interplay between the three HNE N-glycans facilitating a global protein structure that disfavored MBL binding (97). Taken together, this indicates that paucimannosidic ligands, in particular multivalent core fucosylated dimannosylchitobiose structures, are important to facilitate efficient MBL binding. In analogy, high mannosidic protein ligands have been shown to display high avidity binding to various mrCLRs by means of multivalent interactions either facilitated by their multimannosylated antennas or through the assembly in micro-domains (mannose patches) on protein surfaces (98).
Fig. 3.
Glycoform-dependent binding of HNE to human MBL. A, Representative deconvoluted mass spectra of native (top), MBL-unbound (middle) and MBL-bound (bottom) intact HNE. Examples of over- (i.e. M2- and M2F-containing HNE) and under-represented (i.e. FA1G1S1-containing HNE) glycoforms in the MBL-bound relative to the MBL-unbound HNE spectra are highlighted in green and red, respectively. # denotes HNE glycoforms containing paucimannosidic structures that do not demonstrate enhanced/reduced MBL binding. B, The MBL binding affinity was quantified for the fifteen glycoforms of MBL-unbound HNE (dark gray bars) and MBL-bound HNE (black bars) (mean ± S.E., n = 3 technical replicates; ns, not significant (p ≥ 0.05); *, p < 0.05 and ***, p < 0.001 using unpaired two-tailed type 2 Student's t-tests). The glycoform distribution of native HNE is indicated for reference (overlaid, larger light gray bars) and the glycoform structure, nomenclature and any binding preference or discrimination are indicated in green and red, respectively. # denotes HNE glycoforms containing paucimannosidic structures that do not demonstrate enhanced/reduced MBL binding. C, The MBL binding preferences of the individual M2, M2F and FA1G1S1 HNE N-glycans were determined (presented as fold difference of MBL-bound versus MBL-unbound HNE, mean ± S.E., n = 3 technical replicates). D, Comparison of the distribution of native HNE and MBL-bound HNE N-glycoforms. Significantly over- and under-represented HNE glycoforms relative to a perfect regression line (identity line) forced between (0, 0) and (1, 1) are in green diamonds and red diamonds, respectively. The HNE glycoform F carrying tri-paucimannosidic structures demonstrated preferential binding to MBL whereas the sialoglycoforms (Glycoform C, J, K, N, and O) bound to a lesser degree. Paucimannosidic HNE glycoforms that did not demonstrate enhanced/reduced MBL binding are shown as black diamonds.
In conclusion, our data show that HNE interacts with human MBL in vitro and that the N-glycosylation of HNE facilitates binding through paucimannosidic ligand/receptor mechanisms like previously reported interactions of highly mannosylated glycoproteins to MBL (99, 100). It is tempting to speculate that neutrophils synthesize a spectrum of related glycoforms of HNE spanning high-affinity (containing up to three paucimannosidic moieties, two being M2F) to low-affinity (up to two sialoglycans) variants to fine-tune the amount of binding to MBL and possibly to other mrCLRs e.g. SP-A and SP-D present in inflamed airways.
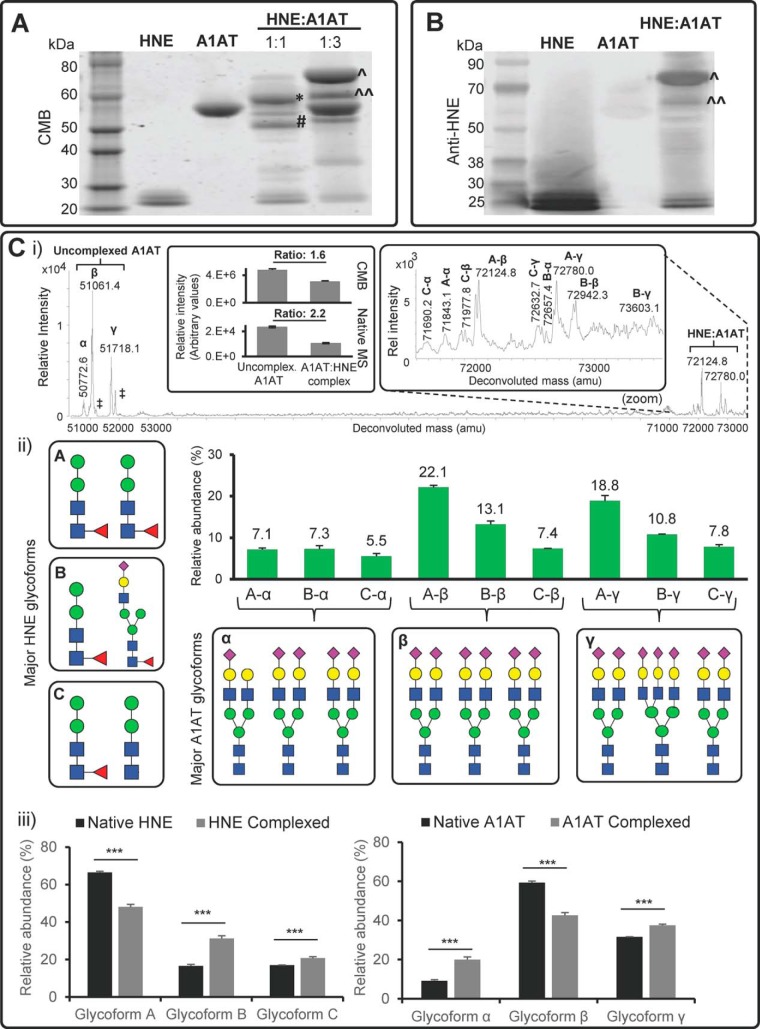
Proteolytic Inhibition of HNE Through Glycoform-dependent Interactions with A1AT
Human A1AT, an abundant 52 kDa sialoglycoprotein in blood, contains three occupied N-glycosylation sites (Asn70, Asn107, Asn271) and is the main physiological inhibitor of HNE (20). The ability of neutrophils to express an enzymatically active cell surface A1AT-resistant form of HNE, represents a nonoxidative mechanism to preserve the potent proteolytic activity of HNE in the presence of equally potent inhibitors such as A1AT in the extracellular milieu (4). As the inhibitory action of A1AT is important to mitigate lung airway damage caused by excessive HNE proteolysis (101, 102), we investigated if the unusual HNE N-glycosylation is involved in regulating the A1AT-based inhibition by interfering with the covalent HNE/A1AT complex formation.
