Abstract
Identification of interactors is a major goal in cell biology. Not only protein-protein but also protein-carbohydrate interactions are of high relevance for signal transduction in biological systems. Here, we aim to identify novel interacting binding partners for the β-galactoside-binding proteins galectin-1 (Gal-1) and galectin-3 (Gal-3) relevant in the context of the eye disease proliferative vitreoretinopathy (PVR). PVR is one of the most common failures after retinal detachment surgeries and is characterized by the migration, adhesion, and epithelial-to-mesenchymal transition of retinal pigment epithelial cells (RPE) and the subsequent formation of sub- and epiretinal fibrocellular membranes. Gal-1 and Gal-3 bind in a dose- and carbohydrate-dependent manner to mesenchymal RPE cells and inhibit cellular processes like attachment and spreading. Yet knowledge about glycan-dependent interactors of Gal-1 and Gal-3 on RPE cells is very limited, although this is a prerequisite for unraveling the influence of galectins on distinct cellular processes in RPE cells. We identify here 131 Gal-3 and 15 Gal-1 interactors by galectin pulldown experiments combined with quantitative proteomics. They mainly play a role in multiple binding processes and are mostly membrane proteins. We focused on two novel identified interactors of Gal-1 and Gal-3 in the context of PVR: the low-density lipoprotein receptor LRP1 and the platelet-derived growth factor receptor β PDGFRB. Addition of exogenous Gal-1 and Gal-3 induced cross-linking with LRP1/PDGFRB and integrin-β1 (ITGB1) on the cell surface of human RPE cells and induced ERK/MAPK and Akt signaling. Treatment with kifunensine, an inhibitor of complex-type N-glycosylation, weakened the binding of Gal-1 and Gal-3 to these interactors and prevented lattice formation. In conclusion, the identified specific glycoprotein ligands shed light into the highly specific binding of galectins to dedifferentiated RPE cells and the resulting prevention of PVR-associated cellular events.
Galectins are widely expressed across different species and organs and share homology in the amino acid sequence of their carbohydrate recognition domain (CRD)1 (1–4). They interact with their protein ligands by binding to β-galactoside-containing moieties on the glycosylated peptide backbones (1, 5, 6). Thereby they decipher the information stored in the glycan chains and can substantially influence many cellular functions, including attachment, spreading, migration, and proliferation (7, 8). Galectins can be subdivided into three classes based on their molecular structure: prototype, chimera type, and tandem-repeat type (3). Galectin-3 (Gal-3) is the only known chimera-type galectin of the human lectin family (1, 9, 10). It consists of a C-terminal domain to bind specific carbohydrate branches and an N-terminal domain, which enables Gal-3 to multimerize (1, 11). Via the C-terminal domain, Gal-3 can cross-link glycoproteins on the plasma membrane and thus induce cell-surface lattice formation (11). Galectin-1 (Gal-1) is assigned to the prototype galectins, consisting of one CRD (9). By hydrophobic interactions at the N-terminal amino acid residues, prototype galectins can self-associate their monomer subunits to form homodimers (4, 12, 13). These dimers can cross-link specific multivalent carbohydrates or glycoconjugates, which results in the formation of cross-linked lattices and the activation of several cellular pathways (4, 14–17).
Proliferative vitreoretinopathy (PVR) is a blinding disease frequently occurring as a complication after rhegmatogenous retinal detachment surgery (18–21). It is characterized by the formation of sub- and epiretinal fibrocellular membranes, which contract and lead to repetitive tractional retinal detachment (20, 22–24). Adhesion, migration, and EMT of retinal pigment epithelial cells (RPE) and retinal Müller glial cells (RMG) are the key cellular events in the onset of PVR (20, 23, 25). During EMT, RPE cells convert from epithelial into mesenchymal cells, lose their epithelial characteristics, and acquire migratory mesenchymal properties (26). Cultured human RPE cells are a well-accepted in vitro model system for early PVR. By cultivating RPE cells on plastic, they begin to dedifferentiate and to transform into a fibroblast-like phenotype (27). Gal-1 and Gal-3 are endogenously expressed in RPE cells. They are present in the cytosol and nucleus, but they are also secreted by a non-classical pathway to the cell surface (28). Extracellularly, Gal-1 and Gal-3 are involved in cell-matrix and cell-cell interactions (8). As shown in previous studies, exogenous Gal-1 and Gal-3 bind carbohydrate-dependent to mesenchymal RPE and inhibit attachment and spreading of these cells (29, 30). We also demonstrated that EMT of RPE cells leads to increased β-1,6-N-glycosylation on the cell surface resulting in increased binding capacity of Gal-3 (31). Most of the recent experimental therapeutic approaches attempted to control PVR development by anti-proliferative or anti-inflammatory agents or by inhibition of single growth factors and their signaling pathways (32–39). However, PVR is a multifactorial, mostly cell-driven process, which requires a multimodal concept (24). Identification of a pharmacological agent that is able to govern several cellular processes simultaneously is necessary to treat PVR. Therefore, Gal-1 and Gal-3 bear a high potential to counteract PVR-associated cellular events.
However, although the cell-surface proteins on RPE cells targeted by specific galectins are largely unknown, this in-depth knowledge is a prerequisite to unravel the possible influence of galectins on the signal transduction mechanisms associated with PVR processes. In many other cell types, several interactors for Gal-1 or Gal-3 have been identified; these include, among others, lysosomal membrane-associated glycoproteins (LAMPs)-1 and -2, neural cell adhesion molecule, cell adhesion molecule L1, CD43, CD45, CD71, mucin-1, and receptors for distinct growth factors like the epidermal growth factor (EGF), transforming growth factor β (TGF-β), or vascular endothelial growth factor (VEGF) (5, 40–50). Extracellular matrix (ECM) proteins like laminin, fibronectin, or vitronectin as well as members of the β1 integrin family are also known Gal-1 and Gal-3 interactors (5, 46, 49, 51–53). Integrins play a major role in cell-matrix interactions. As transmembrane proteins they are able to bind to the ECM by their extracellular part and induce several signal transduction cascades in the cell, e.g. remodeling of the cytoskeleton or proliferation (51, 54). Priglinger et al. (42) showed that Gal-3 induces clustering of CD147 and integrin-β1 (ITGB1) transmembrane glycoprotein receptors on the cell surface of RPE. However, the functional relevance of galectin binding on these different receptors is not explicitly analyzed in the context of PVR.
The purpose of this study was to identify novel and specific glycoprotein ligands for Gal-1 and Gal-3 on the surface of mesenchymal RPE cells by an affinity capture quantitative LC-MS/MS-based approach. From the 131 and 15 specific interactors identified for Gal-3 and Gal-1, respectively, we focused on two novel interactors for functional validation of the PVR-relevant cellular behavior: the low density lipoprotein receptor-related protein (LRP1) and the platelet-derived growth factor receptor β (PDGFRB). Addition of Gal-1 and Gal-3 induced clustering with the identified glycoprotein receptors LRP1 and PDGFRB together with ITGB1 on RPE cell surfaces, validating their potential to influence cellular effects. Relevance of glycosylation of these interactors for the functional galectin binding and the cross-linking activity was also analyzed.
EXPERIMENTAL PROCEDURES
Isolation of Human RPE Cells and Human RPE Cell Culture
Human donor cadaver eyes were received by the Eye Bank of the Department of Ophthalmology at the Linz General Hospital (Linz, Austria) or at the Ludwig-Maximilians-University (LMU) (Munich, Germany) and were processed within 24 h after death as described in Priglinger et al. (42) and Priglinger et al. (31). The securing processes of the human tissue were humane, complied with the Declaration of Helsinki, and were approved by the relatives. The ethics committees of the hospital of the LMU, Munich, and of the Land Oberoesterreich authorized the procedure of isolation of RPE cells from human cadaver eyes, which were enucleated by an ophthalmologist in accordance with the standard operating procedures of the institution. After removal of the cornea for cornea transplantation, the front segment of the eye and the vitreous body were removed. The inner part of the rest of the eye was filled with phosphate-buffered saline (PBS, Gibco), and the retina was aspirated. To get rid of the remaining retina and photoreceptors, the eye was refilled with pre-warmed 1 mm EDTA in PBS (37 °C), pH 7.4, and incubated for 15–20 min at room temperature. PBS, 1 mm EDTA was aspirated, and the eyecup was filled with dissociation buffer (3 mm l-cysteine, 1 μg/μl BSA in PBS, 1 mm EDTA) containing 45 μg of papain (Worthington) per 1 ml of dissociation buffer. After incubation for 23 min at 37 °C, the solution within the eye was gently agitated with a pipette to dispense as many RPE cells as possible. The loosened RPE cells were transferred in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal calf serum (FCS; Gibco) and centrifuged for 5 min at 930 rpm at room temperature. The resuspended RPE cells were cultivated in DMEM, 10% FCS at 37 °C and 5% CO2 with or without 10 μm kifunensine for up to 4 weeks (Sigma-Aldrich). Primary human RPE cells of passage 4–7 were used for experiments. For some experiments, the human ARPE-19 cell line (ATCC® CRL-2302TM) was used (55). ARPE-19 cells were cultivated under the same cell culture conditions like the primary RPE cells. For preparation of protein lysates, RPE cells were washed with ice-cold PBS, collected, and lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1%(w/v) SDS, 0.5% (w/v) sodium deoxycholate, 1% (v/v) Nonidet P-40, Complete 1×).
Expression, Purification, Biotinylation, and Activity Control of Human Galectin-1 and -3
Human Gal-1 and Gal-3 were cloned in the bacterial pETM-11 expression vector as described previously (31, 42). The single modification was that His6-tagged human galectin-1 was dialyzed against PBS containing 5 mm β-mercaptoethanol at the end of the purification process. For biotinylation, 150 mm β-lactose was added to 1 mg of purified Gal-1 and Gal-3, and the proteins were dialyzed for 2 h at 4 °C against 0.1 m sodium hydrogen carbonate with 50 mm β-lactose, pH 9.2, followed by a 1-h biotinylation at room temperature with 100 μg of biotinamidohexanoic acid N-hydroxysuccinimide ester according to the manufacturer's instructions (Sigma-Aldrich). The biotinylated galectins were dialyzed overnight at 4 °C against PBS. Activity of biotinylated galectins was determined semi-quantitatively by hemagglutination assays, adapted from Nowak et al. (56) and St-Pierre et al. (57). Briefly, for preparation of type O red blood cells (RBC), 6 ml of whole blood samples were collected in EDTA tubes and centrifuged at 3500 rpm for 5 min. After removal of the transparent layer, RBCs were washed three times in PBS, diluted 1:10 in PBS, 3% glutaraldehyde, and rotated for 1 h at room temperature. RBCs were washed five times in PBS, 0.0025% NaN3 and resuspended at 4% in PBS, 0.0025 NaN3 to preserve it for 3 months. For hemagglutination assay, serial dilutions of galectins were put in U-shaped 96-well plates, and 10 μl of RBCs were added per well. After incubation at 37 °C for 30 min, the minimum active concentration of each galectin, which prevented sedimentation of the RBCs, was evaluated visually. For further experiments, Gal-1 and Gal-3 were used in sufficiently active concentrations, as specified respectively.
Galectin Pulldown Experiments
For galectin pulldown experiments, 1 mg of Gal-1 or 1 mg of Gal-3 were coupled on 300 mg of cyanogen bromide-activated Sepharose 4B (GE Healthcare). First, the Sepharose beads were activated by washing 15 times with 1 mm HCl, followed by a single washing step in coupling buffer (0.1 m NaHCO3, 0.5 m NaCl, pH 8.3). All washing steps included a centrifugation at 3000 rpm for 1 min. Galectins were mixed 1:1 with coupling solution and were incubated on a rotating wheel with the activated beads at 4 °C overnight. After washing with a 5-fold volume of coupling buffer, unreacted binding sites were blocked with 1 m ethanolamine, pH 8, for 2 h at room temperature. Several subsequent washing steps followed at different pH values (3× coupling solution, 3× acetate buffer (0.1 m NaAc, 0.5 m NaCl, pH 3–4), 3× coupling buffer, 3× acetate buffer, 3× PBS). The galectin beads were stable for several months when stored in 20% ethanol. To test whether the respective galectin, coupled to the beads, was still active, 100 μl of bead slurry (50% beads) was incubated with 200 μg of asialofetuin (Sigma) for 1 h at room temperature in the absence and presence of 0.1 m β-lactose. After washing with PBS, bound proteins were eluted by incubation with 3× Laemmli buffer at 95 °C for 5 min. Eluates were separated by SDS-PAGE (10% gels), and asialofetuin was detected by Coomassie staining (supplemental Fig. 1). For galectin pulldowns, 250 μg of total proteins from human mesenchymal RPE cell lysates were incubated with 20 μl of galectin/bead slurry or Protein G control bead slurry (GE Healthcare) in PBS for 1 h at 37 °C, with gentle mixing every 10 min. After centrifugation, beads were washed four times with PBS followed by elution of bound proteins with 0.5 m β-lactose (Sigma). Eluates were analyzed by label-free quantitative LC-MS/MS. Five independent Gal-3 and five independent Gal-1 as well as the corresponding control Protein G pulldown experiments with 3 or 4 technical replicates each were performed using RPE cell lysates derived from nine different human donors.
Sample Preparation for Mass Spectrometry
β-Lactose eluates of the Gal-1, Gal-3, and Protein G pulldown experiments were proteolyzed with Lys-C and trypsin (Promega) using a modified filter aided sample preparation protocol (58). Briefly, protein eluates were diluted with ammonium bicarbonate buffer (ABC, Sigma) to a final volume of 100 μl, followed by reduction using 1 μl of 1 m dithiothreitol (DTT) for 30 min at 60 °C. After cooling down to room temperature, 100 μl of 8 m Urea Buffer (Sigma), pH 8.5, was added, and proteins were alkylated with 10 μl of 300 mm iodoacetamide (Merck) for 30 min at room temperature in the dark. 2 μl of 1 m DTT was added to quench unreacted iodoacetamide, and protein eluates were transferred to 30-kDa cutoff centrifuge filter (Pall Corp., New York). After washing three times with 400 μl of Urea Buffer and twice with 100 μl of 50 mm ABC, the proteins were subjected to a 2-h digest at room temperature with 1 μg of Lys-C followed by tryptic digest (2 μg of trypsin) overnight at 37 °C. Peptides were collected by centrifugation through the filter and acidified with trifluoroacetic acid (TFA), pH 2.
