To the Editor
Contemporary genomic data sets contain tens of thousands of samples and petabytes of sequencing data1–3. Pipelines to process genomic data sets often comprise dozens of individual steps, each with their own set of parameters4,5. Processing data at this scale and complexity is expensive, can take an unacceptably long time, and requires significant engineering effort. Furthermore, biomedical data sets are often siloed, both for organizational and security considerations and because they are physically difficult to transfer between systems, owing to bandwidth limitations. The solution to better handling these big data problems is twofold: first, we need robust software capable of running analyses quickly and efficiently, and second, we need the software and pipelines to be portable, so that they can be reproduced in any suitable compute environment.
Here, we present Toil, a portable, open-source workflow software that can be used to run scientific workflows on a large scale in cloud or high-performance computing (HPC) environments. Toil was created to include a complete set of features necessary for rapid large-scale analyses across multiple environments. While several other scientific workflow software packages6–8 offer some subset of fault tolerance, cloud support and HPC support, none offers these with the scale and efficiency to process petabyte and larger-scale data sets efficiently. This sets Toil apart in its capacity to produce results faster and for less cost across diverse environments. We demonstrate Toil by processing >20,000 RNA-seq samples (Fig. 1). The resulting meta-analysis of five data sets is available to readers9. The large majority (99%) of these samples were analyzed in under 4 days using a commercial cloud cluster of 32,000 preemptable cores.
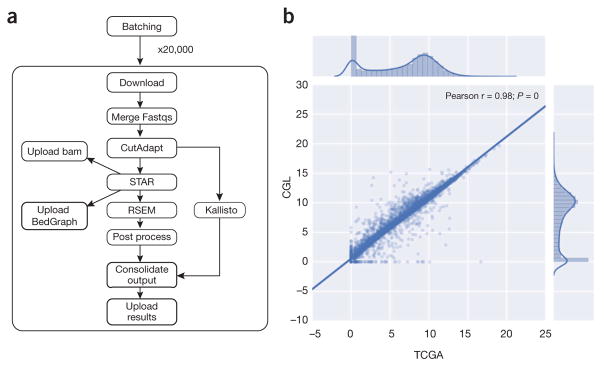
Figure 1.
RNA-seq pipeline and expression concordance. (a) A dependency graph of the RNA-seq pipeline we developed (named CGL). CutAdapt was used to remove extraneous adapters, STAR was used for alignment and read coverage, and RSEM and Kallisto were used to produce quantification data. (b) Scatter plot showing the Pearson correlation between the results of the TCGA best-practices pipeline and the CGL pipeline. 10,000 randomly selected sample and/or gene pairs were subset from the entire TCGA cohort and the normalized counts were plot against each other; this process was repeated five times with no change in Pearson correlation. The unit for counts is: log2(norm_counts+1).
To support the sharing of scientific workflows, we designed Toil to execute common workflow language (CWL; Supplementary Note 1) and provide draft support for workflow description language (WDL). Both CWL and WDL are standards for scientific workflows10,11. A workflow comprises a set of tasks, or ‘jobs’, that are orchestrated by specification of a set of dependencies that map the inputs and outputs between jobs. In addition to CWL and draft WDL support, Toil provides a Python application program interface (API) that allows workflows to be declared statically, or generated dynamically, so that jobs can define further jobs during execution and therefore as needed (Supplementary Note 2 and Supplementary Toil Documentation). The jobs defined in either CWL or Python can consist of Docker containers, which permit sharing of a program without requiring individual tool installation or configuration within a specific environment. Open-source workflows that use containers can be run regardless of environment. We provide a repository of genomic workflows as examples12. Toil supports services, such as databases or servers, that are defined and managed within a workflow. Through this mechanism it integrates with Apache Spark13 (Supplementary Fig. 4), and can be used to rapidly create containerized Spark clusters14 (Supplementary Note 3).
Toil runs in multiple cloud environments including those of Amazon Web Services (AWS; Seattle, WA, USA), Microsoft Azure (Seattle, WA, USA), Google Cloud (Mountain View, CA, USA), OpenStack, and in HPC environments running GridEngine or Slurm and distributed systems running Apache Mesos15–17 (Forest Hill, MD, USA). Toil can run on a single machine, such as a laptop or workstation, to allow for interactive development, and can be installed with a single command. This portability stems from pluggable backend APIs for machine provisioning, job scheduling and file management (Supplementary Note 4). Implementation of these APIs facilitates straightforward extension of Toil to new compute environments. Toil manages intermediate files and checkpointing through a ‘job store’, which can be an object store like AWS’s S3 or a network file-system. The flexibility of the backend APIs allow a single script to be run on any supported compute environment, paired with any job store, without requiring any modifications to the source code.