As indicated by the protein staining and anti-HNE-based immunoblotting of the gel-separated proteins, an enzyme-to-inhibitor molar ratio of 1:3 resulted in an intact HNE/A1AT complex (∼73 kDa), a less intense truncated HNE/A1AT complex (∼60 kDa) and an excess of A1AT inhibitor at ∼52 kDa (Fig. 4A–4B). Truncation of the 60 kDa HNE:A1AT complex was assessed based on the shift in its molecular weight relative to the intact 73 kDa HNE/A1AT complex as previously reported (103). In contrast, equimolar (1:1) A1AT-based inhibition of HNE formed less HNE:A1AT complex and this complex was predominantly truncated (∼60 kDa). In order to study the interaction between HNE and A1AT, an enzyme-to-inhibitor molar ratio of 1:3 was utilized and subjected to native LC-Q-TOF-MS as sufficient amount of the intact 73 kDa HNE:A1AT complex was formed.
Fig. 4.
Glycoform-glycoform enzyme-inhibitor complex formation of HNE and blood plasma-derived human A1AT. A, CMB staining of isolated HNE, isolated human A1AT and HNE:A1AT complexes and uncomplexed proteins after mixing the two glycoprotein binding partners in a 1:1 and 1:3 ratio. *,# denote a truncated HNE:A1AT complex (∼60 kDa) and a truncated uncomplexed (native) A1AT (∼52 kDa), respectively. ∧,∧∧ denote the intact (∼73 kDa) and the truncated form (∼60 kDa) of the HNE:A1AT complex, respectively. B, Immunoblot of HNE:A1AT complexes (1:3 ratio) using anti-HNE antibodies. ∧, ∧∧ as denoted in A. Purified HNE (positive control) and A1AT (negative control) were included. C, i) The deconvoluted ESI mass spectrum shows signals corresponding to various major glycoform-glycoform complexes of HNE (A-C):A1AT(α-γ) at 1:3 ratio (see zoom for magnification) and the coeluting uncomplexed A1AT as analyzed by high resolution native LC-Q-TOF-MS. For example, the signal at 72124.8 Da, labeled as A-β, corresponds to HNE glycoform A (two M2F N-glycans conjugated to HNE) complexed with the biantennary disialylated A1AT glycoform β. The degree of complex formation was assessed using signal intensities from the CMB gels (upper panel) and native MS (lower panel) as shown in the insert. ‡ denotes low abundance A1AT glycoforms carrying fucosylated N-glycans. ii) Glycoform distribution of the observed HNE:A1AT complexes, see inserted boxes for structures and nomenclature of major glycoforms of HNE and A1AT. Mean ± S.E., n = 3 technical replicates. See supplemental Fig. S9 for the complete list of identified HNE:A1AT glycoforms. iii) The glycoforms of the HNE:A1AT complex were compared with the glycoform distribution of native HNE and native A1AT to determine any glycoform-glycoform preferences in the enzyme:inhibitor complex formation (Mean ± S.E., n = 3 technical replicates; ***, p < 0.001 using unpaired two-tailed type 2 Student's t-tests).
Native high-resolution LC-Q-TOF-MS indicated that uncomplexed A1AT (as well as the native form) carried three major glycoforms (here called glycoform α, β and γ) (Fig. 4C i and supplemental Fig. S9). These three glycoforms comprised biantennary mono- and disialylated N-glycans and bi- and triantennary fully sialylated N-glycans in agreement with previously reported site-specific N-glycoprofiles of plasma-derived human A1AT (58, 104). The accurate intact protein mass analysis indicated that all A1AT glycoforms were cysteinylated (Δm = +119.0 Da) as previously reported (58), but otherwise not modified beyond the expected polypeptide chain trimming and the N-glycosylation. Few A1AT glycoforms carrying fucosylated N-glycans (labeled as ‡ in Fig. 4C i) were also observed as previously described (105). However, these structures were not considered further in this glycoanalysis because of their low abundance. Micro-heterogeneous features of the HNE:A1AT complex were observed (Fig. 4C i, insert). The agreement of CMB (Fig. 4A) and native MS-based quantitation of the uncomplexed-to-complexed A1AT ratio (both close to 2 as expected) indicated little, if any, MS-induced gas-phase dissociation of the formed HNE:A1AT complex. In total, nine major HNE:A1AT complex glycoforms were identified and quantified from combinations of already observed glycoforms of native A1AT (α, β, and γ) and HNE (A, B, and C) (Fig. 4C ii). By comparing the HNE:A1AT complex glycoform distribution to the N-glycosylation of native HNE and native A1AT (Fig. 4C iii and supplemental Table S6), it was observed that glycoform B of HNE (sialoglycoform i.e. FA1G1S1) and glycoform α of A1AT (i.e. undersialylated glycoform) were overrepresented in the HNE:A1AT complex (n = 3 technical replicates, p < 0.001). This indicates that sialylated HNE has a higher propensity for engaging in an inhibitor complex with an undersialylated form of A1AT. Although the molecular mechanisms underpinning these HNE:A1AT complex preferences remain unclear, it could be speculated that the negatively charged sialoform of HNE interact strongly with its own highly cationic protein surface and thereby causes less sterical hindrance upon A1AT interaction. Conversely, less sialylated A1AT may more readily bind to HNE because it is sterically less likely to cause interference, while being sufficiently anionic via its sialoglycans and the negatively charged protein surface interface at the exosite of A1AT to interact strongly with HNE. These anionic residues at the A1AT exosite have been shown previously to be important for protease binding (106). This potential glycoform-glycoform preference for protein complex formation is, to the best of our knowledge, the first example indicating that N-glycosylation of both protein partners influences their propensity to engage in a complex.