Mass Spectrometry
LC-MS/MS analysis was performed as described previously on an LTQ OrbitrapXL (Thermo Fisher Scientific Inc.) (59–62). Briefly, every sample was automatically loaded onto an Ultimate3000 nano-HPLC system (Dionex) with a nano-trap column (300 μm inner diameter × 5 mm, packed with Acclaim PepMap100 C18, 5 μm, 100 Å; LC Packings) at a flow rate of 30 μl/min in HPLC buffer containing 0.1% TFA for 5 min. Peptides were separated on a reversed phase chromatography (PepMap, 25 cm, 75 μm inner diameter, 1 μm/100 Å pore size, LC Packings) over 80 or 140 min at a flow rate of 300 nl/min using increasing acetonitrile concentrations in 0.1% formic acid. Maximal injection time for MS spectra was 100 ms and for MS/MS spectra 500 ms. MS data were acquired using from the high resolution MS prescan the 10 most abundant peptide ions for fragmentation in the linear ion trap, if they were at least doubly charged and have an intensity of at least 200 counts, with a dynamic exclusion of 45 s and an isolation width of 2 atomic mass units. MS spectra were recorded within a mass range from 300 to 1500 m/z at a resolution of 60,000 full widths at half-maximum. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (63) partner repository with the dataset identifier PXD005461 and 10.6019/PXD005461.
Protein Identification and Label-free Quantification
The acquired spectra of the different samples were analyzed using Progenesis QI software for proteomics (Version 2.0, Nonlinear Dynamics, Waters) for precursor intensity-based label-free quantification, as described previously (59–62). Peak lists with respective m/z values, intensities, abundances, and m/z width were generated out of the profile data of the MS scans. MS/MS spectra were also transformed and stored in peak lists incorporating m/z and abundance. The retention times of all samples were aligned to the most complex sample as reference. Alignment to a maximal overlay of all 2D features was done both automatically and manually. Only features with charges of at least two and not more than seven were included in further analyses. After alignment and feature exclusion, samples were divided into their respective experimental groups (Gal-1, Gal-3, Protein G lactose eluates), and raw abundances of all features were normalized. Peptide identification was done with Mascot (MatrixScience; version 2.5.1) using the Ensembl human protein database (Homo sapiens, release: 75, 105,287 sequences; release: 80, 100,208 sequences). The parameters for the search were cleavage with trypsin, 0.6-Da fragment mass tolerance, and 10 ppm peptide mass tolerance. Carbamidomethylation was set as fixed modification, and methionine oxidation and deamidation of asparagine and glutamine as variable modifications. One missed cleavage was allowed. A Mascot-integrated decoy database search calculated an average false discovery of <1.25% when searches were performed with a mascot percolator score cutoff of 15 and an appropriate significance threshold p value. Peptide assignments were reimported into Progenesis QI, and all normalized abundances of unique peptides of an identified protein were summed to calculate the total cumulative normalized abundance of the respective protein. These abundances were used to calculate the enrichment factors of the quantified proteins in the lactose eluates of the galectin pulldowns compared with Protein G pulldowns. No minimal thresholds were set neither for the method of peak picking nor selection of data to use for quantification.
Experimental Design and Statistical Rationale
Five independent galectin-1 and galectin-3 as well as the corresponding control Protein G pulldown experiments with nine different biological samples were performed, each including 3 or 4 technical replicates. Statistical analysis was done using normalized abundances of all identified proteins using only unique peptides. For calculation of the enrichment factors of proteins in the lactose eluates of galectin pulldowns compared with unspecific controls, the mean of all technical replicates within one experiment was used. Normal distribution was assumed, and significance was determined by Student's t test. Proteins with p values <0.05, enrichment of ≥2-fold in galectin pulldown eluates (FC ≥2), which were identified in two or more independent experiments, were regarded as potential Gal-1 or Gal-3 interactors.
Volcano Plots, Cellular Component, and Network Analysis
Volcano plot representations of Gal-1 and Gal-3 pulldown experiments were done with Excel. The x axis shows the log2 transformed ratio between normalized abundances of proteins identified in lactose eluates of galectin pulldown compared with unspecific control (Protein G pulldown). The y axis gives the negative log10 transformed p value from the t test of the same two comparisons. p values <0.05 and FC ≥2 were regarded as significant. Infinite fold changes were set to the highest measured ratio plus 1, and fold changes with a value of 0 were equalized with the lowest measured ratio. Cellular component analysis of the 15 identified Gal-1 interactors and the 131 Gal-3 interactors was performed based on the gene ontology (GO) annotation “cellular component” in FunRich (64). For protein network generation, the corresponding gene names of the Gal-1 and Gal-3 interactors were uploaded in the Cytoscape App ClueGO-CluePedia (65) and clustered by the GO term “molecular function” using H. sapiens as the organism for background list. The size of the nodes represents the enrichment significance of the respective terms based on the predefined κ score threshold of 0.4. The ClueGO network reflects the relationship between the respective functional groups represented by their most significant (leading) term. With the identified Gal-1 and Gal-3 interactors, Phobius analysis was done to predict, based on the amino acid sequence of a protein, whether the respective protein has a transmembrane domain and signal peptides (66).
Immunocytochemical Staining
Mesenchymal human RPE cells were cultivated on glass coverslips (VWR Scientific) upon 60–70% confluence in DMEM + 10% FCS with or without 10 μm kifunensine (Sigma-Aldrich) up to passage 4–7. Before fixation with 4% paraformaldehyde, cells were starved for 2 h in serum-free DMEM and treated with biotinylated Gal-1 and Gal-3 with a final concentration of 120 μg/ml Gal-1 or 60 μg/ml Gal-3 for 30 min at 37 °C. As control, no galectin was added. After blocking (Tris-buffered saline with 0.1% Tween 20 (TBS-T) + 1% bovine serum albumin (BSA) + 0.5% goat serum) for 45 min at room temperature, galectin binding was visualized by incubation with streptavidin-AlexaFluor488 (1:500, Thermo Fisher Scientific) for 1 h at room temperature. Coverslips were incubated overnight at 4 °C in rabbit anti-LRP1 (1:50, Abcam), rabbit anti-PDGFRB (1:50, Abcam), or rat anti-ITGB1 (1:60, Developmental Studies Hybridoma Bank (DSHB)) diluted in TBS-T, followed by incubation with goat anti-rabbit AlexaFluor647 or goat anti-rat AlexaFluor568 (1:1000, Dianova) for 1 h at room temperature. Between each staining step at least one washing step was executed with TBS-T for 10 min. After counterstaining with Hoechst (1:5000, Thermo Fisher Scientific) for 8 min at room temperature, coverslips were mounted and photographed on a Leica DMi8 microscope with HCX PL APO ×63/1.20 objective lens. Filter cubes for GFP, Y5, Texas Red, and DAPI detections were used (JH Technologies). All images were captured using a Leica DFC365 FX camera, and constant settings for gain and exposure time were maintained for all samples within an experimental setup. Images were processed by the Leica Application Suite LASX (version 2.0, Leica) and the deconvolution software Huygens Essential using the classic maximum likelihood estimation algorithm with a signal to noise ratio of 40 and 50 iterations (version 16.05, Scientific Volume Imaging B.V., http://svi.nl). As control, cells were stained under equal conditions without primary antibodies and only with secondary antibodies. No unspecific labeling was observed (data not shown). Immunocytochemical staining of human RPE cells was repeated at least three times with cells from three different donors.
Western Blot Analysis
Eluates of the Gal-1 and Gal-3 pulldown experiments with lysates of mesenchymal RPE cells from three different donors, treated or untreated with 10 μm kifunensine for up to 4 weeks, were separated by SDS-PAGE (10% gels) and blotted onto PVDF membranes. Ten micrograms of the whole-cell extracts of the respective cell lysates were used as an input control for the pulldown experiments. After blocking with 5% non-fat dried milk in TBS-T for 1 h at room temperature, blots were incubated with antibodies against rabbit anti-LRP1 (1:20,000, Abcam), rabbit anti-PDGFRB (1:1000, Abcam), or mouse anti-GAPDH (1:10,000, Millipore) at 4 °C overnight. For analysis of phosphorylation changes of ERK after galectin treatment, 15 μg of whole-cell extracts of ARPE-19 cells untreated or treated with 120 μg/ml Gal-1 or 60 μg/ml Gal-3 for 1, 15, or 30 min were separated by SDS-PAGE (10% gels) and blotted onto PVDF membranes. Blocking was performed with 3% BSA in TBS-T for 1 h, and blots were incubated with antibodies against rabbit anti-phospho-ERK p44/p42 (1:2000, Cell Signaling, catalog no. 4370) and mouse anti-GAPDH (1:10,000, Millipore) at 4 °C overnight. After washing three times with TBS-T, blots were incubated with the appropriate HRP-coupled secondary antibodies (1:7500, Jackson ImmunoResearch) for 1 h at room temperature, and binding was visualized by signal development with ECL Plus enhanced chemiluminescence kit (GE Healthcare). Galectin pulldown experiments following Western blot analysis were repeated at least three times on three different RPE lysates.
Analysis of Phosphorylation Profiles
For the simultaneous analysis of relative changes in site-specific phosphorylation profiles of 43 kinases and two related total proteins after galectin treatment, the Human Phospho-Kinase Array (R&D Systems, ARY003B) was used. Selected phospho-specific capture antibodies were spotted in duplicate on nitrocellulose membranes. According to the manufacturer's instructions (R&D Systems), 300 μg of whole-cell extracts of ARPE-19 cells untreated or treated for 15 min with 120 μg/ml Gal-1 or 60 μg/ml Gal-3 were incubated with the nitrocellulose membranes overnight at 4 °C. After subsequent incubation with a mixture of biotinylated detection antibodies and streptavidin-horseradish peroxidase, the amount of phosphorylated protein bound at the respective capture spot was visualized by signal development with chemiluminescent detection reagents. Pixel density of each spot was determined with ImageJ considering background subtraction (67). Mean pixel density, standard deviation, and statistical significance (Student's t test) were determined. p values lower than 0.05 were considered as significant (*) and p values lower than 0.01 as highly significant (**).
RESULTS
Galectin-3 Revealed More Interacting Binding Partners than Galectin-1
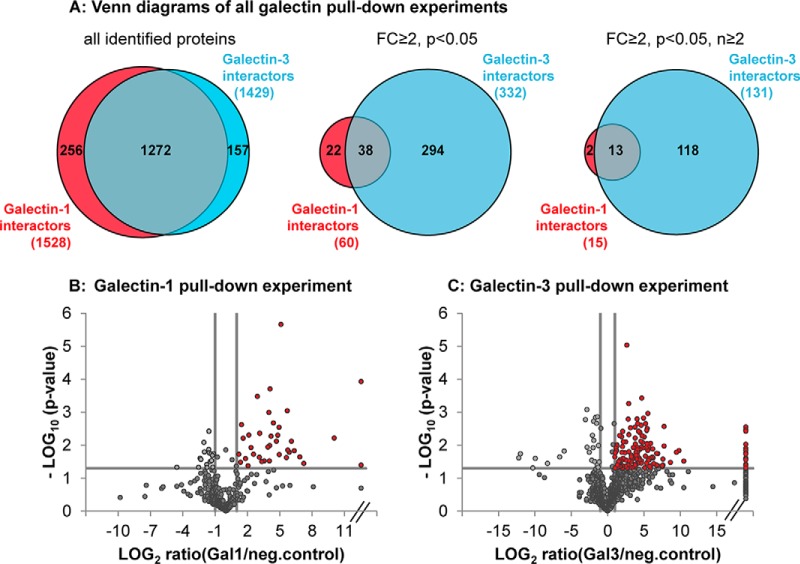
To identify Gal-1- and Gal-3-specific binding proteins in dedifferentiated mesenchymal human RPE cells, the respective galectin was coupled to cyanogen bromide-activated Sepharose beads and incubated with RPE cell lysates. Interacting proteins were eluted by β-lactose as competitor for specific carbohydrate binding. To exclude unspecific binders, the same pulldown experiments were performed with Protein G-coupled Sepharose beads. A subsequent proteomic screening of these lactose eluates revealed identification of 1429 different proteins in all five independent Gal-3 pulldown experiments (with 3 or 4 technical replicates each, supplemental data S1 and S2) and 1528 different proteins in five independent Gal-1 pulldown experiments (3 or 4 technical replicates per experiment, supplemental data S3 and S4, and Fig. 1A, left) with an overlay of 1272 proteins. To identify the most promising Gal-1 and Gal-3 interactors, those proteins that were significantly (p < 0.05) at least 2-fold (fold-change (FC) ≥2) enriched in Gal-1 or Gal-3 eluates, compared with the unspecific control, were selected. 771 proteins in Gal-3 pulldown experiments were specifically enriched in the Gal-3 lactose eluates (FC ≥2), and 332 of these proteins reached significance (FC ≥2, p < 0.05) (Fig. 1A, middle). In contrast to that, although 698 proteins were over 2-fold enriched in Gal-1 eluates (FC ≥2), only 60 proteins could be determined as significantly bound to Gal-1 (FC ≥2, p < 0.05) (Fig. 1A, middle). Consequently, Gal-3 revealed more significant interacting binding partners than Gal-1, which is represented in the volcano plots of one exemplary Gal-1 (Fig. 1B) and one exemplary Gal-3 (Fig. 1C) pulldown experiment. Those proteins that could be detected in two or more independent experiments (FC ≥2, p < 0.05, n ≥2) were determined as the most significant potential interactors of Gal-1 and Gal-3 (Fig. 1A, right). 131 Gal-3 interactors and 15 Gal-1 interactors fulfilling these strict criteria were used for further analysis and validation (Table I and Table II).
Fig. 1.
Galectin-3 revealed more significant interacting binding partners than galectin-1. A, numbers of protein identifications in all Gal-1 (red) and Gal-3 (blue) pulldown experiments (left Venn diagram); numbers of protein identifications of all significantly enriched (p < 0.05) proteins with an enrichment factor over or equal to 2 in the galectin β-lactose eluates compared with negative control (FC ≥2) (central Venn diagram) and of all significantly enriched proteins (p < 0.05, FC ≥2) that could be detected in two or more independent pulldown experiments (right Venn diagram). Numbers of overlapping protein identifications are also represented (gray). B and C, volcano plot representation of one exemplary galectin-1 (B) and one exemplary galectin-3 (C) pulldown experiment (with three replicates each). The log2 transformed ratios between normalized abundances of all proteins identified in lactose eluates of galectin pulldown compared with unspecific control (Protein-G pulldown) is plotted against the respective negative log10 transformed p values of the t test. p values of p < 0.05 and additional regulation of ≥2-fold were regarded as significant (red dots). Infinite fold changes were set to the highest measured ratio plus 1 (dots on the right side of the plot); fold changes with a value of 0 were equalized with the lowest measured ratio (dots on the left site of the plot).