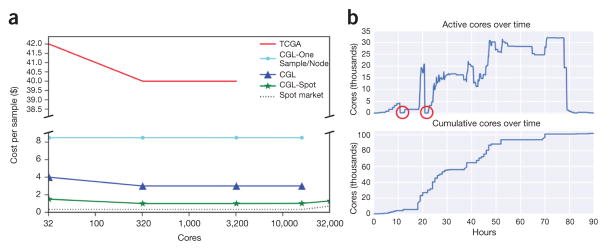
Toil includes numerous performance optimizations to maximize time and cost efficiencies (Supplementary Note 5). Toil implements a leader/worker pattern for job scheduling, in which the leader delegates jobs to workers. To reduce pressure on the leader, workers can decide whether they are capable of running jobs immediately downstream to their assigned task (in terms of resource requirements and workflow dependencies). Frequently, next-generation sequencing workflows are I/O bound, owing to the large volume of data analyzed. To mitigate this, Toil uses file caching and data streaming. Where possible, successive jobs that share files are scheduled on a single node, and caching prevents the need for repeated transfers from the job store. Toil is robust to job failure because workflows can be resumed after any combination of leader and worker failures. This robustness enables workflows to use low-cost machines that can be terminated by the provider at short notice and are currently available at a significant discount on AWS and Google Cloud. We estimate the use of such preemptable machines on AWS lowered the cost of our RNA-seq compute job 2.5-fold, despite encountering over 2,000 premature terminations (Fig. 2). Toil also supports fine-grained resource requirements, enabling each job to specify its core, memory and local storage needs for scheduling efficiency.
Figure 2.
Costs and core usage. (a) Scaling tests were run to ascertain the price per sample at varying cluster sizes for the different analysis methods. TCGA (red) shows the cost of running the TCGA best-practices pipeline as re-implemented as a Toil workflow (for comparison). CGL-One-Sample/Node (cyan) shows the cost of running the revised Toil pipeline, one sample per node. CGL (blue) denotes the pipeline running samples across many nodes. CGL-Spot (green) is the same as CGL, but denotes the pipeline run on the Amazon spot market. The slight rise in cost per sample at 32,000 cores was due to a couple of factors: aggressive instance provisioning directly affected the spot price (dotted line), and saving bam and bedGraph files for each sample. (b) Tracking number of cores during the recompute. The two red circles indicate where all worker nodes were terminated and subsequently restarted shortly thereafter.
Controlled-access data requires appropriate precautions to ensure data privacy and protection. Cloud environments offer measures that ensure stringent standards for protected data. Input files can be securely stored on object stores, using encryption, either transparently or with customer managed keys. Compute nodes can be protected by SSH key pairs. When running Toil, all intermediate data transferred to and from the job store can be optionally encrypted during network transmission and on the compute nodes’ drives using Toil’s cloud-based job store encryption. These and other security measures help ensure protection of the input data, and as part of a broader security plan, can be used to ensure compliance with strict data security requirements.
To demonstrate Toil, we used a single script to compute gene- and isoform-level expression values for 19,952 samples from four studies: The Cancer Genome Atlas (TCGA)1, Therapeutically Applicable Research To Generate Effective Treatments (TARGET; https://ocg.cancer.gov/programs/target), Pacific Pediatric Neuro-Oncology Consortium (PNOC; http://www.pnoc.us/), and the Genotype Tissue Expression Project (GTEx)18. The data set comprised 108 terabytes. The Toil pipeline uses STAR19 to generate alignments and read coverage graphs, and performs quantification using RSEM20 and Kallisto21 (Fig. 1 and Supplementary Note 6). Processing the samples in a single batch on ~32,000 cores on AWS took 90 h of wall time, 368,000 jobs and 1,325,936 core hours. The cost per sample was $1.30, which is an estimated 30-fold reduction in cost, and a similar reduction in time, compared with the TCGA best-practices workflow5. We achieved a 98% gene-level concordance with the previous pipeline’s expression predictions (Figs. 1,2 and Supplementary Fig. 1). Notably, we estimate that the pipeline, without STAR and RSEM, could be used to generate quantifications for $0.19/sample with Kallisto. To illustrate portability, the same pipeline was run on the I-SPY2 data set22 (156 samples) using a private HPC cluster, achieving similar per sample performance (Supplementary Table 1). Expression-level signal graphs (read coverage) of the GTEx data (7,304 samples from 53 tissues, 570 donors) are available from a UCSC Genome Browser23 public track hub (Supplementary Fig. 2). Gene and isoform quantifications for this consistent, union data set are publicly hosted on UCSC Xena9 and are available for direct access through a public AWS bucket (Supplementary Fig. 3 and Supplementary Note 7).