HNE-based cleavage of the A1AT loop is the initial step in the irreversible covalent binding and inhibition of the HNE activity by distorting its active site (107, 108). Oxidation of Met351 or Met358 on the reactive center loop of A1AT has been shown to be an alternative mechanism to reduce the ability of A1AT to inhibit HNE (109). A1AT was not oxidized in our experiments, but the highly oxidative environment at inflammatory sites, which contains large amount of reactive oxygen species secreted during the respiratory burst from activated neutrophils, may efficiently inactivate A1AT to preserve the HNE activity when it is degranulated (110). Restoration of the anti-protease activity to limit the local tissue damage by HNE after A1AT oxidative inactivation has also been reported (111), demonstrating a delicate homeostatic balance involving the inactivation and activation of anti-protease activity during inflammation, which can be easily deregulated in lung diseases such as CF (112). The glycosylation of A1AT was previously suggested to confer resistance toward degradation and extending its half-life in circulation (113). Our observations suggest that protein N-glycosylation in addition serves to modulate the A1AT-based inhibition of the proteolytical efficiency of HNE in intricate ways during innate immune processes.
HNE and Its N-glycans are Bacteriostatic Toward P. aeruginosa
HNE mediates protection against pathogens such as P. aeruginosa by degrading proteins of the outer bacterial membrane (23, 114). However, other potential protective functions mediated by HNE and the importance of HNE N-glycosylation have not been studied in the context of pathogen killing, in part, because of the lack of structural information of the exact N-glycosylation of HNE. We here also investigated whether the unusual N-glycans of HNE influence the growth of a common type of pathogenic bacteria found in inflamed airways.
The clinical P. aeruginosa strain (PASS1) isolated from a CF patient (61) was cultured with physiological relevant concentrations (low micromolar i.e. 1.8 μm and 3.6 μm) of enzymatically active HNE as well as with isolated HNE N-glycans. The enzyme activity of HNE was confirmed by rapid CBG cleavage using gel electrophoresis as shown previously (24). The bacterial growth profile was monitored over 12 h at 37 °C. As expected, enzymatically active HNE significantly inhibited the growth of PASS1 relative to the growth profile of PASS1 without HNE in a concentration-dependent manner (n = 3 technical replicates, p < 0.05) (supplemental Fig. S10A). Interestingly, free HNE N-glycans (3.6 μm) inhibited the bacterial growth comparable with the enzymatically active HNE (1.8 μm). We also observed that the paucimannose-rich nCG, another potent serine protease produced and used by human neutrophils for bacterial defense, and its released N-glycans displayed similar bacteriostatic effects on PASS1 within the same concentration range (supplemental Fig. S10B). Although the underlying mechanisms remain unknown, the apparent bacteriostatic effects at low concentrations of paucimannosidic and monosialylated free N-glycans is intriguing and has to the best of our knowledge not been reported previously. Further work is needed to pin-point which HNE N-glycan or N-glycan subsets display growth inhibitory effects toward P. aeruginosa.
CONCLUSIONS
This is the first report on the detailed structure and function of the N-glycosylation of HNE, a key serine protease in inflammation and bacterial infection. The deep site- and subcellular-specific glycoprofiling of HNE from resting neutrophils showed that HNE carries uniform but highly unusual site-specific N-glycosylation across multiple granular compartments and is undergoing spatiotemporal regulation upon neutrophil activation. The nonconventional paucimannosidic and monosialylated N-glycans abundantly decorating HNE showed intriguing immune-related functions by facilitating and/or modulating the binding of HNE to an immunological relevant mannose receptor, MBL, and to a key inhibitory anti-protease, A1AT, present within inflamed airways. These intriguing observations generate novel insights into the structure and functional relationship of a central player in innate immunity and advance our understanding of the refined molecular and cellular mechanisms and the importance of protein N-glycosylation in the inflamed microenvironment relevant for multiple immune diseases.
DATA AVAILABILITY
The LC-MS/MS data supporting the reported HNE N-glycopeptides and N-glycans including the observed precursor masses (m/z and charge states), annotated MS/MS spectra and LC retention times are available in the supplementary files. The LC-MS data supporting the native glycoprotein analyses and the HNE:MBL and HNE:A1AT binding assays, including the observed deconvoluted masses and their relative abundance are also available in the supplementary files. The LC-MS/MS proteomics data supporting the subcellular- and site-specific N-glycosylation analysis of HNE present across the neutrophil granule compartments have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD005559.
Supplementary Material
Acknowledgments
We thank Ms Charlotte Horne, Dr Simone Diestel, Dr Liisa Kautto, Dr Jodie Abrahams, Dr Zeynep Sumer-Bayraktar, and Mr Christopher Ashwood for providing valuable chemicals and reagents, and for expert technical assistance. Ms Serene Gwee is thanked for proof reading and fruitful discussions. Finally, we would like to sincerely thank Prof Niels Borregaard, University of Copenhagen, Denmark for providing outstanding scientific inspiration in neutrophil biology.
Footnotes
Author contributions: I.L. and M.T.-A. designed the research. I.L., N.H., N.P., and M.T-A. supplied reagents and access to instrumentation. I.L., O.∅. performed the experiments. I.L., O.∅. and M.T-A. analysed the data. I.L., O.∅., N.H, N.P., and M.T.-A. wrote the paper.
* I.L. was supported by an international Macquarie University Research Scholarship (iMQRES) and an Australian Cystic Fibrosis postgraduate studentship. M.T.-A. was supported by a fellowship from the Cancer Institute NSW, Australia, and a Macquarie University Research Development Grant (MQRDG).
 This article contains supplemental material.
This article contains supplemental material.
The authors declare no conflict of interest.
1 The abbreviations used are:
- HNE
- human neutrophil elastase
- A1AT
- α1-antitrypsin
- ACN
- acetonitrile
- CBG
- corticosteroid binding globulin
- CF
- cystic fibrosis
- CMB
- coomassie brilliant blue
- dHex
- deoxyhexose
- DTT
- dithiothreitol
- ETD
- electron transfer dissociation
- FA
- formic acid
- fMLP
- N-formylmethionine-leucyl-phenylalanine
- Fuc
- fucose
- GlcNAc
- N-acetylglucosamine
- HBSS
- Hank's Balanced Salt Solution
- Hex
- hexose
- HexNAc
- N-acetylhexosamine
- HILIC
- hydrophilic interaction liquid chromatography
- LPS
- lipopolysaccharide
- Man
- mannose
- MBL
- mannose binding lectin
- MPO
- myeloperoxidase
- mrCLR
- mannose-recognising C-type lectin receptor
- MWCO
- molecular weight cut off
- nCG
- neutrophil cathepsin G
- NET
- neutrophil extracellular trap
- NHS
- N-hydroxysuccinimide ester
- NeuAc
- N-acetylneuraminic acid
- OD
- optical density
- PDB
- Protein Data Bank
- PGC
- porous graphitised carbon
- PMA
- phorbol myristate acetate
- RP
- reversed phase
- SPE
- solid phase extraction
- TBST
- tris buffered saline with Tween 20
- TFA
- trifluoroacetic acid
- XIC
- extracted ion chromatogram.