Table I. List of RPE cell proteins with high affinity to galectin-1.
15 identified Gal-1 interactors in order of their enrichment factors in the β-Lactose eluates of galectin pulldowns compared with unspecific controls. For calculation the mean of all technical replicates within one experiment was used. Significance was determined by Student's t test. Proteins with p values of p < 0.05, enrichment of ≥2-fold in galectin pulldown eluates (FC ≥ 2), and additional identification in two or more independent pulldown experiments (n ≥ 2) were regarded as significant. The ratios represent the maximum fold changes of all experiments and the corresponding p values. TMD, transmembrane domain; SP, signal peptide; FC, fold change.
| 1st accession no. | Gene name | Ratio Gal-1/negative control | p value | TMD | SP | Protein name |
|---|---|---|---|---|---|---|
| ENSP00000333298 | LAMP1 | 1065.8 | 0.006 | 1 | Y | Lysosome-associated membrane protein 1 |
| ENSP00000307513 | MRC2 | 136.4 | 0.049 | 1 | 0 | Mannose receptor, C-type 2 |
| ENSP00000007722 | ITGA3 | 104.6 | 0.021 | 1 | Y | Integrin, α3 |
| ENSP00000261023 | ITGAV | 59.2 | 0.016 | 1 | Y | Integrin, αV |
| ENSP00000303351 | ITGB1 | 53.9 | 0.014 | 1 | Y | Integrin, β1 |
| ENSP00000233714 | LANCL1 | 52.2 | 0.001 | 0 | 0 | LanC l antibiotic synthetase component C-like 1 (bacterial) |
| ENSP00000314508 | GBA | 34.8 | 0.000 | 0 | Y | Glucosidase, β, acid |
| ENSP00000331544 | FBLN1 | 34.1 | 0.018 | 0 | Y | Fibulin 1 |
| ENSP00000243077 | LRP1 | 31.1 | 0.019 | 1 | Y | Low-density lipoprotein receptor-related protein 1 |
| ENSP00000258341 | LAMC1 | 21.3 | 0.002 | 0 | Y | Laminin, γ1 (formerly LAMB2) |
| ENSP00000262776 | LGALS3BP | 19.0 | 0.043 | 0 | Y | Lectin, galactoside-binding, soluble, three binding proteins |
| ENSP00000228506 | MLEC | 18.1 | 0.037 | 1 | Y | Malectin |
| ENSP00000265304 | SSBP1 | 6.1 | 0.045 | 0 | 0 | Single-stranded DNA- binding protein 1, mitochondrial |
| ENSP00000200639 | LAMP2 | 5.9 | 0.001 | 1 | Y | Lysosome-associated membrane protein 2 |
| ENSP00000258733 | GPNMB | 5.3 | 0.049 | 1 | Y | Glycoprotein (transmembrane) nmb |
Table II. List of RPE cell proteins with high affinity to galectin-3.
131 identified Gal-3 interactors in order of their enrichment factors in the lactose eluates of galectin pulldowns compared with unspecific controls. For calculation, the mean of all technical replicates within one experiment was used. Significance was determined by Student's t test. Proteins with p values of p < 0.05, enrichment of ≥2-fold in galectin pulldown eluates (FC ≥ 2), and additional identification in two or more independent pulldown experiments (n ≥) were regarded as significant. The ratios represent the maximum fold changes of all experiments and the corresponding p values. TMD, transmembrane domain; SP, signal peptide; FC, fold change.
| 1st accession no. | Gene name | Ratio Gal3/neg. control | p value | TMD | SP | Protein name |
|---|---|---|---|---|---|---|
| ENSP00000273784 | AHSG | Infinity | 0.016 | 0 | Y | α2-HS-glycoprotein |
| ENSP00000222374 | CADM4 | Infinity | 0.047 | 1 | Y | Cell adhesion molecule 4 |
| ENSP00000312435 | DAG1 | Infinity | 0.004 | 1 | Y | Dystroglycan 1 (dystrophin-associated glycoprotein 1) |
| ENSP00000321573 | DCBLD2 | Infinity | 0.007 | 3 | Y | Discoidin, CUB, and LCCL domain containing 2 |
| ENSP00000334145 | F3 | Infinity | 0.003 | 1 | 0 | Coagulation factor III (thromboplastin, tissue factor) |
| ENSP00000314508 | GBA | Infinity | 0.014 | 0 | Y | Glucosidase, β, acid |
| ENSP00000282588 | ITGA1 | Infinity | 0.002 | 1 | Y | Integrin, α1 |
| ENSP00000264106 | ITGA6 | Infinity | 0.015 | 1 | Y | Integrin, α6 |
| ENSP00000266041 | ITIH4 | Infinity | 0.014 | 0 | Y | Inter-α-trypsin inhibitor heavy chain family, member 4 |
| ENSP00000258341 | LAMC1 | Infinity | 0.016 | 0 | Y | Laminin, γ1 (formerly LAMB2) |
| ENSP00000231004 | LOX | Infinity | 0.046 | 0 | Y | Lysyl oxidase |
| ENSP00000374135 | LRP1B | Infinity | 0.029 | 1 | Y | Low density lipoprotein receptor-related protein 1B |
| ENSP00000199940 | MAP2 | Infinity | 0.035 | 0 | 0 | Microtubule-associated protein 2 |
| ENSP00000217939 | MXRA5 | Infinity | 0.012 | 0 | Y | Matrix-remodeling associated 5 |
| ENSP00000294785 | NCSTN | Infinity | 0.009 | 1 | Y | Nicastrin |
| ENSP00000324270 | OXTR | Infinity | 0.041 | 7 | 0 | Oxytocin receptor |
| ENSP00000319782 | PODXL | Infinity | 0.020 | 1 | Y | Podocalyxin-like |
| ENSP00000356572 | QSOX1 | Infinity | 0.007 | 0 | Y | Quiescin Q6 sulfhydryl oxidase 1 |
| ENSP00000266771 | SLC15A4 | Infinity | 0.014 | 13 | 0 | Solute carrier family 15 (oligopeptide transporter), member 4 |
| ENSP00000444408 | SLC1A5 | Infinity | 0.017 | 9 | 0 | Solute carrier family 1 (neutral amino acid transporter), member 5 |
| ENSP00000004531 | SLC7A2 | Infinity | 0.042 | 14 | 0 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| ENSP00000335300 | TPCN1 | Infinity | 0.002 | 12 | 0 | Two-pore segment channel 1 |
| ENSP00000351190 | ITIH2 | 527,961.6 | 0.006 | 0 | Y | Inter-α-trypsin inhibitor heavy chain 2 |
| ENSP00000418725 | ITGB1 | 20,223.9 | 0.011 | 0 | Y | Integrin, β1 (fibronectin receptor, β-polypeptide, antigen CD29 includes MDF2, MSK12) |
| ENSP00000269141 | CDH2 | 4103.6 | 0.003 | 1 | Y | Cadherin 2, type 1, N-cadherin (neuronal) |
| ENSP00000257857 | CD63 | 3186.1 | 0.032 | 4 | 0 | CD63 molecule |
| ENSP00000329797 | CADM1 | 2304.6 | 0.003 | 1 | Y | Cell adhesion molecule 1 |
| ENSP00000333298 | LAMP1 | 1591.0 | 0.003 | 1 | Y | Lysosome-associated membrane protein 1 |
| ENSP00000231368 | LNPEP | 1439.2 | 0.031 | 1 | 0 | Leucyl/cystinyl aminopeptidase |
| ENSP00000257879 | ITGA7 | 1433.6 | 0.000 | 1 | 0 | Integrin, α7 |
| ENSP00000256689 | SLC38A2 | 910.1 | 0.015 | 11 | 0 | Solute carrier family 38, member 2 |
| ENSP00000268613 | CDH13 | 900.6 | 0.008 | 0 | 0 | Cadherin 13 |
| ENSP00000368752 | PRNP | 737.3 | 0.024 | 2 | Y | Prion protein |
| ENSP00000263398 | CD44 | 581.2 | 0.017 | 1 | Y | CD44 molecule (Indian blood group) |
| ENSP00000382340 | ABCC1 | 521.0 | 0.002 | 10 | 0 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| ENSP00000293379 | ITGA5 | 477.1 | 0.002 | 1 | Y | Integrin, α5 (fibronectin receptor, α-polypeptide) |
| ENSP00000230418 | PTK7 | 231.3 | 0.005 | 1 | Y | Protein-tyrosine kinase 7 |
| ENSP00000261978 | LTBP2 | 219.4 | 0.002 | 0 | Y | Latent transforming growth factor β-binding protein 2 |
| ENSP00000280527 | CRIM1 | 218.6 | 0.003 | 1 | Y | Cysteine-rich transmembrane BMP regulator 1 (chordin-like) |
| ENSP00000308727 | SUSD5 | 218.1 | 0.001 | 1 | Y | Sushi domain containing 5 |
| ENSP00000336888 | SLC44A2 | 208.8 | 0.006 | 10 | 0 | Solute carrier family 44 (choline transporter), member 2 |
| ENSP00000258733 | GPNMB | 202.7 | 0.007 | 1 | Y | Glycoprotein (transmembrane) nmb |
| ENSP00000256997 | ACP2 | 198.2 | 0.026 | 1 | Y | Acid phosphatase 2, lysosomal |
| ENSP00000357190 | PTPRK | 197.0 | 0.040 | 1 | Y | Protein-tyrosine phosphatase, receptor type, K |
| ENSP00000356787 | ATP1B1 | 188.5 | 0.044 | 1 | 0 | ATPase, Na+/K+-transporting, β1 polypeptide |
| ENSP00000310206 | SEZ6L2 | 184.0 | 0.041 | 1 | Y | Seizure-related 6 homolog (mouse)-like 2 |
| ENSP00000306864 | VASN | 180.9 | 0.024 | 1 | Y | Vasorin |
| ENSP00000348307 | SIRPA | 146.7 | 0.044 | 1 | Y | Signal-regulatory protein α |
| ENSP00000421922 | LRPAP1 | 141.9 | 0.001 | 0 | Y | Low density lipoprotein receptor-related protein-associated protein 1 |
| ENSP00000320084 | CD276 | 121.5 | 0.010 | 1 | Y | CD276 molecule |
| ENSP00000305988 | ALCAM | 121.5 | 0.007 | 1 | Y | Activated leukocyte cell adhesion molecule |
| ENSG00000125730 | C3 | 119.1 | 0.042 | 0 | Y | Complement component 3 |
| ENSP00000290401 | NPTN | 115.6 | 0.002 | 1 | Y | Neuroplastin |
| ENSP00000378392 | PSAP | 111.3 | 0.002 | 0 | Y | Prosaposin |
| ENSP00000264036 | MCAM | 109.0 | 0.022 | 1 | Y | Melanoma cell adhesion molecule |
| ENSP00000311502 | HEG1 | 101.8 | 0.003 | 0 | 0 | Heart development protein with EGF-like domains 1 |
| ENSP00000331544 | FBLN1 | 100.3 | 0.015 | 0 | Y | Fibulin 1 |
| ENSP00000311402 | SLC4A2 | 96.9 | 0.046 | 13 | 0 | Solute carrier family 4 (anion exchanger), member 2 |
| ENSP00000228506 | MLEC | 96.6 | 0.000 | 1 | Y | Malectin |
| ENSP00000053867 | GRN | 85.3 | 0.013 | 0 | Y | Granulin |
| ENSP00000296181 | ITGB5 | 84.8 | 0.001 | 1 | Y | Integrin, β5 |
| ENSP00000265077 | VCAN | 83.4 | 0.003 | 0 | Y | Versican |
| ENSP00000295633 | FSTL1 | 77.8 | 0.008 | 0 | Y | Follistatin-like 1 |
| ENSP00000413922 | ITIH3 | 76.7 | 0.024 | 0 | Y | Inter-α-trypsin inhibitor heavy chain 3 |
| ENSP00000352288 | PLXNB2 | 75.0 | 0.005 | 1 | Y | Plexin B2 |
| ENSP00000318557 | SLC12A4 | 74.5 | 0.013 | 14 | 0 | Solute carrier family 12 (potassium/chloride transporter), member 4 |
| ENSP00000324101 | CD151 | 72.1 | 0.028 | 4 | 0 | CD151 molecule (Raph blood group) |
| ENSP00000266718 | LUM | 65.3 | 0.001 | 0 | Y | Lumican |
| ENSP00000273258 | ARL6IP5 | 60.8 | 0.049 | 4 | 0 | ADP-ribosylation-like factor 6 interacting protein 5 |
| ENSP00000269228 | NPC1 | 60.7 | 0.009 | 13 | Y | Niemann-Pick disease, type C1 |
| ENSP00000347596 | EFEMP1 | 57.1 | 0.004 | 0 | Y | EGF-containing fibulin-like extracellular matrix protein 1 |
| ENSP00000333697 | TMEM179B | 52.2 | 0.007 | 3 | Y | Transmembrane protein 179B |
| ENSP00000312506 | CSPG4 | 44.1 | 0.003 | 1 | Y | Chondroitin sulfate proteoglycan 4 |
| ENSP00000188790 | FAP | 43.6 | 0.022 | 0 | Y | Fibroblast activation protein, α |
| ENSP00000323534 | FN1 | 43.3 | 0.002 | 0 | Y | Fibronectin 1 |
| ENSP00000262776 | LGALS3BP | 41.9 | 0.004 | 0 | Y | Lectin, galactoside-binding, soluble, 3 binding protein |
| ENSP00000243077 | LRP1 | 41.5 | 0.001 | 1 | Y | Low density lipoprotein receptor-related protein 1 |
| ENSP00000315130 | CLU | 39.7 | 0.002 | 0 | Y | Clusterin |
| ENSP00000206423 | CCDC80 | 38.8 | 0.001 | 0 | Y | Coiled-coil domain containing 80 |
| ENSP00000318646 | RPS15A | 37.8 | 0.003 | 0 | 0 | Ribosomal protein S15a |
| ENSP00000200639 | LAMP2 | 37.6 | 0.004 | 1 | Y | Lysosome-associated membrane protein 2 |
