Summary
Immune cell function and fate is intimately linked to engagement of metabolic pathways. The contribution of core metabolic pathways to immune cell bioenergetics has been vigorously investigated in recent years. However, precisely how other peripheral metabolic pathways support immune cells beyond energy generation is less well understood. Here we survey the literature and highlight recent advances in our understanding of several ancillary metabolic pathways and how they support processes beyond ATP production and ultimately contribute to protective immunity.
Introduction
In union there is strength - Aesop
The immune system protects the host against infection and cancer, and it does so by coordinating cell activation and proliferation with effector functions such as cytokine secretion and cell-mediated cytotoxicity. A key feature of the immune system is the plasticity to an ongoing challenge that allows immune cells to adapt to changing circumstances. Interacting metabolic pathways underlie this plasticity and have been the focus of intense interest as therapeutic targets to harness the full potential of the immune system. Cellular metabolism is often oversimplified to the mere production of ATP or the maintenance of redox balance by controlling NADH and NAD+ levels. Glycolysis, fatty acid oxidation (FAO) and amino acid oxidation feed carbons into the tricarboxylic acid (TCA) cycle that in turn produces reducing equivalents in form of NADH (or FADH2). NADH donates electrons to the mitochondrial electron transport chain that is ultimately coupled to ATP production. However, the plasticity of the cell requires the metabolism to be flexible to meet the cellular demands not only in terms of energy, but also in terms of biosynthesis.
The involvement of core metabolic pathways in defining various immune functions has been extensively reviewed (O'Neill et al., 2016; Pearce and Pearce, 2013). We focus here on several ‘peripheral’ pathways that are linked to core metabolism and shape the complexity of the immune response (Figure 1), but have been less explored in immune cells. Here we highlight the immunological implications of pathways leading to the biosynthesis of polyamines, cholesterol, hexosamines, and nucleotides, with a unifying theme that the convergence of these pathways in immune cells establishes their plasticity, revealing that in metabolism, where there is union there is strength.
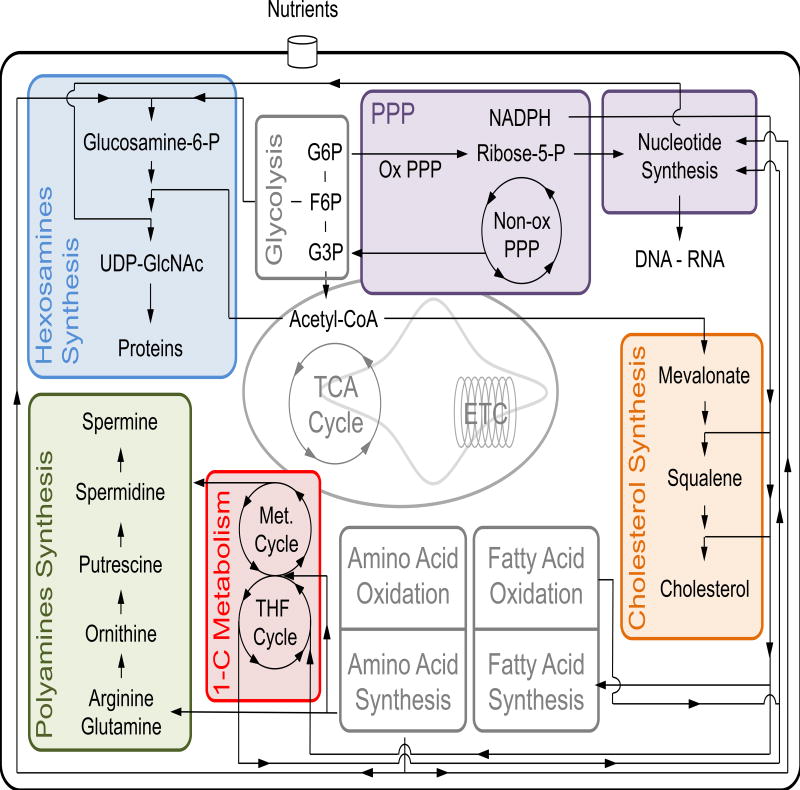
Figure 1. Ancillary metabolic pathways are intimately intertwined with core metabolism.