REFERENCES
- 1. Borregaard N., Sorensen O. E., and Theilgaard-Monch K. (2007) Neutrophil granules: a library of innate immunity proteins. Trends Immunol. 28, 340–345 [DOI] [PubMed] [Google Scholar]
- 2. Rorvig S., Honore C., Larsson L. I., Ohlsson S., Pedersen C. C., Jacobsen L. C., Cowland J. B., Garred P., and Borregaard N. (2009) Ficolin-1 is present in a highly mobilizable subset of human neutrophil granules and associates with the cell surface after stimulation with fMLP. J. Leukoc. Biol. 86, 1439–1449 [DOI] [PubMed] [Google Scholar]
- 3. Rorvig S., Ostergaard O., Heegaard N. H., and Borregaard N. (2013) Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J. Leukoc. Biol. 94, 711–721 [DOI] [PubMed] [Google Scholar]
- 4. Owen C. A., Campbell M. A., Sannes P. L., Boukedes S. S., and Campbell E. J. (1995) Cell surface-bound elastase and cathepsin G on human neutrophils: a novel, non-oxidative mechanism by which neutrophils focus and preserve catalytic activity of serine proteinases. J. Cell Biol. 131, 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nauseef W. M., and Borregaard N. (2014) Neutrophils at work. Nat. Immunol. 15, 602–611 [DOI] [PubMed] [Google Scholar]
- 6. Reece S. T., Loddenkemper C., Askew D. J., Zedler U., Schommer-Leitner S., Stein M., Mir F. A., Dorhoi A., Mollenkopf H. J., Silverman G. A., and Kaufmann S. H. (2010) Serine protease activity contributes to control of Mycobacterium tuberculosis in hypoxic lung granulomas in mice. J. Clin. Invest. 120, 3365–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gombart A. F., and Koeffler H. P. (2002) Neutrophil specific granule deficiency and mutations in the gene encoding transcription factor C/EBP(epsilon). Curr. Opin. Hematol. 9, 36–42 [DOI] [PubMed] [Google Scholar]
- 8. Le Gars M., Descamps D., Roussel D., Saussereau E., Guillot L., Ruffin M., Tabary O., Hong S. S., Boulanger P., Paulais M., Malleret L., Belaaouaj A., Edelman A., Huerre M., Chignard M., and Sallenave J. M. (2013) Neutrophil elastase degrades cystic fibrosis transmembrane conductance regulator via calpains and disables channel function in vitro and in vivo. Am. J. Respir. Crit. Care Med. 187, 170–179 [DOI] [PubMed] [Google Scholar]
- 9. Tidwell T., Wechsler J., Nayak R. C., Trump L., Salipante S. J., Cheng J. C., Donadieu J., Glaubach T., Corey S. J., Grimes H. L., Lutzko C., Cancelas J. A., and Horwitz M. S. (2014) Neutropenia-associated ELANE mutations disrupting translation initiation produce novel neutrophil elastase isoforms. Blood 123, 562–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horwitz M. S., Duan Z., Korkmaz B., Lee H. H., Mealiffe M. E., and Salipante S. J. (2007) Neutrophil elastase in cyclic and severe congenital neutropenia. Blood 109, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nayak R. C., Trump L. R., Aronow B. J., Myers K., Mehta P., Kalfa T., Wellendorf A. M., Valencia C. A., Paddison P. J., Horwitz M. S., Grimes H. L., Lutzko C., and Cancelas J. A. (2015) Pathogenesis of ELANE-mutant severe neutropenia revealed by induced pluripotent stem cells. J. Clin. Invest. 125, 3103–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kettritz R. (2016) Neutral serine proteases of neutrophils. Immunol. Rev. 273, 232–248 [DOI] [PubMed] [Google Scholar]
- 13. Lindmark A., Persson A. M., and Olsson I. (1990) Biosynthesis and processing of cathepsin G and neutrophil elastase in the leukemic myeloid cell line U-937. Blood 76, 2374–2380 [PubMed] [Google Scholar]
- 14. Gullberg U., Lindmark A., Lindgren G., Persson A. M., Nilsson E., and Olsson I. (1995) Carboxyl-terminal prodomain-deleted human leukocyte elastase and cathepsin G Are efficiently targeted to granules and enzymatically activated in the rat basophilic/mast cell Line RBL. J. Biol. Chem. 270, 12912–12918 [DOI] [PubMed] [Google Scholar]
- 15. Benson K. F., Li F. Q., Person R. E., Albani D., Duan Z., Wechsler J., Meade-White K., Williams K., Acland G. M., Niemeyer G., Lothrop C. D., and Horwitz M. (2003) Mutations associated with neutropenia in dogs and humans disrupt intracellular transport of neutrophil elastase. Nat. Genet. 35, 90–96 [DOI] [PubMed] [Google Scholar]
- 16. Niemann C. U., Abrink M., Pejler G., Fischer R. L., Christensen E. I., Knight S. D., and Borregaard N. (2007) Neutrophil elastase depends on serglycin proteoglycan for localization in granules. Blood 109, 4478–4486 [DOI] [PubMed] [Google Scholar]
- 17. Cowland J. B., and Borregaard N. (2016) Granulopoiesis and granules of human neutrophils. Immunol. Rev. 273, 11–28 [DOI] [PubMed] [Google Scholar]
- 18. Sinha S., Watorek W., Karr S., Giles J., Bode W., and Travis J. (1987) Primary structure of human neutrophil elastase. Proc. Natl. Acad. Sci. U.S.A. 84, 2228–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korkmaz B., Horwitz M. S., Jenne D. E., and Gauthier F. (2010) Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol. Rev. 62, 726–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lussier B., and Wilson A. A. (2016) Alpha-1 Antitrypsin: The Protein. pp. 17–30, Humana Press, New York [Google Scholar]
- 21. Grigorieva D. V., Gorudko I. V., Sokolov A. V., Kostevich V. A., Vasilyev V. B., Cherenkevich S. N., and Panasenko O. M. (2016) Myeloperoxidase Stimulates Neutrophil Degranulation. Bull. Exp. Biol. Med. 161, 495–500 [DOI] [PubMed] [Google Scholar]
- 22. Clemmensen S. N., Jacobsen L. C., Rorvig S., Askaa B., Christenson K., Iversen M., Jorgensen M. H., Larsen M. T., van Deurs B., Ostergaard O., Heegaard N. H., Cowland J. B., and Borregaard N. (2011) Alpha-1-antitrypsin is produced by human neutrophil granulocytes and their precursors and liberated during granule exocytosis. Eur. J. Haematol. 86, 517–530 [DOI] [PubMed] [Google Scholar]