| ENSP00000307513 | MRC2 | 37.5 | 0.002 | 1 | 0 | Mannose receptor, C type 2 |
| ENSP00000261023 | ITGAV | 32.1 | 0.015 | 1 | Y | Integrin, αV |
| ENSP00000341730 | RPL10 | 31.6 | 0.003 | 0 | 0 | Ribosomal protein L10 |
| ENSP00000371626 | TRA2B | 31.4 | 0.006 | 0 | 0 | Transformer 2 βhomolog (Drosophila) |
| ENSP00000323929 | A2M | 30.9 | 0.002 | 0 | Y | α2-Macroglobulin |
| ENSP00000333769 | BSG | 26.8 | 0.000 | 1 | Y | Basigin (Ok blood group) |
| ENSP00000007722 | ITGA3 | 26.7 | 0.001 | 1 | Y | Integrin, α3 (antigen CD49C, α3 subunit of VLA-3 receptor) |
| ENSP00000349437 | IGF2R | 26.5 | 0.003 | 1 | Y | Insulin-like growth factor 2 receptor |
| ENSP00000084795 | RPL18 | 25.6 | 0.000 | 0 | 0 | Ribosomal protein L18 |
| ENSP00000222399 | LAMB1 | 24.9 | 0.000 | 0 | Y | Laminin, β1 |
| ENSP00000226359 | AFP | 22.1 | 0.013 | 0 | Y | α-Fetoprotein |
| ENSP00000252804 | PXDN | 21.3 | 0.006 | 0 | Y | Peroxidasin |
| ENSP00000275730 | SLC12A9 | 20.8 | 0.003 | 13 | 0 | Solute carrier family 12, member 9 |
| ENSP00000359602 | LMBRD1 | 19.6 | 0.013 | 7 | 0 | LMBR1 domain containing 1 |
| ENSP00000366460 | PLXDC2 | 19.2 | 0.007 | 1 | Y | Plexin domain containing 2 |
| ENSP00000286371 | ATP1B3 | 18.8 | 0.035 | 1 | 0 | ATPase, Na+/K+-transporting, β3 polypeptide |
| ENSP00000264896 | SCARB2 | 15.7 | 0.026 | 1 | Y | Scavenger receptor class B, member 2 |
| ENSP00000340815 | SLC3A2 | 15.6 | 0.012 | 1 | 0 | Solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| ENSP00000327290 | ITGA11 | 15.2 | 0.016 | 1 | Y | Integrin, α11 |
| ENSP00000260356 | THBS1 | 15.2 | 0.002 | 0 | Y | Thrombospondin 1 |
| ENSP00000341861 | SERPING1 | 13.7 | 0.041 | 0 | Y | Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| ENSP00000376899 | PTGFRN | 13.3 | 0.016 | 1 | Y | Prostaglandin F2 receptor inhibitor |
| ENSP00000377047 | PTPRZ1 | 13.1 | 0.048 | 1 | Y | Protein-tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| ENSP00000296585 | ITGA2 | 12.1 | 0.003 | 1 | Y | Integrin, α2 (CD49B, α2 subunit of VLA-2 receptor) |
| ENSP00000261799 | PDGFRB | 11.7 | 0.012 | 1 | Y | Platelet-derived growth factor receptor, β-polypeptide |
| ENSP00000355330 | TGM2 | 11.4 | 0.001 | 0 | 0 | Transglutaminase 2 |
| ENSP00000222247 | RPL18A | 10.2 | 0.001 | 0 | 0 | Ribosomal protein L18a |
| ENSP00000344456 | CTNNB1 | 9.1 | 0.000 | 0 | 0 | Catenin (cadherin-associated protein), β1, 88 kDa |
| ENSP00000272317 | RPS27A | 8.8 | 0.006 | 0 | 0 | Ribosomal protein S27a |
| ENSP00000264832 | ICAM1 | 6.7 | 0.000 | 1 | Y | Intercellular adhesion molecule 1 |
| ENSP00000296674 | RPS23 | 6.4 | 0.016 | 0 | 0 | Ribosomal protein S23 |
| ENSP00000346015 | RPL27A | 5.9 | 0.000 | 0 | 0 | Ribosomal protein L27a |
| ENSP00000369743 | RPS6 | 4.8 | 0.001 | 0 | 0 | Ribosomal protein S6 |
| ENSP00000253788 | RPL27 | 4.8 | 0.003 | 0 | 0 | Ribosomal protein L27 |
| ENSP00000416293 | SLC2A1 | 4.5 | 0.010 | 12 | 0 | Solute carrier family 2 (facilitated glucose transporter), member 1 |
| ENSP00000311430 | RPL4 | 4.3 | 0.027 | 0 | 0 | Ribosomal protein L4 |
| ENSP00000277865 | GLUD1 | 4.2 | 0.036 | 0 | Y | Glutamate dehydrogenase 1 |
| ENSP00000345689 | RAB5C | 4.2 | 0.026 | 0 | 0 | RAB5C, member RAS oncogene family |
| ENSP00000363018 | RPL10A | 3.8 | 0.006 | 0 | 0 | Ribosomal protein L10a |
| ENSP00000348849 | RPS26 | 3.8 | 0.024 | 0 | 0 | Ribosomal protein S26 |
| ENSP00000225698 | C1QBP | 3.4 | 0.027 | 0 | 0 | Complement component 1, q subcomponent binding protein |
| ENSP00000346022 | RPL9 | 3.4 | 0.011 | 0 | 0 | Ribosomal protein L9 |
| ENSP00000295598 | ATP1A1 | 3.3 | 0.003 | 8 | 0 | ATPase, Na+/K+-transporting, α1 polypeptide |
| ENSP00000379888 | RPS8 | 3.2 | 0.026 | 0 | 0 | Ribosomal protein S8 |
| ENSP00000305920 | GLB1 | 3.1 | 0.047 | 0 | Y | Galactosidase, β1 |
| ENSP00000325136 | HADHB | 3.1 | 0.030 | 0 | Y | Hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), β subunit |
| ENSP00000346027 | RPL21 | 3.1 | 0.036 | 0 | 0 | Ribosomal protein L21 |
| ENSP00000366156 | SRM | 3.1 | 0.038 | 0 | 0 | Spermidine synthase |
| ENSP00000251453 | RPS16 | 2.5 | 0.040 | 0 | 0 | Ribosomal protein S16 |
| ENSP00000342070 | CTSB | 2.5 | 0.004 | 0 | Y | Cathepsin B |
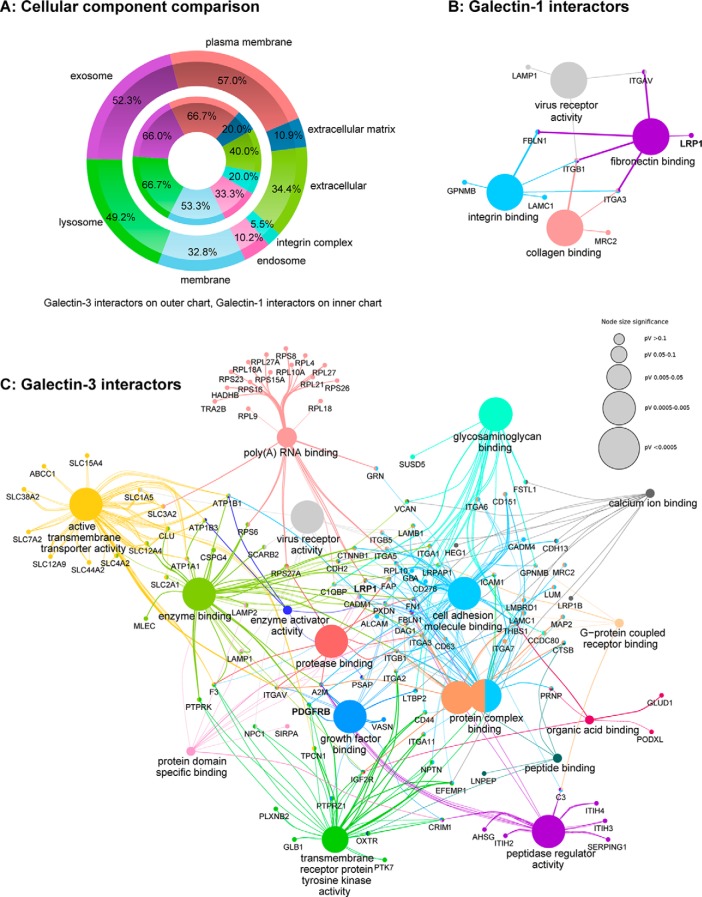
Galectin-1 and Galectin-3 Interactors Play a Role in Multiple Binding Processes and Are Mainly Localized in Membranes
GeneRanker analysis of the 131 Gal-3- and the 15 Gal-1-interacting proteins based on the Gene Ontology (GO) term “cellular component” (supplemental Table S1) and visualized by FunRich classifications (64) revealed that both Gal-1 and Gal-3 interactors were mainly localized in membranes (Fig. 2A). Gal-3 interactors are represented on the outer ring in Fig. 2A and Gal-1 interactors on the inner ring, illustrating that classifications to subcellular localizations of interactors of both galectins are equally spread. Phobius analysis assigned 9 of the 15 Gal-1 interactors and 71 Gal-3 interactors to have at least one transmembrane domain (Tables I and II). Based on GeneRanker analysis, 7 Gal-1 interactors and 67 of the 131 identified Gal-3 interactors are localized to the plasma membrane or on the cell surface. Gal-1 and Gal-3 interactors were also analyzed by the GO term “molecular functions” (supplemental Table S1) and clustered based on this term in the Cytoscape ClueGo-CluePedia (65) network (Fig. 2, B and C). Fig. 2, B and C, shows that Gal-1 and Gal-3 interactors play a role in multiple binding processes. Whereas Gal-1 interactors are involved in integrin, collagen, and fibronectin-binding processes (Fig. 2B), Gal-3 interactors include functions like glycosaminoglycan and growth factor binding among others (Fig. 2C). With this approach, we could identify known Gal-3 interactors like for example LAMP1, BSG, or different members of the integrin family. Two novel identified interactors of great interest are the low density lipoprotein receptor-related protein 1 (LRP1) and the platelet-derived growth factor receptor β (PDGFRB). LRP1 is an endocytotic receptor involved in processes of cell migration and invasion, as well as in the regulation of growth factor homeostasis (68–71), and is overexpressed in RPE and RMG cells in PVR (71). In this pulldown approach, enrichment factors of LRP1 of 31.1 as Gal-1 interactor and of 41.5 as Gal-3 interactor could be identified (Tables I and II), which lead to the hypothesis that LRP1 could be a context-relevant ligand for both galectins. LRP1 also associates with the PDGF receptor in endosomal compartments and modulates its signaling properties affecting the MAPK and Akt/phosphatidylinositol 3-kinase pathways (72). PDGF and PDGFR are key regulators of cell migration and proliferation and play a significant role in PVR development (73). PDGFRB could be identified as a Gal-3 interactor with an enrichment factor of 11.7. To validate LRP1 and PDGFRB as potential targets for Gal-1 and Gal-3, further functional experiments were performed.
Fig. 2.
Galectin interactors play a role in multiple binding processes and are mainly localized in membranes. 15 proteins could be identified as potential Gal-1 interactors and 131 proteins as potential Gal-3 interactors. A, comparison of the 15 galectin-1 interactors (inner chart) and the 131 galectin-3 interactors (outer chart) is based on the gene ontology (GO) annotation “cellular component” in FunRich (64). B and C, network of galectin-1 (B) and galectin-3 (C) interactors, clustered by the GO term “molecular function” in the cytoscape app ClueGo-CluePedia (65). The size of the nodes represents the statistical significance of the enrichment of the terms. The group heading term is the most significant within a group (default). The color code reflects the functional groups. The edges show the connection of distinct genes to specific molecular functions. Identified Gal-1 interactors that were not connected to other proteins in the Gal-1 network are as follows: LGALS3BP, LAMP2, GBA, LANCL1, MLEC, and SSBP1. Identified Gal-3 interactors that were not connected to other proteins in the Gal-3 network are as follows: PLXDC2, TGM2, BSG, SRM, RAB5C, QSOX1, PTGFRN, ACP2, MCAM, LOX, LGALS3BP, SEZ6L2, MXRA5, NCSTN, AFP, DCBLD2, ARL6IP5, and TMEM179B.
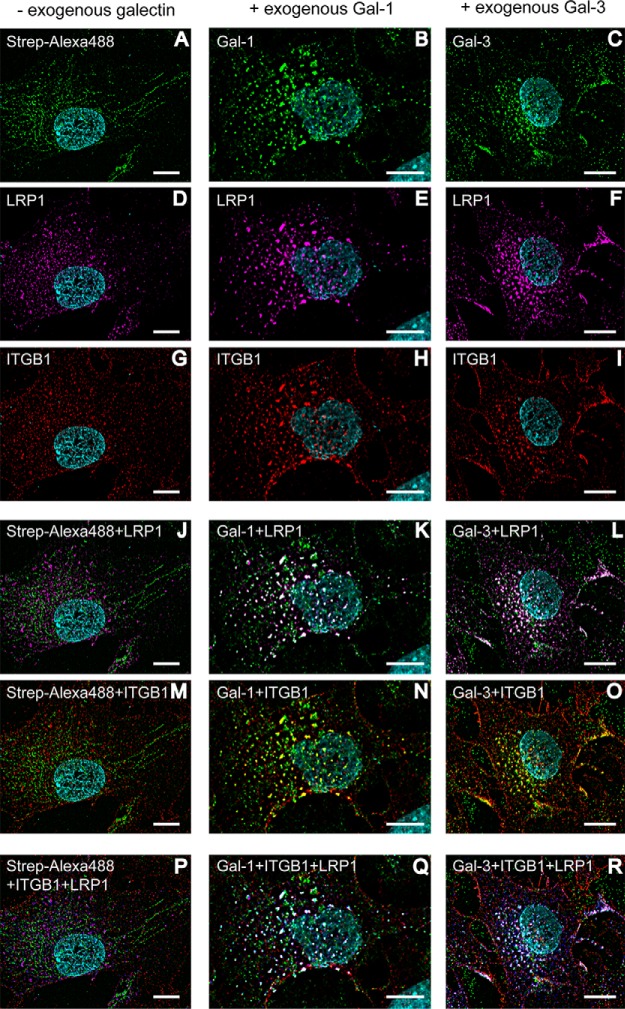
Galectin-1 Induces Cross-linking of LRP1 and Galectin-3 Induces Cross-linking of LRP1 and PDGFRB, Including ITGB1 on the Surface of Retinal Pigment Epithelial Cells
It is known that Gal-1 and Gal-3 can cross-link and cluster transmembrane glycoproteins and additionally reduce their mobility (11, 14, 15). Dynamic lattices on the cell surface are built, and several cellular processes can be influenced (14, 15). To check whether Gal-1 and Gal-3 are able to cross-link with LRP1 and PDGFRB as well as with the known galectin interactor ITGB1, mesenchymal human RPE cells were treated with biotinylated Gal-1 or Gal-3 for 30 min before fixation. The fixed cells were stained with streptavidin-coupled AlexaFluor488 to visualize galectin binding and subsequently with the respective antibodies against LRP1, PDGFRB, and ITGB1. Immunofluorescence analysis revealed large speckle staining patterns of both galectins (Fig. 3, B and C), LRP1 (Fig. 3, E and F), and ITGB1 (Fig. 3, H and I) in cells treated with Gal-1 and Gal-3 compared with diffuse staining patterns in untreated cells (Fig. 3, A, D and G). Visualization of double staining with Gal-1/Gal-3 and LRP1 as well as with Gal-1/Gal-3 and ITGB1 indicated a clear overlay of both staining patterns, visible by white (Fig. 3, K and L) and yellow (Fig. 3, N and O) speckles. Additionally, Gal-1 and Gal-3 induced cross-linking of LRP1 and ITGB1, verified by clear overlay of the respective galectin, LRP1- and ITGB1-staining patterns (white speckles, Fig. 3, Q and R). Cross-linking of Gal-3 with PDGFRB and ITGB1 could also be verified (supplemental Fig. 2). Whereas exogenous galectin treatment resulted in clear co-localization of LRP1/PDGFRB and ITGB1 on the cell surface of human RPE cells, no obvious cluster formation could be seen without addition of exogenous galectin (Fig. 3, J, M, and P).