Core metabolic pathways (grey shaded) use most of the carbon equivalents derived from nutrients for the production of energy, to control redox balance and to generate biomass. The ‘peripheral’ pathways we describe in this review are also intertwined with core metabolism. In this figure we focus our attention on some of the documented interactions between ‘peripheral’ and core metabolic pathways. The pentose phosphate pathway (PPP, purple shaded) branches off glycolysis, feeds ribose-5-phosphate (ribose-5-P) to nucleotide synthesis and represents a source of reducing equivalents in the form of NADPH. NADPH is involved in fatty acid and cholesterol synthesis, and also enters the 1-carbon (1-C) metabolism to balance redox state. The 1-C metabolism (red shaded), together with amino acids and ribose-5-P, supports de novo nucleotide biosynthesis. Furthermore, the 1-C metabolism is the key source of S-adenosylmethionine required for spermidine and spermine synthesis (green shaded). The polyamine pathway also utilizes the amino acids arginine, glutamine and ornithine as precursors for polyamine synthesis. The production of hexosamines (blue shaded) integrates fructose-6-P, glutamine, nucleotides and acetyl-CoA to generate the amino sugar UDP-GlcNAc, involved in the post-transcriptional modification of proteins. Finally, acetyl-CoA is used to synthesize cholesterol (orange shaded) that together with its intermediates and derivatives coordinates intracellular signaling.
ETC: electron transport chain; F-6-P: fructose-6-phosphate; G-3-P: glycerol-3-phosphate; G-6-P: glucose-6-phosphate; Met: methionine; Non-ox PPP: non-oxidative branch of PPP; Ox PPP: oxidative branch of PPP; TCA: tricarboxylic acid; THF: tetrahydrofolate; UDP-GlcNAc: uridine diphosphate N-actetylglucosamine.
Polyamine Biosynthesis
Polyamines are highly conserved small cationic metabolites ubiquitously present in all eukaryotic cells. In mammals, polyamines comprise diamine putrescine, triamine spermidine and tetraamine spermine. Ornithine, the precursor to polyamines, is synthesised as part of the urea cycle where it is converted from arginine by arginase. Ornithine synthesis may also receive input from glutamine via the production of carbamoyl phosphate that subsequently feeds into the urea cycle, or from proline and glutamate through Δ1-Pyrroline 5-carboxylate that ultimately forms ornithine by way of ornithine acetyltransferase (OAT) (Figure 2). For polyamine synthesis, ornithine is converted to putrescine by ornithine decarboxylase (ODC), after which spermidine and spermine may be synthesised by spermidine synthase and spermine synthase, respectively (Figure 2). Positively charged at physiological pH, polyamines bind to acidic sites on proteins and nucleic acids (Pegg, 2016). By binding DNA, polyamines influence gene expression through multiple mechanisms (reviewed by (Childs et al., 2003)), including by controlling the expression of transcription factors such as Myc (Celano et al., 1989; Patel and Wang, 1997). Cellular uptake and efflux of polyamines is poorly understood. In mammals, there are endocytic and caveolin-regulated pathways for polyamine transport (Roy et al., 2008; Soulet et al., 2004) whilst members of the SLC superfamily of membrane transporters have also been implicated in polyamine influx (reviewed in (Abdulhussein and Wallace, 2014)). Proliferating cells synthesize polyamines and inhibition of this process retards cell growth, however precisely how polyamines promote cell growth remains unclear (Pegg, 2016). Intracellular polyamine levels are tightly controlled by the expression of the catabolic enzymes spermidine-spermine acetyltransferase (SAT1) and polyamine oxidase (PAO) while the biosynthetic enzymes face intricate regulation that strictly dictates their expression and activity. This level of control facilitates the highly inducible nature of the polyamine synthesis pathway that many immune cells engage during cell activation and differentiation.
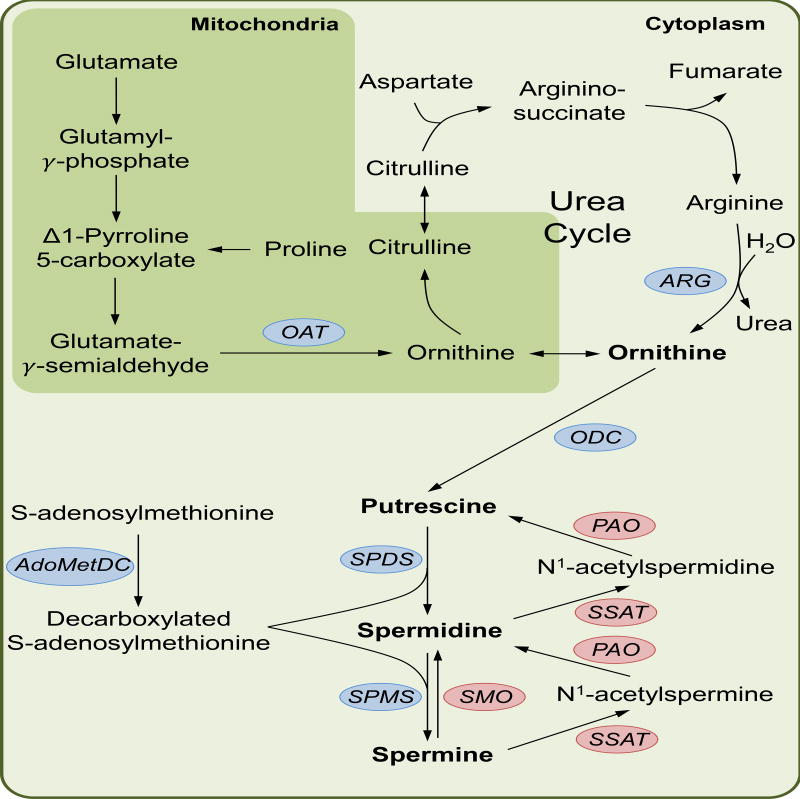
Figure 2. The polyamine biosynthesis pathway.