- 23. Hirche T. O., Benabid R., Deslee G., Gangloff S., Achilefu S., Guenounou M., Lebargy F., Hancock R. E., and Belaaouaj A. (2008) Neutrophil elastase mediates innate host protection against Pseudomonas aeruginosa. J. Immunol. 181, 4945–4954 [DOI] [PubMed] [Google Scholar]
- 24. Sumer-Bayraktar Z., Grant O. C., Venkatakrishnan V., Woods R. J., Packer N. H., and Thaysen-Andersen M. (2016) Asn347 glycosylation of corticosteroid-binding globulin fine-tunes the host Immune response by modulating proteolysis by pseudomonas aeruginosa and neutrophil elastase. J. Biol. Chem. 291, 17727–17742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sinden N. J., Baker M. J., Smith D. J., Kreft J. U., Dafforn T. R., and Stockley R. A. (2015) alpha-1-antitrypsin variants and the proteinase/antiproteinase imbalance in chronic obstructive pulmonary disease. Am. J. Physiol. Lung Cell Mol. Physiol. 308, L179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoenderdos K., and Condliffe A. (2013) The neutrophil in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 48, 531–539 [DOI] [PubMed] [Google Scholar]
- 27. Papayannopoulos V., Metzler K. D., Hakkim A., and Zychlinsky A. (2010) Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 191, 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Metzler K. D., Goosmann C., Lubojemska A., Zychlinsky A., and Papayannopoulos V. (2014) A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Reports 8, 883–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sorensen O. E., and Borregaard N. (2016) Neutrophil extracellular traps - the dark side of neutrophils. J. Clin. Invest. 126, 1612–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watorek W., Halbeek H., and Travis J. (1993) The isoforms of human neutrophil elastase and cathepsin G differ in their carbohydrate side chain structures. Bio Chem Hoppe Seyler 374, 385–393 [DOI] [PubMed] [Google Scholar]
- 31. Hansen G., Gielen-Haertwig H., Reinemer P., Schomburg D., Harrenga A., and Niefind K. (2011) Unexpected active-site flexibility in the structure of human neutrophil elastase in complex with a new dihydropyrimidone inhibitor. J. Mol. Biol. 409, 681–691 [DOI] [PubMed] [Google Scholar]
- 32. Kollner I., Sodeik B., Schreek S., Heyn H., von Neuhoff N., Germeshausen M., Zeidler C., Kruger M., Schlegelberger B., Welte K., and Beger C. (2006) Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood 108, 493–500 [DOI] [PubMed] [Google Scholar]
- 33. Kawata J., Yamaguchi R., Yamamoto T., Ishimaru Y., Sakamoto A., Aoki M., Kitano M., Umehashi M., Hirose E., and Yamaguchi Y. (2016) Human neutrophil elastase induce interleukin-10 expression in peripheral blood mononuclear cells through protein kinase C theta/delta and phospholipase pathways. Cell J. 17, 692–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chawla A., Alatrash G., Philips A. V., Qiao N., Sukhumalchandra P., Kerros C., Diaconu I., Gall V., Neal S., Peters H. L., Clise-Dwyer K., Molldrem J. J., and Mittendorf E. A. (2016) Neutrophil elastase enhances antigen presentation by upregulating human leukocyte antigen class I expression on tumor cells. Cancer Immunol. Immunother. 65, 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salazar V. A., Rubin J., Moussaoui M., Pulido D., Nogues M. V., Venge P., and Boix E. (2014) Protein post-translational modification in host defense: the antimicrobial mechanism of action of human eosinophil cationic protein native forms. FEBS J. 281, 5432–5446 [DOI] [PubMed] [Google Scholar]
- 36. Hulsmeier A. J., Tobler M., Burda P., and Hennet T. (2016) Glycosylation site occupancy in health, congenital disorder of glycosylation and fatty liver disease. Sci. Rep. 6, 33927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jensen P. H., Karlsson N. G., Kolarich D., and Packer N. H. (2012) Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 7, 1299–1310 [DOI] [PubMed] [Google Scholar]
- 38. Loke I., Packer N. H., and Thaysen-Andersen M. (2015) Complementary LC-MS/MS-based N-Glycan, N-Glycopeptide, and intact N-glycoprotein profiling reveals unconventional Asn71-glycosylation of human neutrophil cathepsin G. Biomolecules 5, 1832–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 40. Cooper C. A., Gasteiger E., and Packer N. H. (2001) GlycoMod–a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1, 340–349 [DOI] [PubMed] [Google Scholar]
- 41. Mysling S., Palmisano G., Hojrup P., and Thaysen-Andersen M. (2010) Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 82, 5598–5609 [DOI] [PubMed] [Google Scholar]
- 42. Parker B. L., Thaysen-Andersen M., Fazakerley D. J., Holliday M., Packer N. H., and James D. E. (2016) Terminal galactosylation and sialylation switching on membrane glycoproteins upon TNF-alpha-induced insulin resistance in adipocytes. Mol. Cell. Proteomics 15, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stavenhagen K., Hinneburg H., Thaysen-Andersen M., Hartmann L., Varon Silva D., Fuchser J., Kaspar S., Rapp E., Seeberger P. H., and Kolarich D. (2013) Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom. 48, 627–639 [DOI] [PubMed] [Google Scholar]
- 44. Sethi M. K., Kim H., Park C. K., Baker M. S., Paik Y. K., Packer N. H., Hancock W. S., Fanayan S., and Thaysen-Andersen M. (2015) In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology 25, 1064–1078 [DOI] [PubMed] [Google Scholar]
- 45. Everest-Dass A. V., Abrahams J. L., Kolarich D., Packer N. H., and Campbell M. P. (2013) Structural feature ions for distinguishing N- and O-linked glycan isomers by LC-ESI-IT MS/MS. J. Am. Soc. Mass Spectrom. 24, 895–906 [DOI] [PubMed] [Google Scholar]