Fig. 3.
Exogenous galectin-1 and galectin-3 induce cross-linking of LRP1 and ITGB1 on the cell surface of human mesenchymal RPE cells. Immunocytochemical staining of human RPE cells, pretreated before fixation with or without biotinylated Gal-1 or Gal-3 for 30 min. Galectin binding was visualized by streptavidin-AlexaFluor488 (green), LRP1 by AlexaFluor647 (magenta), and ITGB1 by AlexaFluor568 (red). Gal-1 and Gal-3 (B and C), LRP1 (E and F) and ITGB1 (H and I) stainings show a pronounced punctate staining pattern compared with diffuse staining patterns in untreated cells (A, D and G). Double staining of RPE cells with Gal-1/Gal-3 and LRP1 as well as with Gal-1/Gal-3 and ITGB1 indicated a clear overlay of both staining patterns, visible by white (K and L) and yellow (N and O) spots. For visualization of the clustering of galectin, LRP1 and ITGB1, LRP1 staining was changed in silico to blue, and the overlay is seen in white (Q and R). Whereas exogenous addition of galectin led to clear co-localization of LRP1 and ITGB1 on human RPE cells, no cross-linking could be seen without exogenous galectin (J, M and P). Representative images from four independent experiments are shown. Scale bar, 10 μm.
Binding of Galectin-1 and Galectin-3 on LRP1 and PDGFRB Is Glycosylation-dependent
Crucial for galectin binding and lattice formation are the numbers of glycoprotein ligands and the branching of their N-glycans (74). In detail, the amount and the branching of N-acetyllactosamine (LacNAc) residues in the glycan pattern are decisive for affinity. To find out whether binding and cross-linking of Gal-1 and Gal-3 with LRP1 and PDGFRB are dependent on specific glycan structures, human RPE cells were treated for up to 4 weeks with 10 μm kifunensine to prevent complex-type N1,6-glycosylation of proteins. Kifunensine is a potent and selective inhibitor of Golgi class I α-mannosidases (75–77). When it is added to cell culture medium at concentrations of 4 mm or higher, a complete shift of N-glycan patterns from complex structures to Man9(GlcNAc)2 is induced (76, 77). Galectin pulldown experiments were repeated with RPE cell lysates of cells, treated with or without 10 μm kifunensine. Western blot analysis revealed that LRP1 and PDGFRB bound weaker to Gal-1 and Gal-3 when cells were treated with kifunensine (Fig. 4). This indicates that complex-type N-glycosylation of LRP1 and PDGFRB is necessary for galectin binding. Inhibition of complex-type N-glycosylation led also to less galectin binding on the cell surface (Fig. 5, A and C) and no cross-linking of the respective galectins with LRP1 (Fig. 5, A4 and C4), PDGFRB (supplemental Fig. 3I), and ITGB1 (Fig. 5, A3 and C3) was induced, in contrast to the untreated cells as described before.
Fig. 4.
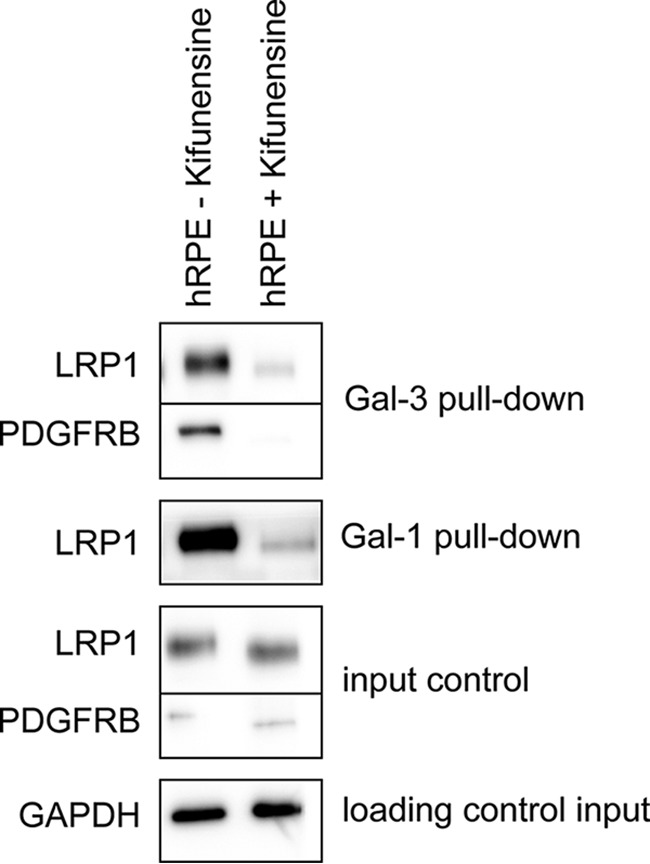
Complex-type N-glycosylation of galectin-interactors is required for galectin binding. Gal-1 and Gal-3 pulldown experiments with lysates of human RPE cells treated or untreated with 10 μm kifunensine. The eluates were analyzed by Western blotting with antibodies against PDGFRB and LRP1. 10 μg of the whole-cell extracts of the respective cell types were used as an input control for the pulldown experiments and probed for GAPDH as loading control. Representative blots from three independent experiments are shown.
Fig. 5.
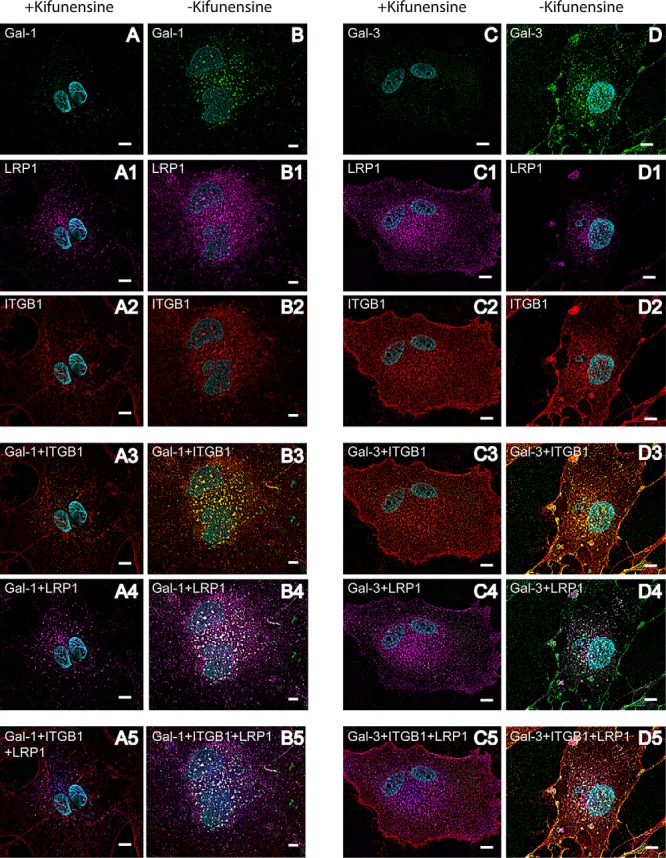
Complex-type N-glycosylation of galectin-interactors is necessary for galectin-induced cross-linking of LRP1 and ITGB1 on the cell surface of mesenchymal RPE cells. Immunocytochemical staining of human RPE cells, pretreated with 10 μm kifunensine. Before fixation cells were pretreated with biotinylated Gal-1 or Gal-3 for 30 min. Galectin binding was visualized with streptavidin-AlexaFluor488 (green) (A–D), LRP1 by AlexaFluor647 (magenta) (A1–D1), and ITGB1 by AlexaFluor568 (red) (A2–D2). Overlay of LRP1 and galectin staining patterns is visible in white (A4–D4) on the overlay of ITGB1 and galectin staining patterns are in yellow (A3–D3). For visualization of the clustering of galectin, LRP1 and ITGB1, LRP1 staining was changed in silico to blue, and the overlay is seen in white (A5–D5). Whereas addition of exogenous galectin led to clear co-localization of LRP1 and ITGB1 on human RPE cells not treated with kifunensine (B–B5 and D–D5), no cross-linking could be observed in RPE cells treated with kifunensine (A–A5 and C–C5). Representative images from two independent experiments are shown. Scale bar, 10 μm.
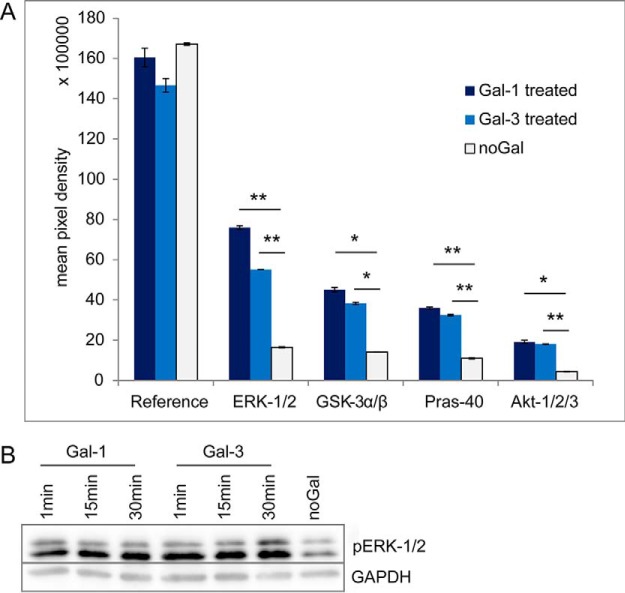
Enhanced Phosphorylation of ERK-1/2 and AKT-1/2/3 by Binding of Gal-1 and Gal-3
To analyze functional and signal modulating effects of galectins on RPE cells, simultaneous screening of changes in phosphorylation profiles in response to galectin binding was performed. ARPE-19 cells were untreated or treated with Gal-1 and Gal-3 for 1, 15, or 30 min. Both Gal-1 and Gal-3 treatment significantly enhanced phosphorylation of the extracellular signal-regulated kinase ERK-1/2 and the serine/threonine protein kinase AKT-1/2/3 (Ser-473) (Fig. 6A). ERK-1 was phosphorylated at the threonine residue Thr-202 and at the tyrosine residue Tyr-204, ERK-2 at Thr-185 and Tyr-187. The corresponding Western blot analysis (Fig. 6B) showed that phosphorylation of ERK-1/2 was already enhanced after 1 min of Gal-1 and Gal-3 treatment and was still detectable after 30 min. Phosphorylation of glycogen synthase kinase GSK-3α/β (S21/S9) and proline-rich protein Pras-40 (Thr-246) were also induced by galectin treatment (Fig. 6A).
Fig. 6.
Enhanced phosphorylation of ERK-1/2 and AKT-1/2/3 by binding of Gal-1 and Gal-3. Analysis of phosphorylation levels induced by Gal-1 or Gal-3 treatment by Western blotting. A, 300 μg of ARPE-19 cell extracts untreated or treated with Gal-1 or Gal-3 for 15 min were incubated with a nitrocellulose membrane of the human phosphokinase array. Profiling of phosphorylation levels revealed significant induction of ERK-1/2, GSK-3α/β, Pras-40, and Akt-1/2/3 phosphorylation after treatment with Gal-1 or Gal-3. The amount of phosphorylated protein bound at the respective capture spot was visualized by signal development with chemiluminescent detection reagents, and mean pixel density was determined in duplicate. p values lower than 0.05 were considered as significant (*), and p values lower than 0.01 as highly significant (**). B, 15 μg of whole-cell extracts of ARPE-19 cells untreated or treated with Gal-1 or Gal-3 for 1, 15, or 30 min were separated by SDS-PAGE (10% gels) and blotted onto PVDF membranes. Blots were incubated with antibodies against phospho-ERK p44/p42 and GAPDH. Representative blots from three independent experiments are shown.
DISCUSSION
This study was designed to identify specific interactors for Gal-1 and Gal-3 on mesenchymal RPE cell surfaces to get deeper insight in the functional effects of galectins on RPE cells in context of PVR. For interactome screening, we used whole-cell extracts of cultured human RPE cells that dedifferentiated and transformed into a fibroblast-like phenotype under the used cell culture conditions. Cultivation of RPE cells on plastic in the presence of serum is a widely accepted in vitro model system for early PVR (27), and because we wanted to identify galectin interactors on RPE cells mimicking status of early PVR, we used primary human RPE cells of nine different human donors. Besides, the human ARPE-19 cell line is often used in RPE cell research (55, 78). In this approach, we used them for the analysis of signal modulating effects of galectins on RPE cells due to the required high amount of whole-cell extracts.