Polyamine synthesis proceeds with the decarboxylation of ornithine to form putrescine via the action of ornithine decarboxylase (ODC). Spermidine and spermine are then formed in step-wise fashion by spermidine synthase (SPDS) and spermine synthase (SPMS). Both SPDS and SPMS utilize decarboxylated S-adenosylmethionine, a product of S-adenosylmethionine decarboxylase (AdoMetDC), as a substrate for spermidine and spermine synthesis. Together with ODC, AdoMetDC comprise the rate-limiting steps of polyamine synthesis. During catabolism, spermine can be directly oxidized to spermidine by spermine oxidase (SMO). Induction of Spermidine/Spermine N1-acetyltransferase (SSAT), the rate-limiting step of polyamine catabolism, results in N1-acetylated forms of spermine and spermidine. Through the subsequent activity of polyamine oxidase (PAO) spermine can be converted to spermidine and spermidine to putrescine. The precursor to polyamine synthesis, ornithine, is synthesised by multiple routes. As an intermediate of the urea cycle, ornithine is synthesised from arginine by arginase. The urea cycle may also act as a vehicle to link the TCA cycle to polyamine synthesis as aspartate is used to form argininosuccinate that is subsequently combined with fumarate to make arginine. Arginine may also be taken up as a substrate and synthesised directly to ornithine. Both proline and glutamate can feed into ornithine synthesis through Δ1-Pyrroline 5-carboxylate that once catalyzed to Glutamate-γ-semialdehyde can be converted to ornithine by ornithine acetyltransferase (OAT). Finally, glutamine can also feed into the urea cycle via carbamoyl phosphate that enters the cycle at citrulline (not shown).
The bulk of polyamine research to date has been in cancer where upregulation of the polyamine synthesis pathway is known to drive cell proliferation (Casero and Marton, 2007). The requirement for polyamines in proliferating cells indicates that this pathway is likely to be engaged during lymphocyte activation and clonal expansion. This idea is supported by numerous older reports in T cells showing a direct link between polyamine synthesis and T cell proliferation (Bowlin et al., 1987; Kay and Pegg, 1973; Schall et al., 1991; Scott et al., 1985a). In naïve T cells, the activity and production of polyamine synthesis enzymes are maintained at low levels (Kay and Lindsay, 1973; Kay and Pegg, 1973), but this changes with T cell activation as ODC and S-adenosylmethionine decarboxylase activity, the rate limiting steps in polyamine synthesis, increase within hours of stimulation (Fidelus et al., 1984; Kay and Lindsay, 1973; Kay and Pegg, 1973; Scott et al., 1985b). Although the induction of this pathway early after T cell activation has been noted, few recent studies have tackled the role of polyamines in lymphocytes. Green and colleagues demonstrated using modern mass spectrometry techniques that increased levels of polyamines can be detected 24 hours post-activation in T cells and that ODC inhibition with difluoromethylornithine (DFMO) is sufficient to inhibit activation-induced proliferation (Wang et al., 2011). Together with previous observations, the kinetics of these data suggest that polyamines are important in T cell clonal expansion. How these metabolites control this process mechanistically requires further investigation. Regarding their production, reported data suggest that the synthesis of polyamines in T cells is under the direction of Myc (Wang et al., 2011). The ODC promoter is known to contain Myc consensus binding sequences (Packham and Cleveland, 1997) and ODC gene expression is regulated by Myc (Wagner et al., 1993). Indeed, Myc-deficient T cells fail to induce Odc and other polyamine synthesis genes leading to decreased polyamine production after activation (Wang et al., 2011).