Although there is an extensive history of open-source workflow-execution software6–8, the shift to cloud platforms and the advent of standard workflow languages is changing the scale of analyses. Toil is a portable workflow software that supports open community standards for workflow specification and enables researchers to move their computation according to cost, time and data location. For example, in our analysis the sample data were intentionally co-located in the same region as the compute servers in order to provide optimal bandwidth when scaling to thousands of simultaneous jobs (Supplementary Note 8). This type of flexibility enables larger, more comprehensive analyses. Further, it means that results can be reproduced using the original computation’s set of tools and parameters. If we had run the original TCGA best-practices RNA-seq pipeline with one sample per node, it would have cost ~$800,000. Through the use of efficient algorithms (STAR and Kallisto) and Toil, we were able to reduce the final cost to $26,071 (Supplementary Note 9).
We have demonstrated the utility of Toil by creating one of the single largest, consistently analyzed, public human RNA-seq expression repositories, which we hope the community will find useful.
Supplementary Material
Acknowledgments
This work was supported by (BD2K) the National Human Genome Research Institute of the National Institutes of Health award no. 5U54HG007990 and (Cloud Pilot) the National Cancer Institute of the National Institutes of Health under the Broad Institute subaward no. 5417071-5500000716. Work was also supported by NIH grants 4U24CA180951 and 1U24CA210974. The UCSC Genome Browser work was supported by the NHGRI award 5U41HG002371 (Corporate Sponsors). Dr. David Haussler is an investigator of the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or our corporate sponsors.
Footnotes
Editor’s note: This article has been peer-reviewed.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Data availability. Data are available from this project at the Toil xena hub (https://genome-cancer.soe.ucsc.edu/proj/site/xena/datapages/?host=https://toil.xenahubs.net).
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.V., A.A.R. and B.P wrote the manuscript. J.V., A.A.R., A.N., J.A., C.K., J.N., H.S., P.A., J.P., A.D.D., B.O. and B.P. contributed to Toil development. F.A.N. and A.M. contributed to Toil-Spark integration. J.V. wrote the RNA-seq pipeline and automation software. M.H. and C.B. contributed WDL and cloud support. P.A. and S.Z. contributed CWL support. J.Z., B.C. and M.G. hosted quantification results on UCSC Xena. K.R. hosted GTEx results in UCSC Genome Browser. W.J.K., J.Z., S.Z., G.G., D.A.P., A.D.J., M.C., D.H. and B.P. provided scientific leadership and project oversight.
References
- 1.Weinstein JN, et al. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, et al. Database. 2011 http://dx.doi.org/10.1093/database/bar026.
- 3.Siva N. Lancet. 2015;385:103–104. doi: 10.1016/S0140-6736(14)62453-3. [DOI] [PubMed] [Google Scholar]
- 4.McKenna A, et al. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UNC Bioinformatics. TCGA mRNA-seq pipeline for UNC data. 2013 https://webshare.bioinf.unc.edu/public/mRNAseq_TCGA/UNC_mRNAseq_summary.pdf.
- 6.Albrecht M, Michael A, Patrick D, Peter B, Douglas T. Proceedings of the 1st ACM SIGMOD Workshop on Scalable Workflow Execution Engines and Technologies (SWEET ’12); ACM (Association of Computing Machinery; 2012. p. 1. http://dx.doi.org/10.1145/2443416.2443417. [Google Scholar]
- 7.Bernhardsson E, Frieder E Luigi. Github. 2016 https://github.com/spotify/luigi.
- 8.Goecks J, Nekrutenko A, Taylor J. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.UCSC. Xena. 2016 http://xena.ucsc.edu.
- 10.Amstutz P. Common workflow language. Github. 2016 https://github.com/common-workflow-language/common-workflow-language.
- 11.Frazer S. Workflow description language. Github. 2014 https://github.com/broadinstitute/wdl.
- 12.Vivian J. Toil scripts. Github. 2016 https://github.com/BD2KGenomics/toil-scripts/tree/master/src/toil_scripts.
- 13.Apache Software Foundation. Apache Spark. http://spark.apache.org/ (2017)
- 14.Massie M, et al. ADAM: genomics formats and processing patterns for cloud scale computing. University of California; Berkeley: 2013. Technical Report No. UCB/EECS-2013-207. [Google Scholar]
- 15.Gentzsch W. Proceedings First IEEE/ACM International Symposium on Cluster Computing and the Grid. IEEE; 2001. pp. 35–36. http://dx.doi.org/10.1109/ccgrid.2001.923173. [Google Scholar]
- 16.Yoo AB, Jette MA, Mark G. Lecture Notes in Computer Science. Springer; Berlin, Heidelberg: 2003. pp. 44–60. [Google Scholar]
- 17.Apache Software Foundation. Apache Mesos. http://mesos.apache.org/
- 18.GTEx Consortium. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, et al. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Dewey CN. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray NL, Pimentel H, Melsted P, Pachter L. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 22.Barker AD, et al. Clin Pharmacol Ther. 2009;86:97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 23.Kent WJ, et al. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.