Gal-1 and Gal-3 pulldown experiments and subsequent quantitative LC-MS/MS analysis with strict filtering to avoid unspecific binders resulted in a total of 131 significant Gal-3 interacting binding partners, whereas only 15 proteins remained as significant Gal-1 ligands. The unequal number of interactors can be explained by structural differences of Gal-1 and Gal-3 in their CRD domains. Gal-3 is the only known chimera-type galectin, and it cross-links glycoproteins by its C-terminal domain and multimerizes by its N-terminal domain after binding to saccharide ligands (9, 11). In contrast, Gal-1 consists of one CRD and can form homodimers by its N-terminal domain (12, 13, 16, 17). By coupling of galectins to Sepharose beads, physiological multimerization is presumably hindered, and this conformational change might influence the ability of the CRD domain to bind glycoproteins. However, the binding activity of Gal-1 and Gal-3 after coupling to the beads was confirmed by incubation with asialofetuin, a known interactor via β-galactoside moieties (supplemental Fig. 1). In vivo, ligand binding occurs at the CRD domain, whereas multimerization of galectins and formation of cross-linked lattices is triggered by the N-terminal domain (Figs. 3 and 5) (11, 79). Consequently, multimerization is in vivo not important for recognition of specific ligands, and coupling of galectins to beads should not generally hinder identification of galectin interactors. However, we cannot exclude a different behavior of Gal-1 and Gal-3 with respect to forced monomerization through bead coupling, but Gal-1 and Gal-3 pulldown experiments were done in parallel under the same conditions with the same RPE cell lysates, and multimerization is at least equally important for Gal-3 as compared with Gal-1. Thus, we assume that the lower numbers of Gal-1 interactors are not due to technical limitations, but rather reflect a reduced spectrum of interactors. In vivo, the pentameric form of Gal-3 may facilitate to bind more different glycoprotein receptors on the RPE cell surface. Stillman et al. (40) showed, for instance, that lower concentrations of Gal-3 are required to trigger T cell death than of Gal-1, suggesting that Gal-3 is able to bind more ligands simultaneously. Even though all members of the galectin family bind to galactose-β1,4-N-acetylglucosamine, it is assumed that the structural differences in their CRD domains not only lead to different specificities for distinct glycoproteins, but also to distinct biological activities (40, 80, 81). Whereas Gal-3, for example, is associated with antiapoptotic effects, Gal-1 induces apoptosis in several cell types (82, 83). In contrast, binding to different interactors does not necessarily mean that different downstream mechanisms are influenced. In the literature it is shown that Gal-1 and Gal-3 can bind to distinct receptors but converge on similar downstream signaling in several analyses for induction of T cell death (40) or of neutrophil respiratory burst (84, 85). This assumption is further underscored by the finding that Gal-1 and Gal-3 do not show synergistic effects with respect to inhibition of RPE cell attachment and spreading when added simultaneously (30).
Nevertheless, the identified Gal-1 and Gal-3 interactors have many common features. They are distributed in the same cellular components, mainly in membranes, and play a role in multiple binding processes (Fig. 2). Analysis of galectin interactors in whole RPE cell lysates with combined pulldown experiments and quantitative MS screening revealed both intra- and extracellular interactors. It is known that Gal-1 and Gal-3 are present both inside and outside the cell and that Gal-1 and Gal-3 interact also with intracellular proteins. For example, they are involved in processes like pre-mRNA splicing (86, 87), but these processes are assumed to be based on protein-protein interactions rather than carbohydrate-lectin interactions on the cell surface or ECM (88). In this approach, interacting proteins were eluted with β-lactose to verify carbohydrate-dependent binding on galectins. Thus, we are focusing on the Gal-1 and Gal-3 interactors on the cell surface and ECM. Seven Gal-1 interactors and 67 of the 131 identified Gal-3 interactors are localized in plasma membrane or on the cell surface, based on GeneRanker analysis. Fig. 2A shows the distribution of the identified interactors on cellular components. Most of the interactors are localized in membranes, the extracellular space, or intraluminal parts, validating that this approach mainly pulled down the carbohydrate-dependent interacting ligands. GeneRanker analysis also showed that both Gal-1 and Gal-3 interactors are involved in cell adhesion processes (supplemental Table 1). Gal-3 interactors also play a role in ECM organization and cell migration, whereas Gal-1 interactors are involved in integrin-mediated signaling pathways. With respect to PVR, these biological processes are the key cellular processes in disease development, which is characterized by adhesion, migration and EMT of RPE and RMG cells (20, 23, 25). The data monitored by our GeneRanker analysis are of particular interest because target proteins of both Gal-1 and Gal-3 may play a role in the pathogenesis of PVR and thus be instrumental for influencing the disease process in terms of a therapeutic approach. From recent studies it became evident that PVR is a multifactorial cellular process that cannot be attenuated by inhibition of single growth factors and their downstream signaling pathways or by anti-inflammatory or anti-proliferative approaches alone (32–39). As seen from our data, Gal-1 and Gal-3 may have the ability to orchestrate several cellular processes involved in PVR development simultaneously by interacting with a variety of distinct cell surface interactors, and thus provide a multimodal therapeutic concept.
Most galectin ligands are branched N-glycans on transmembrane proteins (89). Accordingly, we found that complex-type N-glycosylation of the galectin interactors LRP1 and PDGFRB is required for galectin binding and lattice formation on the RPE cell surface (Figs. 4 and 5). Of note, these glycans are up-regulated during EMT of human RPE cells, and these changes lead to increased binding of Gal-3 to mesenchymal RPE cells (31). Consequently, it can be assumed that EMT sensitizes the susceptibility of cell-surface receptors to galectins and that complex-type N-glycan structures are very important for galectin binding. Analyzing the mechanisms of these glycan-galectin interactions will provide evidence whether these glycan structures are a prerequisite for galectin binding and how they influence interaction processes.
Several galectin interactors in different cell types are known so far (90). Gal-3 interacts for instance with EGFR and TGF-βR in tumor cells (45, 91). Both Gal-1 and Gal-3 bind to β1-integrins (42, 92, 93) and extracellular matrix molecules like fibronectin and laminin (52, 94, 95). Besides, Gal-1 and Gal-3 can also bind to immune cell glycoproteins and to neural recognition molecules (40, 43, 82, 96–99). ITGB1 and CD147 (BSG), the two previously identified counter-receptors for Gal-3 in RPE cells, were confirmed with this approach (42). Additionally, known interactors of Gal-1 or Gal-3 like laminin (52, 94), LAMP1 and LAMP2 (100), and integrins (101) among others were confirmed. Most of these interactors were identified in distinct cell types but not in RPE cells. Thus, we could on the one hand confirm several of the known interactors in RPE cells, and on the other hand we identify many new so far unknown interactors.
For validation, we focused on two novel identified Gal-1 and Gal-3 interactors, LRP1 and PDGFRB. LRP1 (or CD91) is a 600-kDa glycoprotein, consisting of an extracellular and a transmembrane domain (102, 103). LRP1 recognizes at least 30 different ligands, among others lipoproteins (104), the β-amyloid precursor protein (68), and the protease inhibitor α2-macroglobulin (α2M), which is responsible for the clearance of several growth factors and cytokines like, for example, TGF-β (105–107). LRP1 recognizes extracellular ligands and induces endocytosis for degradation by lysosomes (70). Thus, it is assumed that LRP1 also plays a significant role in the clearance of α2M-associated growth factors and could potentially be involved in pathologic events during PVR development (71). Hollborn et al. (71) found that LRP1 mRNA levels are up-regulated in human RPE cells, stimulated with TGF-β1, TGF-β2, or VEGF. They hypothesize that protease treatment aiming to induce α2M-mediated clearance of growth factors accompanied by increased LRP1-mediated endocytosis is a potential treatment strategy for PVR (71). However, in PVR RPE and RMG cells are exposed to high amounts of growth factors and cytokines. Although Milenkovic et al. (108) found that α2M inhibited RMG cell proliferation, it remains unclear whether it is possible to clear most of these growth factors by α2M activation or by intravitreal addition of α2M. It is also known that several signaling pathways, including ERK/MAPK, Akt, and NF-κB, are activated by binding of α2M to LRP1 in distinct cell types, including macrophages and RMG cells. Bonacci et al. (109) found that proliferation and MAPK–ERK-1/2 activation in a macrophage-derived cell line is induced by binding of α2M to LRP1. They could verify that α2M promotes expression and secretion of matrix-metalloproteinase MMP-9, which was also mediated by MAPK–ERK-1/2 and NF-κB (110). α2M activates also glial fibrillary acidic protein expression in RMG cells induced by LRP1, which is assumed to be mediated by the JAK/STAT-signaling pathway (111). Barcelona et al. (112) found that α2M-mediated by LRP1 induces RMG cell migration by regulating MT1-MMP activity. We show here that α2M is a significant Gal-3 interactor with an enrichment factor over 30.9 and a p value of 0.002 (Table II). Which effect the binding of Gal-1 and Gal-3 on LRP1 or α2M has on cellular processes of RPE cells remains to be solved. Interestingly, LRP1 can be tyrosine-phosphorylated by the growth factor PDGFR, which in turn regulates its activity by endocytosis and intracellular trafficking of LRP1 (113–115). Thus, LRP1 is predicted to interact as a co-receptor that modulates PDGFR initiated signal transduction pathways, like for example the control of cell migration (113–115). In-depth characterization of downstream signals influenced by Gal-1 and Gal-3 interaction with the identified glycan-dependent interactors will provide more insight into how galectins modify RPE cell behavior. By simultaneous screening of changes in phosphorylation profiles of distinct protein kinases, we could show that both ERK/MAPK and Akt signaling pathways are affected by galectin binding (Fig. 6). ERK phosphorylation was stable up to 30 min after galectin treatment. Akt, which is also called protein kinase B, is one of the main downstream targets of the phosphatidylinositol 3-kinase pathway (116, 117). GSK-3α/β and Pras-40 are substrates of Akt (116–118), and accordingly, both were also phosphorylated after galectin treatment. Because it is known that LRP1 and PDGFRB associate in endosomal compartments and affect MAPK and Akt/phosphatidylinositol 3-kinase pathways (72), we assume that galectin-induced cluster formation of LRP1, ITGB1, and PDGFRB on the cell surface have an influence on those signaling pathways in RPE cells. Interestingly, Gal-1 also induced clustering of PDGFRB and ITGB1 (data not shown), even though PDGFRB was not identified as a Gal-1 interactor by the pulldown experiments, giving a hint that Gal-1 not only forms lattices with specific interactors but that larger interacting protein complexes might be included. The experimental setup with the galectin pulldown assays results in an initial set of galectin interactors, but it does not allow us to distinguish between direct and indirect galectin interactors, which is a general problem in interactome studies based on pulldown approaches. In vivo, Gal-1 and Gal-3 interact due to their multimerization with several glycoproteins simultaneously, and formation of cross-linked lattices takes place. In many biological systems, it is described that clustering of ordered arrays of galectins and their glycoprotein ligands on the cell surface is required for cellular signaling and adhesion processes. The interplay between the different ligands, direct or indirect, is very important to get deeper insights in the functional effects of galectins on RPE cells.
In conclusion, not only known interactors of Gal-1 and Gal-3 were confirmed, but also many novel interactors could be detected in this first proteome-wide comprehensive Gal-1 and Gal-3 interactome screening. Two of the identified interactors, namely LRP1 and PDGFRB, could be validated as Gal-1 and Gal-3 interactors by showing that exogenously added Gal-1 and Gal-3 induce cross-linking on the RPE cell surface with LRP1 and PDGFRB together with the transmembrane protein ITGB1 in a complex-type N-glycan-binding-dependent manner.
DATA AVAILABILITY
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (63) partner repository with the dataset identifier PXD005461 and 10.6019/PXD005461. Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD005461; FTP Download: ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2017/06/PXD005461 PubMed ID: 28576849.
Supplementary Material
Acknowledgments
We thank Nicole Senninger, Jennifer Behler, and Fabian Gruhn for excellent technical assistance and Uli Ohmayer and Christine von Törne for helpful discussions.
Footnotes
Author contributions: J. O., C. S. P., J. M., M. G., and S. M. H. designed research; J. O. performed research; C. S. P., A. G., S. P., and S. M. H. contributed new reagents or analytic tools; J. O., J. M., M. G., and S. M. H. analyzed data; J. O., M. G., and S. M. H. wrote the paper.
* This work was supported by Deutsche Forschungsgemeinschaft Grants (DFG) HA6014/2-2 (to S. M. H.) and PR 1248/2-2 (to C. S. P.). The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CRD
- carbohydrate recognition domain
- α2M
- α2-macroglobulin
- ABC
- ammonium bicarbonate
- CD
- cluster of differentiation
- ECM
- extracellular matrix
- EMT
- epithelial-to-mesenchymal transition
- FC
- fold change
- Gal-1
- galectin-1
- Gal-3
- galectin-3
- GO
- gene ontology
- ITGB1
- integrin β1
- LAMP
- lysosomal-membrane-associated glycoprotein
- LRP1
- low-density lipoprotein receptor
- NCAM
- neural cell adhesion molecule
- PDGFRB
- platelet-derived growth factor receptor β
- PVR
- proliferative vitreoretinopathy
- RMG
- retinal Müller glial cells
- RPE
- retinal pigment epithelial cell
- RT
- room temperature
- TGF-β
- transforming growth factor beta
- TXR
- texas red
- UA
- urea
- VEGF
- vascular endothelial growth factor.