A study by Monticelli et al. reports a pivotal role for arginine metabolism in group 2 innate lymphoid cells (ILC2s) (Monticelli et al., 2016). In the absence of Arg1, lung ILC2s fail to proliferate in vivo during inflammation. Interestingly, a significant proportion of arginine is metabolized to spermidine in ILC2s, hinting at a role for spermidine in driving ILC2 proliferation. Of note, the scavenging of arginine from extracellular environments by cells such as myeloid-derived suppressor cells (MSDCs), macrophages and tumor cells limits T cell proliferation. It would be interesting to explore the role of polyamines within this paradigm and whether arginine scavenging is in fact a ploy to restrict polyamine synthesis that appears to be an absolute requirement for T cell expansion. Therefore, while polyamine inhibition has thus far failed as a chemotherapeutic, it might yield success in controlling immune cell proliferation in the context of autoimmunity.
In macrophages, the functional importance of polyamines is also unclear. In the steady state, polyamine levels are thought to be relatively low (Van den Bossche et al., 2012) but IL-4 is likely to induce polyamine synthesis through its ability to augment Arg1, suggesting that polyamines might be particularly important for macrophage alternative activation (herein referred to as M2). Reports that polyamines have anti-inflammatory effects on macrophages may intimate that polyamine synthesis acts as a metabolic regulator of macrophage differentiation, restraining classical activation (M1), while promoting M2 formation. In support of this, LPS-induced expression of TNF, IL-1, IL-6 and CD80 is dampened by polyamines (Yang et al., 2016a). Similarly, both spermine and spermidine can suppress LPS-induced IL-12 production and nitric oxide formation in mouse macrophages (Bussiere et al., 2005; Chaturvedi et al., 2010; Hasko et al., 2000; Szabo et al., 1994; Yang et al., 2016a). In light of these findings, polyamine production in M1 cells might function to temper inflammation and promote resolution. Hardbower et al. have addressed this question using Odcflox/flox LysM-Cre mice (Hardbower et al., 2017). During H.pylori and C. rodentium infection Odcflox/flox LysM-Cre mice display increased gastritis and colitis, respectively, and enhanced M1 activation. These observations add weight to the idea that ODC expression and polyamine production restrain M1 responses during infection to limit inflammation. In M2 macrophages, arginase is upregulated along with ornithine production (Jha et al., 2015), begging the question of functional relevance of polyamines in this subset. Pharmacological depletion of polyamines in vitro has a significant effect on the phenotype of IL-4-treated macrophages (Van den Bossche et al., 2012) in which they fail to upregulate signature M2 genes. In vivo, spermidine treatment can alleviate experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis, and this effect was partially dependent on spermidine’s ability to promote Arg1 in macrophages, another prototypic M2 marker. Macrophages from spermidine-treated EAE mice could confer the protective effect when adoptively transferred into new EAE mice (Yang et al., 2016a). However, bone marrow-derived macrophages from Odcflox/flox LysM-Cre mice stimulated with IL-4 show enhanced expression of the M2 markers Arg1 and Ym1 (Hardbower et al., 2017), leaving the status of polyamines in alternative activation unclear.
In line with a generally suppressive role for polyamines in the myeloid lineage, recent work from Grohman and colleagues reports an important function for spermidine in conferring immunomodulatory properties to dendritic cells (DCs) (Mondanelli et al., 2017). Specific subsets of immune cells express indoleamine 2,3-dioxygenase 1 (IDO1), an immunosuppressive enzyme that degrades tryptophan, an amino acid essential for effective T cell responses. In TGFβ-stimulated DCs, IDO1 activity requires Arg1-dependent synthesis of spermidine that in turn induces the activation of Src kinase – a factor important for IDO1 function. Remarkably, MSDCs could confer an IDO1-dependent immunosuppressive phenotype to DCs in a cell contact-independent manner through their release of polyamines into the extracellular milieu (Mondanelli et al., 2017).
Polyamines are also critical for the replication of Zika and Chikungunya virus through their contribution to viral RNA transcription and translation. A noteworthy observation of this work was the induction of the polyamine degradation enzyme SAT1 by type I interferon as a novel anti-viral response (Mounce et al., 2016). This raises the possibility that polyamine metabolism in the immune system might be under cytokine control.