- 46. Stadlmann J., Pabst M., Kolarich D., Kunert R., and Altmann F. (2008) Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics 8, 2858–2871 [DOI] [PubMed] [Google Scholar]
- 47. Winchester B. (2005) Lysosomal metabolism of glycoproteins. Glycobiology 15:1R–15R [DOI] [PubMed] [Google Scholar]
- 48. Aebi M. (2013) N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 [DOI] [PubMed] [Google Scholar]
- 49. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., Etzler M. E., Frank M., Vliegenthart J. F., Lutteke T., Perez S., Bolton E., Rudd P., Paulson J., Kanehisa M., Toukach P., Aoki-Kinoshita K. F., Dell A., Narimatsu H., York W., Taniguchi N., and Kornfeld S. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dalpathado D. S., and Desaire H. (2008) Glycopeptide analysis by mass spectrometry. Analyst 133, 731–738 [DOI] [PubMed] [Google Scholar]
- 51. Leymarie N., and Zaia J. (2012) Effective use of mass spectrometry for glycan and glycopeptide structural analysis. Anal. Chem. 84, 3040–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee L. Y., Moh E. S., Parker B. L., Bern M., Packer N. H., and Thaysen-Andersen M. (2016) Toward automated N-glycopeptide identification in glycoproteomics. J. Proteome Res. 15, 3904–3915 [DOI] [PubMed] [Google Scholar]
- 53. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., and Mesirov J. P. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 [DOI] [PubMed] [Google Scholar]
- 54. Hubber S. J., and Thornton J. M. (1993) NACCESS computer program. Department of Biochemistry and Molecular Biology, University College, London [Google Scholar]
- 55. Hormann K., Stukalov A., Muller A. C., Heinz L. X., Superti-Furga G., Colinge J., and Bennett K. L. (2016) A surface biotinylation strategy for reproducible plasma membrane protein purification and tracking of genetic and drug-induced alterations. J. Proteome Res. 15, 647–658 [DOI] [PubMed] [Google Scholar]
- 56. Zipser B., Bello-DeOcampo D., Diestel S., Tai M. H., and Schmitz B. (2012) Mannitou monoclonal antibody uniquely recognizes paucimannose, a marker for human cancer, stemness, and inflammation. J. Carbohydr. Chem. 31, 504–518 [Google Scholar]
- 57. Eaton S. L., Roche S. L., Llavero Hurtado M., Oldknow K. J., Farquharson C., Gillingwater T. H., and Wishart T. M. (2013) Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting. PLoS ONE 8, e72457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kolarich D., Weber A., Turecek P. L., Schwarz H. P., and Altmann F. (2006) Comprehensive glyco-proteomic analysis of human alpha1-antitrypsin and its charge isoforms. Proteomics 6, 3369–3380 [DOI] [PubMed] [Google Scholar]
- 59. Kolarich D., Turecek P. L., Weber A., Mitterer A., Graninger M., Matthiessen P., Nicolaes G. A., Altmann F., and Schwarz H. P. (2006) Biochemical, molecular characterization, and glycoproteomic analyses of alpha(1)-proteinase inhibitor products used for replacement therapy. Transfusion 46, 1959–1977 [DOI] [PubMed] [Google Scholar]
- 60. Thaysen-Andersen M., Venkatakrishnan V., Loke I., Laurini C., Diestel S., Parker B. L., and Packer N. H. (2015) Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J. Biol. Chem. 290, 8789–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Penesyan A., Kumar S. S., Kamath K., Shathili A. M., Venkatakrishnan V., Krisp C., Packer N. H., Molloy M. P., and Paulsen I. T. (2015) Genetically and phenotypically distinct pseudomonas aeruginosa cystic fibrosis isolates share a core proteomic signature. PLoS ONE 10, e0138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Venkatakrishnan V., Thaysen-Andersen M., Chen S. C., Nevalainen H., and Packer N. H. (2015) Cystic fibrosis and bacterial colonization define the sputum N-glycosylation phenotype. Glycobiology 25, 88–100 [DOI] [PubMed] [Google Scholar]
- 63. Olczak M., and Watorek W. (2002) Structural analysis of N-glycans from human neutrophil azurocidin. Biochem. Biophys. Res. Commun. 293, 213–219 [DOI] [PubMed] [Google Scholar]
- 64. Zoega M., Ravnsborg T., Hojrup P., Houen G., and Schou C. (2012) Proteinase 3 carries small unusual carbohydrates and associates with alpha-defensins. J. Proteomics 75, 1472–1485 [DOI] [PubMed] [Google Scholar]
- 65. Ishizuka A., Hashimto Y., Naka R., Kinoshita M., Kakehi K., Seino J., Funakoshi Y., Suzuki T., Kameyama A., and Narimatsu H. (2008) Accumulation of free complex-type N-glycans in MKN7 and MKN45 stomach cancer cells. Biochem. J. 413, 227–237 [DOI] [PubMed] [Google Scholar]
- 66. Seino J., Wang L., Harada Y., Huang C., Ishii K., Mizushima N., and Suzuki T. (2013) Basal autophagy is required for the efficient catabolism of sialyloligosaccharides. J. Biol. Chem. 288, 26898–26907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Korkmaz B., Moreau T., and Gauthier F. (2008) Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie 90, 227–242 [DOI] [PubMed] [Google Scholar]
- 68. Huang W., Yamamoto Y., Li Y., Dou D., Alliston K. R., Hanzlik R. P., Williams T. D., and Groutas W. C. (2008) X-ray snapshot of the mechanism of inactivation of human neutrophil elastase by 1,2,5-thiadiazolidin-3-one 1,1-dioxide derivatives. J. Med. Chem. 51, 2003–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bourgoin-Voillard S., Leymarie N., and Costello C. E. (2014) Top-down tandem mass spectrometry on RNase A and B using a Qh/FT-ICR hybrid mass spectrometer. Proteomics 14, 1174–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Twumasi D. Y., and Liener I. E. (1977) Proteases from purulent sputum. Purification and properties of the elastase and chymotrypsin-like enzymes. J. Biol. Chem. 252, 1917–1926 [PubMed] [Google Scholar]