REFERENCES
- 1. Barondes S. H., Castronovo V., Cooper D. N., Cummings R. D., Drickamer K., Feizi T., Gitt M. A., Hirabayashi J., Hughes C., and Kasai K. (1994) galectins: a family of animal β-galactoside-binding lectins. Cell 76, 597–598 [DOI] [PubMed] [Google Scholar]
- 2. Cooper D. N., and Barondes S. H. (1999) God must love galectins; he made so many of them. Glycobiology 9, 979–984 [DOI] [PubMed] [Google Scholar]
- 3. Hirabayashi J., and Kasai K. (1993) The family of metazoan metal-independent β-galactoside-binding lectins: structure, function and molecular evolution. Glycobiology 3, 297–304 [DOI] [PubMed] [Google Scholar]
- 4. Cooper D. N. (2002) Galectinomics: finding themes in complexity. Biochim. Biophys. Acta 1572, 209–231 [DOI] [PubMed] [Google Scholar]
- 5. Römer C. E., and Elling L. (2011) Galectins: Structures, Binding Properties and Function in Cell Adhesion (Pignatello R., ed.) pp. 1–28, INTECH Open Access Publisher [Google Scholar]
- 6. Gabius H.-J. (2011) The Sugar Code: Fundamentals of Glycosciences (Gabius H. J., ed.) pp. 456–459, John Wiley & Sons, Inc., New York [Google Scholar]
- 7. Hughes R. C. (2001) Galectins as modulators of cell adhesion. Biochimie 83, 667–676 [DOI] [PubMed] [Google Scholar]
- 8. Elola M. T., Wolfenstein-Todel C., Troncoso M. F., Vasta G. R., and Rabinovich G. A. (2007) Galectins: matricellular glycan-binding proteins linking cell adhesion, migration, and survival. Cell. Mol. Life Sci. 64, 1679–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gabius H. J. (1997) Animal lectins. Eur. J. Biochem. 243, 543–576 [DOI] [PubMed] [Google Scholar]
- 10. Leffler H., Carlsson S., Hedlund M., Qian Y., and Poirier F. (2002) Introduction to galectins. Glycoconj. J. 19, 433–440 [DOI] [PubMed] [Google Scholar]
- 11. Ahmad N., Gabius H.-J., André S., Kaltner H., Sabesan S., Roy R., Liu B., Macaluso F., and Brewer C. F. (2004) galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. J. Biol. Chem. 279, 10841–10847 [DOI] [PubMed] [Google Scholar]
- 12. Cho M., and Cummings R. D. (1997) Galectin-1: oligomeric structure and interactions with polylactosamine. Trends Glycosci. Glycotechnol. 9, 47–56 [Google Scholar]
- 13. Lobsanov Y. D., Gitt M. A., Leffler H., Barondes S. H., and Rini J. M. (1993) X-ray crystal structure of the human dimeric S-Lac lectin, L-14-II, in complex with lactose at 2.9-A resolution. J. Biol. Chem. 268, 27034–27038 [DOI] [PubMed] [Google Scholar]
- 14. Fred Brewer C. (2002) Binding and cross-linking properties of galectins. Biochim. Biophys. Acta 1572, 255–262 [DOI] [PubMed] [Google Scholar]
- 15. Brewer C. F. (1997) Cross-linking activities of galectins and other multivalent lectins. Trends Glycosci. Glycotechnol. 9, 155–165 [Google Scholar]
- 16. Bourne Y., Bolgiano B., Liao D.-I., Strecker G., Cantau P., Herzberg O., Feizi T., and Cambillau C. (1994) Cross-linking of mammalian lectin (galectin-1) by complex biantennary saccharides. Nat. Struct. Mol. Biol. 1, 863–870 [DOI] [PubMed] [Google Scholar]
- 17. Gupta D., and Brewer C. F. (1994) Homogeneous aggregation of the 14-kDa. β-Galactoside specific vertebrate lectin complex with asialofetuin in mixed systems. Biochemistry 33, 5526–5530 [DOI] [PubMed] [Google Scholar]
- 18. (1983) The classification of retinal detachment with proliferative vitreoretinopathy. Ophthalmology 90, 121–125 [DOI] [PubMed] [Google Scholar]
- 19. Cardillo J. A., Stout J. T., LaBree L., Azen S. P., Omphroy L., Cui J. Z., Kimura H., Hinton D. R., and Ryan S. J. (1997) Post-traumatic proliferative vitreoretinopathy. Ophthalmology 104, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 20. Machemer R. (1988) Proliferative vitreoretinopathy (PVR): a personal account of its pathogenesis and treatment. Invest. Ophthalmol. Vis. Sci. 29, 1771–1783 [PubMed] [Google Scholar]
- 21. Pastor J. C. (1998) Proliferative vitreoretinopathy: an overview. Surv. Ophthalmol. 43, 3–18 [DOI] [PubMed] [Google Scholar]
- 22. Kampik A., Kenyon K. R., Michels R. G., Green W. R., and de la Cruz Z. C. (1981) Epiretinal and vitreous membranes: comparative study of 56 cases. Arch. Ophthalmol. 99, 1445–1454 [DOI] [PubMed] [Google Scholar]
- 23. Machemer R. (1978) Pathogenesis and classification of massive periretinal proliferation. Br. J. Ophthalmol. 62, 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pastor J. C., de la Rúa E. R., Martín F. (2002) Proliferative vitreoretinopathy: risk factors and pathobiology. Prog. Retin. Eye Res. 21, 127–144 [DOI] [PubMed] [Google Scholar]
- 25. Hiscott P., Sheridan C., Magee R. M., and Grierson I. (1999) Matrix and the retinal pigment epithelium in proliferative retinal disease. Prog. Retin. Eye Res. 18, 167–190 [DOI] [PubMed] [Google Scholar]
- 26. Yang S., Li H., Li M., and Wang F. (2015) Mechanisms of epithelial-mesenchymal transition in proliferative vitreoretinopathy. Discov. Med. 20, 207–217 [PubMed] [Google Scholar]
- 27. Grisanti S., and Guidry C. (1995) Transdifferentiation of retinal pigment epithelial cells from epithelial to mesenchymal phenotype. Invest. Ophthalmol. Vis. Sci. 36, 391–405 [PubMed] [Google Scholar]
- 28. Hughes R. C. (1999) Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim. Biophys. Acta 1473, 172–185 [DOI] [PubMed] [Google Scholar]
- 29. Alge-Priglinger C. S., André S., Kreutzer T. C., Deeg C. A., Kampik A., Kernt M., Schöffl H., Priglinger S. G., and Gabius H.-J. (2009) Inhibition of human retinal pigment epithelial cell attachment, spreading, and migration by the human lectin galectin-1. Mol. Vis. 15, 2162–2173 [PMC free article] [PubMed] [Google Scholar]
- 30. Alge-Priglinger C. S., André S., Schoeffl H., Kampik A., Strauss R. W., Kernt M., Gabius H.-J., and Priglinger S. G. (2011) Negative regulation of RPE cell attachment by carbohydrate-dependent cell surface binding of galectin-3 and inhibition of the ERK–MAPK pathway. Biochimie 93, 477–488 [DOI] [PubMed] [Google Scholar]
- 31. Priglinger C. S., Obermann J., Szober C. M., Merl-Pham J., Ohmayer U., Behler J., Gruhn F., Kreutzer T. C., Wertheimer C., Geerlof A., Priglinger S. G., and Hauck S. M. (2016) Epithelial-to-mesenchymal transition of RPE cells in vitro confers increased β-1,6-N-glycosylation and increased susceptibility to galectin-3 binding. PLoS One 11, e0146887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pennock S., Rheaume M.-A., Mukai S., and Kazlauskas A. (2011) A novel strategy to develop therapeutic approaches to prevent proliferative vitreoretinopathy. Am. J. Pathol. 179, 2931–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Velez G., Weingarden A. R., Lei H., Kazlauskas A., and Gao G. (2013) SU9518 Inhibits proliferative vitreoretinopathy in fibroblast and genetically modified Müller cell-induced rabbit models. Invest. Ophthalmol. Vis. Sci. 54, 1392–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Priglinger C. S., and Priglinger S. (2013) Pharmacological approach to treatment of proliferative vitreoretinopathy. Ophthalmologe 110, 948–959 [DOI] [PubMed] [Google Scholar]
- 35. Wiedemann P., Hilgers R. D., Bauer P., and Heimann K. (1998) Adjunctive daunorubicin in the treatment of proliferative vitreoretinopathy: results of a multicenter clinical trial. Am. J. Ophthalmol. 126, 550–559 [DOI] [PubMed] [Google Scholar]
- 36. Charteris D. G., Aylward G. W., Wong D., Groenewald C., Asaria R. H., Bunce C., and PVR Study Group. (2004) A randomized controlled trial of combined 5-fluorouracil and low-molecular-weight heparin in management of established proliferative vitreoretinopathy. Ophthalmology 111, 2240–2245 [DOI] [PubMed] [Google Scholar]
- 37. Turgut B., Uyar F., Ustundag B., Celiker U., Akpolat N., and Demir T. (2012) The impact of tacrolimus on growth factors in experimental proliferative vitreoretinopathy. Retina 32, 232–241 [DOI] [PubMed] [Google Scholar]
- 38. Kita T., Hata Y., Arita R., Kawahara S., Miura M., Nakao S., Mochizuki Y., Enaida H., Goto Y., Shimokawa H., Hafezi-Moghadam A., and Ishibashi T. (2008) Role of TGF-β in proliferative vitreoretinal diseases and ROCK as a therapeutic target. Proc. Natl. Acad. Sci. U.S.A. 105, 17504–17509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng Y., Ikuno Y., Ohj M., Kusaka S., Jiang R., Cekiç O., Sawa M., and Tano Y. (2003) Platelet-derived growth factor receptor kinase inhibitor AG1295 and inhibition of experimental proliferative vitreoretinopathy. Jpn. J. Ophthalmol. 47, 158–165 [DOI] [PubMed] [Google Scholar]
- 40. Stillman B. N., Hsu D. K., Pang M., Brewer C. F., Johnson P., Liu F.-T., and Baum L. G. (2006) Galectin-3 and galectin-1 bind distinct cell surface glycoprotein receptors to induce T cell death. J. Immunol. 176, 778–789 [DOI] [PubMed] [Google Scholar]
- 41. Yu L.-G., Andrews N., Zhao Q., McKean D., Williams J. F., Connor L. J., Gerasimenko O. V., Helens J., Hirabayashi J., Kasai K., and Rhodes J. M. (2007) Galectin-3 interaction with Thomsen-Friedenreich disaccharide on cancer-associated MUC1 causes increased cancer cell endothelial adhesion. J. Biol. Chem. 282, 773–781 [DOI] [PubMed] [Google Scholar]
- 42. Priglinger C. S., Szober C. M., Priglinger S. G., Merl J., Euler K. N., Kernt M., Gondi G., Behler J., Geerlof A., Kampik A., Ueffing M., and Hauck S. M. (2013) Galectin-3 induces clustering of CD147 and integrin-β1 transmembrane glycoprotein receptors on the RPE cell surface. PLoS One 8, e70011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Probstmeier R., Montag D., and Schachner M. (1995) Galectin-3, a β-galactoside-binding animal lectin, binds to neural recognition molecules. J. Neurochem. 64, 2465–2472 [DOI] [PubMed] [Google Scholar]
- 44. Markowska A. I., Jefferies K. C., and Panjwani N. (2011) Galectin-3 protein modulates cell surface expression and activation of vascular endothelial growth factor receptor 2 in human endothelial cells. J. Biol. Chem. 286, 29913–29921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., and Dennis J. W. (2004) Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science 306, 120–124 [DOI] [PubMed] [Google Scholar]
- 46. Rapoport E. M., Kurmyshkina O. V., and Bovin N. V. (2008) Mammalian Galectins: structure, carbohydrate specificity, and functions. Biochemistry 73, 393–405 [DOI] [PubMed] [Google Scholar]
- 47. Markowska A. I., Liu F.-T., and Panjwani N. (2010) Galectin-3 is an important mediator of VEGF-and bFGF-mediated angiogenic response. J. Exp. Med. 207, 1981–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pace K. E., Lee C., Stewart P. L., and Baum L. G. (1999) Restricted receptor segregation into membrane microdomains occurs on human T cells during apoptosis induced by galectin-1. J. Immunol. 163, 3801–3811 [PubMed] [Google Scholar]
- 49. Dumic J., Dabelic S., and Flögel M. (2006) Galectin-3: an open-ended story. Biochim. Biophys. Acta 1760, 616–635 [DOI] [PubMed] [Google Scholar]
- 50. Dong S., and Hughes R. C. (1997) Macrophage surface glycoproteins binding to galectin-3 (Mac-2-antigen). Glycoconj. J. 14, 267–274 [DOI] [PubMed] [Google Scholar]
- 51. Furtak V., Hatcher F., and Ochieng J. (2001) Galectin-3 mediates the endocytosis of β-1 integrins by breast carcinoma cells. Biochem. Biophys. Res. Commun. 289, 845–850 [DOI] [PubMed] [Google Scholar]
- 52. Kuwabara I., and Liu F.-T. (1996) Galectin-3 promotes adhesion of human neutrophils to laminin. J. Immunol. 156, 3939–3944 [PubMed] [Google Scholar]
- 53. Ochieng J., Leite-Browning M. L., and Warfield P. (1998) Regulation of cellular adhesion to extracellular matrix proteins by galectin-3. Biochem. Biophys. Res. Commun. 246, 788–791 [DOI] [PubMed] [Google Scholar]
- 54. Alberts B., Johnson A., Lewis J., Walter P., Raff M., and Roberts K. (2002) Molecular Biology Cell, 4th Ed., International Student Edition, Routledge, London, UK [Google Scholar]
- 55. Kuznetsova A. V., Kurinov A. M., and Aleksandrova M. A. (2014) Cell models to study regulation of cell transformation in pathologies of retinal pigment epithelium. J. Ophthalmol. 2014, 801787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nowak T. P., Haywood P. L., and Barondes S. H. (1976) Developmentally regulated lectin in embryonic chick muscle and a myogenic cell line. Biochem. Biophys. Res. Commun. 68, 650–657 [DOI] [PubMed] [Google Scholar]
- 57. St-Pierre C., Ouellet M., Giguère D., Ohtake R., Roy R., Sato S., and Tremblay M. J. (2012) Galectin-1-specific inhibitors as a new class of compounds to treat HIV-1 infection. Antimicrob. Agents Chemother. 56, 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 59. Hauck S. M., Dietter J., Kramer R. L., Hofmaier F., Zipplies J. K., Amann B., Feuchtinger A., Deeg C. A., and Ueffing M. (2010) Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol. Cell. Proteomics 9, 2292–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Merl J., Deeg C. A., Swadzba M. E., Ueffing M., and Hauck S. M. (2013) Identification of autoantigens in body fluids by combining pull-downs and organic precipitations of intact immune complexes with quantitative label-free mass spectrometry. J. Proteome Res. 12, 5656–5665 [DOI] [PubMed] [Google Scholar]
- 61. Grosche A., Hauser A., Lepper M. F., Mayo R., von Toerne C., Merl-Pham J., and Hauck S. M. (2016) The proteome of native adult Muller glial cells from murine retina. Mol. Cell. Proteomics 15, 462–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ly A., Merl-Pham J., Priller M., Gruhn F., Senninger N., Ueffing M., and Hauck S. M. (2016) Proteomic profiling suggests central role of STAT signaling during retinal degeneration in the rd10 mouse model. J. Proteome Res. 15, 1350–1359 [DOI] [PubMed] [Google Scholar]
- 63. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pathan M., Keerthikumar S., Ang C. S., Gangoda L., Quek C. Y., Williamson N. A., Mouradov D., Sieber O. M., Simpson R. J., Salim A., Bacic A., Hill A. F., Stroud D. A., Ryan M. T., Agbinya J. I., et al. (2015) FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597–2601 [DOI] [PubMed] [Google Scholar]
- 65. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.-H., Pagès F., Trajanoski Z., and Galon J. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Käll L., Krogh A., and Sonnhammer E. L. (2004) A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 67. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kounnas M. Z., Moir R. D., Rebeck G. W., Bush A. I., Argraves W. S., Tanzi R. E., Hyman B. T., and Strickland D. K. (1995) LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted β-amyloid precursor protein and mediates its degradation. Cell 82, 331–340 [DOI] [PubMed] [Google Scholar]
- 69. Ulery P. G., Beers J., Mikhailenko I., Tanzi R. E., Rebeck G. W., Hyman B. T., and Strickland D. K. (2000) Modulation of β-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that LRP contributes to the pathogenesis of Alzheimer's disease. J. Biol. Chem. 275, 7410–7415 [DOI] [PubMed] [Google Scholar]
- 70. Li Y., Lu W., Marzolo M. P., and Bu G. (2001) Differential functions of members of the low density lipoprotein receptor family suggested by their distinct endocytosis rates. J. Biol. Chem. 276, 18000–18006 [DOI] [PubMed] [Google Scholar]
- 71. Hollborn M., Birkenmeier G., Saalbach A., Iandiev I., Reichenbach A., Wiedemann P., and Kohen L. (2004) Expression of LRP1 in retinal pigment epithelial cells and its regulation by growth factors. Invest. Ophthalmol. Vis. Sci. 45, 2033–2038 [DOI] [PubMed] [Google Scholar]
- 72. Muratoglu S. C., Mikhailenko I., Newton C., Migliorini M., and Strickland D. K. (2010) Low density lipoprotein receptor-related protein 1 (LRP1) forms a signaling complex with platelet-derived growth factor receptor-β in endosomes and regulates activation of the MAPK pathway. J. Biol. Chem. 285, 14308–14317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cui J., Lei H., Samad A., Basavanthappa S., Maberley D., Matsubara J., and Kazlauskas A. (2009) PDGF receptors are activated in human epiretinal membranes. Exp. eye Res. 88, 438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nabi I. R., Shankar J., and Dennis J. W. (2015) The galectin lattice at a glance. J. Cell Sci. 128, 2213–2219 [DOI] [PubMed] [Google Scholar]
- 75. Kayakiri H., Kasahara C., Oku T., and Hashimoto M. (1990) Synthesis of kifunensine, an immunomodulating substance isolated from microbial source. Tetrahedron Lett. 31, 225–226 [Google Scholar]
- 76. Elbein A. D. (1991) Glycosidase inhibitors: inhibitors of N-linked oligosaccharide processing. FASEB J. 5, 3055–3063 [DOI] [PubMed] [Google Scholar]
- 77. Elbein A. D., Tropea J. E., Mitchell M., and Kaushal G. P. (1990) kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. J. Biol. Chem. 265, 15599–15605 [PubMed] [Google Scholar]
- 78. Kang G.-Y., Bang J. Y., Choi A. J., Yoon J., Lee W.-C., Choi S., Yoon S., Kim H. C., Baek J.-H., Park H. S., Lim H. J., and Chung H. (2014) Exosomal proteins in the aqueous humor as novel biomarkers in patients with neovascular age-related macular degeneration. J. Proteome Res. 13, 581–595 [DOI] [PubMed] [Google Scholar]
- 79. Vijayakumar S., Peng H., and Schwartz G. J. (2013) Galectin-3 mediates oligomerization of secreted hensin using its carbohydrate-recognition domain. Am. J. Physiol. Renal Physiol. 305, F90–F99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kopitz J., von Reitzenstein C., André S., Kaltner H., Uhl J., Ehemann V., Cantz M., and Gabius H.-J. (2001) Negative regulation of neuroblastoma cell growth by carbohydrate-dependent surface binding of galectin-1 and functional divergence from galectin-3. J. Biol. Chem. 276, 35917–35923 [DOI] [PubMed] [Google Scholar]
- 81. Ahmad N., Gabius H.-J., Sabesan S., Oscarson S., and Brewer C. F. (2004) Thermodynamic binding studies of bivalent oligosaccharides to galectin-1, galectin-3, and the carbohydrate recognition domain of galectin-3. Glycobiology 14, 817–825 [DOI] [PubMed] [Google Scholar]
- 82. Pace K. E., Hahn H. P., Pang M., Nguyen J. T., and Baum L. G. (2000) Cutting edge: CD7 delivers a pro-apoptotic signal during galectin-1-induced T cell death. J. Immunol. 165, 2331–2334 [DOI] [PubMed] [Google Scholar]
- 83. Yang R.-Y., Hsu D. K., and LIu F.-T. (1996) Expression of galectin-3 modulates T-cell growth and apoptosis. Proc. Natl. Acad. Sci. U.S.A. 93, 6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Almkvist J., and Karlsson A. (2004) Galectins as inflammatory mediators. Glycoconj. J. 19, 575–581 [DOI] [PubMed] [Google Scholar]
- 85. Almkvist J., Dahlgren C., Leffler H., and Karlsson A. (2002) Activation of the neutrophil nicotinamide adenine dinucleotide phosphate oxidase by galectin-1. J. Immunol. 168, 4034–4041 [DOI] [PubMed] [Google Scholar]
- 86. Dagher S. F., Wang J. L., and Patterson R. J. (1995) Identification of galectin-3 as a factor in pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 92, 1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vyakarnam A., Dagher S. F., Wang J. L., and Patterson R. J. (1997) Evidence for a role for galectin-1 in pre-mRNA splicing. Mol. Cell. Biol. 17, 4730–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu F.-T., and Rabinovich G. A. (2005) Galectins as modulators of tumour progression. Nat. Rev. Cancer 5, 29–41 [DOI] [PubMed] [Google Scholar]
- 89. Patnaik S. K., Potvin B., Carlsson S., Sturm D., Leffler H., and Stanley P. (2006) Complex N-glycans are the major ligands for galectin-1,-3, and-8 on Chinese hamster ovary cells. Glycobiology 16, 305–317 [DOI] [PubMed] [Google Scholar]
- 90. Boscher C., Dennis J. W., and Nabi I. R. (2011) Glycosylation, galectins and cellular signaling. Curr. Opin. Cell Biol. 23, 383–392 [DOI] [PubMed] [Google Scholar]
- 91. Lajoie P., Partridge E. A., Guay G., Goetz J. G., Pawling J., Lagana A., Joshi B., Dennis J. W., and Nabi I. R. (2007) Plasma membrane domain organization regulates EGFR signaling in tumor cells. J. Cell Biol. 179, 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Saravanan C., Liu F.-T., Gipson I. K., and Panjwani N. (2009) Galectin-3 promotes lamellipodia formation in epithelial cells by interacting with complex N-glycans on α3β1 integrin. J. Cell Sci. 122, 3684–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Moiseeva E. P., Williams B., Goodall A. H., and Samani N. J. (2003) Galectin-1 interacts with β-1 subunit of integrin. Biochem. Biophys. Res. Commun. 310, 1010–1016 [DOI] [PubMed] [Google Scholar]
- 94. Moiseeva E. P., Spring E. L., Baron J. H., and de Bono D. (1999) Galectin 1 modulates attachment, spreading and migration of cultured vascular smooth muscle cells via interactions with cellular receptors and components of extracellular matrix. J. Vasc. Res. 36, 47–58 [DOI] [PubMed] [Google Scholar]
- 95. Kariya Y., Kawamura C., Tabei T., and Gu J. (2010) Bisecting GlcNAc residues on laminin-332 down-regulate galectin-3-dependent keratinocyte motility. J. Biol. Chem. 285, 3330–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Perillo N. L., Pace K. E., Seilhamer J. J., and Baum L. G. (1995) Apoptosis of T cells mediated by galectin-1. Nature 378, 736–739 [DOI] [PubMed] [Google Scholar]
- 97. Hernandez J. D., Nguyen J. T., He J., Wang W., Ardman B., Green J. M., Fukuda M., and Baum L. G. (2006) Galectin-1 binds different CD43 glycoforms to cluster CD43 and regulate T cell death. J. Immunol. 177, 5328–5336 [DOI] [PubMed] [Google Scholar]
- 98. Walzel H., Blach M., Hirabayashi J., Kasai K.-I., and Brock J. (2000) Involvement of CD2 and CD3 in galectin-1 induced signaling in human Jurkat T-cells. Glycobiology 10, 131–140 [DOI] [PubMed] [Google Scholar]
- 99. Fukumori T., Takenaka Y., Yoshii T., Kim H.-R., Hogan V., Inohara H., Kagawa S., and Raz A. (2003) CD29 and CD7 mediate galectin-3-induced type II T-cell apoptosis. Cancer Res. 63, 8302–8311 [PubMed] [Google Scholar]
- 100. Sarafian V., Jadot M., Foidart J. M., Letesson J. J., Van den Brûle F., Castronovo V., Wattiaux R., and Coninck S. W. (1998) Expression of Lamp-1 and Lamp-2 and their interactions with galectin-3 in human tumor cells. Int. J. Cancer 75, 105–111 [DOI] [PubMed] [Google Scholar]
- 101. Gu M., Wang W., Song W. K., Cooper D. N., and Kaufman S. J. (1994) Selective modulation of the interaction of α7β1 integrin with fibronectin and laminin by L-14 lectin during skeletal muscle differentiation. J. Cell Sci. 107, 175–181 [DOI] [PubMed] [Google Scholar]
- 102. Herz J., Kowal R. C., Goldstein J. L., and Brown M. S. (1990) Proteolytic processing of the 600 kd low density lipoprotein receptor-related protein (LRP) occurs in a trans-Golgi compartment. EMBO J. 9, 1769–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Herz J., Hamann U., Rogne S., Myklebost O., Gausepohl H., and Stanley K. K. (1988) Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 7, 4119–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Beisiegel U., Weber W., Ihrke G., Herz J., and Stanley K. K. (1989) The LDL-receptor-related protein, LRP, is an apolipoprotein E-binding protein. Nature 341, 162–164 [DOI] [PubMed] [Google Scholar]
- 105. Kristensen T., Moestrup S. K., Gliemann J., Bendtsen L., Sand O., and Sottrup-Jensen L. (1990) Evidence that the newly cloned low-density-lipoprotein receptor related protein (LRP) is the α2-macroglobulin receptor. FEBS Lett. 276, 151–155 [DOI] [PubMed] [Google Scholar]
- 106. Strickland D. K., Ashcom J. D., Williams S., Burgess W. H., Migliorini M., and Argraves W. S. (1990) Sequence identity between the α2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J. Biol. Chem. 265, 17401–17404 [PubMed] [Google Scholar]
- 107. Herz J., and Strickland D. K. (2001) LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 108, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Milenkovic I., Birkenmeier G., Wiedemann P., Reichenbach A., and Bringmann A. (2005) Effect of α2-macroglobulin on retinal glial cell proliferation. Graefe Arch. Clin. Exp. Ophthalmol. 243, 811–816 [DOI] [PubMed] [Google Scholar]
- 109. Bonacci G. R., Cáceres L. C., Sánchez M. C., and Chiabrando G. A. (2007) Activated α2-macroglobulin induces cell proliferation and mitogen-activated protein kinase activation by LRP-1 in the J774 macrophage-derived cell line. Arch. Biochem. Biophys. 460, 100–106 [DOI] [PubMed] [Google Scholar]
- 110. Cáceres L. C., Bonacci G. R., Sánchez M. C., and Chiabrando G. A. (2010) Activated α2 macroglobulin induces matrix metalloproteinase 9 expression by low-density lipoprotein receptor-related protein 1 through MAPK-ERK1/2 and NF-κB activation in macrophage-derived cell lines. J. Cell. Biochem. 111, 607–617 [DOI] [PubMed] [Google Scholar]
- 111. Barcelona P. F., Ortiz S. G., Chiabrando G. A., and Sánchez M. C. (2011) α2-Macroglobulin induces glial fibrillary acidic protein expression mediated by low-density lipoprotein receptor-related protein 1 in Müller cells. Invest. Ophthalmol. Vis. Sci. 52, 778–786 [DOI] [PubMed] [Google Scholar]
- 112. Barcelona P. F., Jaldín-Fincati J. R., Sánchez M. C., and Chiabrando G. A. (2013) Activated α2-macroglobulin induces Müller glial cell migration by regulating MT1-MMP activity through LRP1. FASEB J. 27, 3181–3197 [DOI] [PubMed] [Google Scholar]
- 113. Boucher P., Gotthardt M., Li W.-P., Anderson R. G., and Herz J. (2003) LRP: role in vascular wall integrity and protection from atherosclerosis. Science 300, 329–332 [DOI] [PubMed] [Google Scholar]
- 114. Boucher P., Liu P., Gotthardt M., Hiesberger T., Anderson R. G., and Herz J. (2002) Platelet-derived growth factor mediates tyrosine phosphorylation of the cytoplasmic domain of the low density lipoprotein receptor-related protein in caveolae. J. Biol. Chem. 277, 15507–15513 [DOI] [PubMed] [Google Scholar]
- 115. Loukinova E., Ranganathan S., Kuznetsov S., Gorlatova N., Migliorini M. M., Loukinov D., Ulery P. G., Mikhailenko I., Lawrence D. A., and Strickland D. K. (2002) Platelet-derived growth factor (PDGF)-induced tyrosine phosphorylation of the low density lipoprotein receptor-related protein (LRP). Evidence for integrated co-receptor function between LRP and the PDGF. J. Biol. Chem. 277, 15499–15506 [DOI] [PubMed] [Google Scholar]
- 116. Kovacina K. S., Park G. Y., Bae S. S., Guzzetta A. W., Schaefer E., Birnbaum M. J., and Roth R. A. (2003) Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J. Biol. Chem. 278, 10189–10194 [DOI] [PubMed] [Google Scholar]
- 117. Cantley L. C. (2002) The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 118. Wang H., Zhang Q., Wen Q., Zheng Y., Jiang H., Lin J., Lazarovici P., Philip L., and Zheng W. (2012) Proline-rich Akt substrate of 40 kDa (PRAS40): a novel downstream target of PI3K/Akt signaling pathway. Cell. Signal. 24, 17–24 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (63) partner repository with the dataset identifier PXD005461 and 10.6019/PXD005461. Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD005461; FTP Download: ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2017/06/PXD005461 PubMed ID: 28576849.