- 71. Thaysen-Andersen M., and Packer N. H. (2012) Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology 22, 1440–1452 [DOI] [PubMed] [Google Scholar]
- 72. Mellquist J. L., Kasturi L., Spitalnik S. L., and Shakin-Eshleman S. H. (1998) The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37, 6833–6837 [DOI] [PubMed] [Google Scholar]
- 73. Lee L. Y., Lin C. H., Fanayan S., Packer N. H., and Thaysen-Andersen M. (2014) Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front. Immunol. 5, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Borregaard N., and Cowland J. B. (1997) Granules of the human neutrophilic polymorphonuclear leukocyte. Blood 89, 3503–3521 [PubMed] [Google Scholar]
- 75. Le Cabec V., Cowland J. B., Calafat J., and Borregaard N. (1996) Targeting of proteins to granule subsets is determined by timing and not by sorting: The specific granule protein NGAL is localized to azurophil granules when expressed in HL-60 cells. Proc. Natl. Acad. Sci. U.S.A. 93, 6454–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lominadze G., Powell D. W., Luerman G. C., Link A. J., Ward R. A., and McLeish K. R. (2005) Proteomic analysis of human neutrophil granules. Mol. Cell. Proteomics 4, 1503–1521 [DOI] [PubMed] [Google Scholar]
- 77. Campbell E. J., and Owen C. A. (2007) The sulfate groups of chondroitin sulfate- and heparan sulfate-containing proteoglycans in neutrophil plasma membranes are novel binding sites for human leukocyte elastase and cathepsin G. J. Biol. Chem. 282, 14645–14654 [DOI] [PubMed] [Google Scholar]
- 78. Korkmaz B., Attucci S., Juliano M. A., Kalupov T., Jourdan M. L., Juliano L., and Gauthier F. (2008) Measuring elastase, proteinase 3 and cathepsin G activities at the surface of human neutrophils with fluorescence resonance energy transfer substrates. Nat. Protoc. 3, 991–1000 [DOI] [PubMed] [Google Scholar]
- 79. Loke I., Kolarich D., Packer N. H., and Thaysen-Andersen M. (2016) Emerging roles of protein mannosylation in inflammation and infection. Mol. Aspects Med. 51, 31–55 [DOI] [PubMed] [Google Scholar]
- 80. Robajac D., Vanhooren V., Masnikosa R., Mikovic Z., Mandic V., Libert C., and Nedic O. (2016) Preeclampsia transforms membrane N-glycome in human placenta. Exp. Mol. Pathol. 100, 26–30 [DOI] [PubMed] [Google Scholar]
- 81. Lollike K., Lindau M., Calafat J., and Borregaard N. (2002) Compound exocytosis of granules in human neutrophils. J. Leukoc. Biol. 71, 973–980 [PubMed] [Google Scholar]
- 82. Reeves E. P., Lu H., Jacobs H. L., Messina C. G., Bolsover S., Gabella G., Potma E. O., Warley A., Roes J., and Segal A. W. (2002) Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416, 291–297 [DOI] [PubMed] [Google Scholar]
- 83. Potera R. M., Jensen M. J., Hilkin B. M., South G. K., Hook J. S., Gross E. A., and Moreland J. G. (2016) Neutrophil azurophilic granule exocytosis is primed by TNF-alpha and partially regulated by NADPH oxidase. Innate Immun. 8, 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sengelov H., Follin P., Kjeldsen L., Lollike K., Dahlgren C., and Borregaard N. (1995) Mobilization of granules and secretory vesicles during in vivo exudation of human neutrophils. J. Immunol. 154, 4157–4165 [PubMed] [Google Scholar]
- 85. Petrescu A. J., Petrescu S. M., Dwek R. A., and Wormald M. R. (1999) A statistical analysis of N- and O-glycan linkage conformations from crystallographic data. Glycobiology 9, 343–352 [DOI] [PubMed] [Google Scholar]
- 86. Hajjar E., Mihajlovic M., Witko-Sarsat V., Lazaridis T., and Reuter N. (2008) Computational prediction of the binding site of proteinase 3 to the plasma membrane. Proteins 71, 1655–1669 [DOI] [PubMed] [Google Scholar]
- 87. van Liempt E., Bank C. M., Mehta P., Garcia-Vallejo J. J., Kawar Z. S., Geyer R., Alvarez R. A., Cummings R. D., Kooyk Y., and van Die I. (2006) Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 580, 6123–6131 [DOI] [PubMed] [Google Scholar]
- 88. Lai J., Bernhard O. K., Turville S. G., Harman A. N., Wilkinson J., and Cunningham A. L. (2009) Oligomerization of the macrophage mannose receptor enhances gp120-mediated binding of HIV-1. J. Biol. Chem. 284, 11027–11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Coffelt S. B., Wellenstein M. D., and de Visser K. E. (2016) Neutrophils in cancer: neutral no more. Nat. Rev. Cancer 16, 431–446 [DOI] [PubMed] [Google Scholar]
- 90. Singel K. L., and Segal B. H. (2016) Neutrophils in the tumor microenvironment: trying to heal the wound that cannot heal. Immunol. Rev. 273, 329–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Moore K. L., Patel K. D., Bruehl R. E., Li F., Johnson D. A., Lichenstein H. S., Cummings R. D., Bainton D. F., and McEver R. P. (1995) P-selectin glycoprotein ligand-1 mediates rolling of human neutrophils on P-selectin. J. Cell Biol. 128, 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pham C. T. (2006) Neutrophil serine proteases: specific regulators of inflammation. Nat. Rev. Immunol. 6, 541–550 [DOI] [PubMed] [Google Scholar]
- 93. Ip W. K., Takahashi K., Ezekowitz R. A., and Stuart L. M. (2009) Mannose-binding lectin and innate immunity. Immunol. Rev. 230, 9–21 [DOI] [PubMed] [Google Scholar]
- 94. Arnold J. N., Wormald M. R., Suter D. M., Radcliffe C. M., Harvey D. J., Dwek R. A., Rudd P. M., and Sim R. B. (2005) Human serum IgM glycosylation: identification of glycoforms that can bind to mannan-binding lectin. J. Biol. Chem. 280, 29080–29087 [DOI] [PubMed] [Google Scholar]
- 95. Fidler K. J., Hilliard T. N., Bush A., Johnson M., Geddes D. M., Turner M. W., Alton E. W., Klein N. J., and Davies J. C. (2009) Mannose-binding lectin is present in the infected airway: a possible pulmonary defence mechanism. Thorax 64, 150–155 [DOI] [PubMed] [Google Scholar]
- 96. Butler G. S., Sim D., Tam E., Devine D., and Overall C. M. (2002) Mannose-binding lectin (MBL) mutants are susceptible to matrix metalloproteinase proteolysis: potential role in human MBL deficiency. J. Biol. Chem. 277, 17511–17519 [DOI] [PubMed] [Google Scholar]
- 97. Gabius H.-J., Kaltner H., Kopitz J., and André S. (2015) The glycobiology of the CD system: a dictionary for translating marker designations into glycan/lectin structure and function. Trends Biochem. Sci. 40, 360–376 [DOI] [PubMed] [Google Scholar]
- 98. Cohen M. (2015) Notable aspects of glycan-protein interactions. Biomolecules 5, 2056–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mazumder P., and Mukhopadhyay C. (2012) Conformations, dynamics and interactions of di-, tri- and pentamannoside with mannose binding lectin: a molecular dynamics study. Carbohydr. Res. 349, 59–72 [DOI] [PubMed] [Google Scholar]
- 100. Weis W. I., Drickamer K., and Hendrickson W. A. (1992) Structure of a c-type mannose-binding protein complexed with an oligosaccharide. Nature 360, 127–134 [DOI] [PubMed] [Google Scholar]
- 101. Garratt L. W., Sutanto E. N., Ling K. M., Looi K., Iosifidis T., Martinovich K. M., Shaw N. C., Buckley A. G., Kicic-Starcevich E., Lannigan F. J., Knight D. A., Stick S. M., and Kicic A. (2016) Alpha-1 antitrypsin mitigates the inhibition of airway epithelial cell repair by neutrophil elastase. Am. J. Respir. Cell Mol. Biol. 54, 341–349 [DOI] [PubMed] [Google Scholar]
- 102. Janciauskiene S., and Welte T. (2016) Well-known and less well-known functions of alpha-1 antitrypsin. Its role in chronic obstructive pulmonary disease and other disease developments. Ann Am Thorac Soc 13, S280–288 [DOI] [PubMed] [Google Scholar]
- 103. Ogushi F., Fells G. A., Hubbard R. C., Straus S. D., and Crystal R. G. (1987) Z-type alpha 1-antitrypsin is less competent than M1-type alpha 1-antitrypsin as an inhibitor of neutrophil elastase. J. Clin. Invest. 80, 1366–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. McCarthy C., Saldova R., Wormald M. R., Rudd P. M., McElvaney N. G., and Reeves E. P. (2014) The role and importance of glycosylation of acute phase proteins with focus on alpha-1 antitrypsin in acute and chronic inflammatory conditions. J. Proteome Res. 13, 3131–3143 [DOI] [PubMed] [Google Scholar]
- 105. McCarthy C., Saldova R., O'Brien M. E., Bergin D. A., Carroll T. P., Keenan J., Meleady P., Henry M., Clynes M., Rudd P. M., Reeves E. P., and McElvaney N. G. (2014) Increased outer arm and core fucose residues on the N-glycans of mutated alpha-1 antitrypsin protein from alpha-1 antitrypsin deficient individuals. J. Proteome Res. 13, 596–605 [DOI] [PubMed] [Google Scholar]
- 106. Rashid Q., Kapil C., Singh P., Kumari V., and Jairajpuri M. A. (2015) Understanding the specificity of serpin-protease complexes through interface analysis. J. Biomol. Struct. Dyn. 33, 1352–1362 [DOI] [PubMed] [Google Scholar]
- 107. Elliott P. R., Lomas D. A., Carrell R. W., and Abrahams J. P. (1996) Inhibitory conformation of the reactive loop of alpha 1-antitrypsin. Nat. Struct. Biol. 3, 676–681 [DOI] [PubMed] [Google Scholar]
- 108. Dementiev A., Dobo J., and Gettins P. G. (2006) Active site distortion is sufficient for proteinase inhibition by serpins: structure of the covalent complex of alpha1-proteinase inhibitor with porcine pancreatic elastase. J. Biol. Chem. 281, 3452–3457 [DOI] [PubMed] [Google Scholar]
- 109. Taggart C., Cervantes-Laurean D., Kim G., McElvaney N. G., Wehr N., Moss J., and Levine R. L. (2000) Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J. Biol. Chem. 275, 27258–27265 [DOI] [PubMed] [Google Scholar]
- 110. Luisetti M., and Travis J. (1996) Bioengineering: α1-proteinase inhibitor site-specific mutagenesis. Chest 110, 278S–283S [DOI] [PubMed] [Google Scholar]
- 111. Carp H., Janoff A., Abrams W., Weinbaum G., Drew R. T., Weissbach H., and Brot N. (1983) Human methionine sulfoxide-peptide reductase, an enzyme capable of reactivating oxidized alpha-1-proteinase inhibitor in vitro. Am. Rev. Respir. Dis. 127, 301–305 [DOI] [PubMed] [Google Scholar]
- 112. Laval J., Ralhan A., and Hartl D. (2016) Neutrophils in cystic fibrosis. Biol. Chem. 397, 485–496 [DOI] [PubMed] [Google Scholar]
- 113. Sarkar A., and Wintrode P. L. (2011) Effects of glycosylation on the stability and flexibility of a metastable protein: the human serpin alpha(1)-antitrypsin. Int J Mass Spectrom 302, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wasiluk K. R., Skubitz K. M., and Gray B. H. (1991) Comparison of granule proteins from human polymorphonuclear leukocytes which are bactericidal toward Pseudomonas aeruginosa. Infect. Immun. 59, 4193–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The LC-MS/MS data supporting the reported HNE N-glycopeptides and N-glycans including the observed precursor masses (m/z and charge states), annotated MS/MS spectra and LC retention times are available in the supplementary files. The LC-MS data supporting the native glycoprotein analyses and the HNE:MBL and HNE:A1AT binding assays, including the observed deconvoluted masses and their relative abundance are also available in the supplementary files. The LC-MS/MS proteomics data supporting the subcellular- and site-specific N-glycosylation analysis of HNE present across the neutrophil granule compartments have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD005559.