Cholesterol biosynthesis
Cholesterol is well-known for its role in the progression of cardiovascular disease, however, cholesterol is also an essential component of biological membranes as it regulates fluidity and allows organization of transmembrane proteins into signaling platforms named lipid rafts (Lange et al., 1989). Cholesterol and the intermediates of its biosynthesis are key for the generation of steroid hormones, vitamins, bile acids and for protein prenylation, a post-translational modification that tethers proteins to biological membranes. Cholesterol is synthesized starting from the precursor acetyl-CoA. The enzyme HMG-CoA reductase uses acetyl-CoA and NADPH to generate mevalonate, the committing step of the pathway. Mevalonate undergoes several phosphorylation steps that generate activated isoprenes, later condensed to form the 30-carbon molecule squalene. A series of reactions allows cyclization of squalene to form the four fused rings characteristic of cholesterol and other sterols (Figure 3).
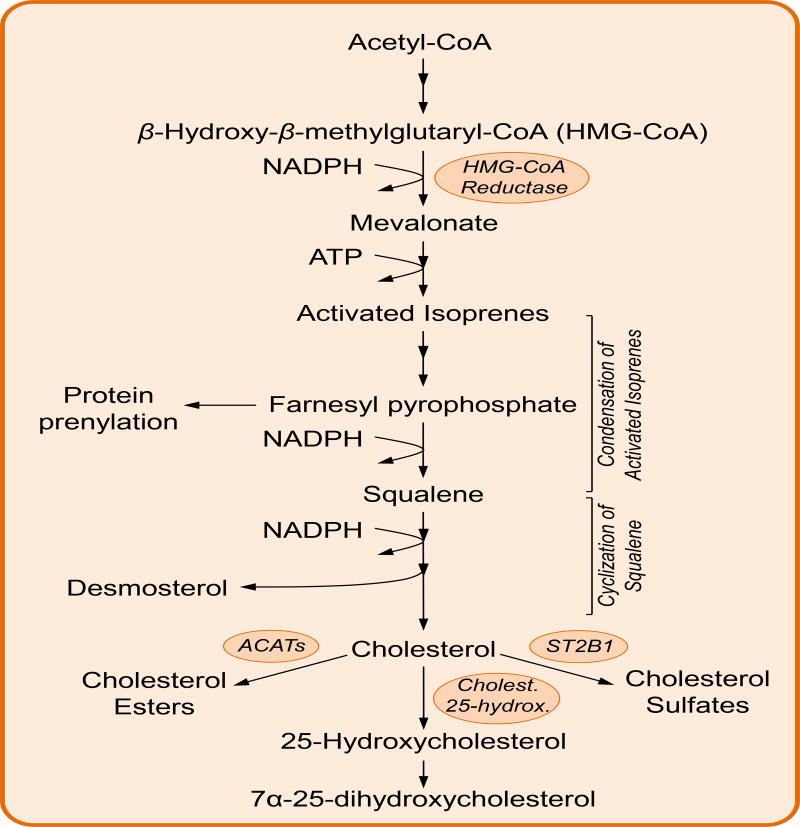
Figure 3. The cholesterol biosynthesis pathway.
HMG-CoA reductase combines 3 molecules of acetyl-CoA with NADPH to synthesize mevalonate, the precursor of cholesterol biosynthesis. Several steps of phosphorylation generate activated isoprenes, later condensed to form squalene. Cyclization of squalene occurs in several steps that finally generate the four fused rings characteristic of cholesterol and other sterols. Intermediates of cholesterol biosynthesis, such as farnesyl pyrophosphate are used to post-translational modify proteins (ie: protein prenylation). Similarly, many cholesterol derivatives such as cholesterol esters, hydroxycholesterols and cholesterol sulfates play important roles in modulating the immune function.
Intracellular cholesterol levels are tightly controlled by the opposite action of two transcription factors, sterol regulatory element-binding protein 2 (SREBP2) and liver-X receptor (LXR). Low cholesterol levels activate SREBP2 that controls expression of HMG-CoA reductase and the enzymes involved in cyclization of squalene (Jeon and Osborne, 2012). Conversely, high cholesterol levels activate LXR, which coordinates cholesterol efflux via ABC transporters, or cholesterol esterification for intracellular storage by acyl-CoA cholesterol acyl transferases (ACATs).
The importance of cholesterol regulation is evident in activated T cells, which reprogram their metabolism by simultaneously inducing SREBP2 and repressing LXR signalling (Bensinger et al., 2008; Kidani et al., 2013). T cell progression into the cell cycle is sustained by SREBP2 activation and cholesterol biosynthesis, whereas LXR signaling inhibition is mediated by ST2B1 (encoded by Sult2b1) that degrades LXR agonists. Down-modulation of cholesterol biosynthesis leaves T cell activation unaffected, but compromises cell cycle progression and metabolic reprogramming of activated T cells. Mice deficient for the chaperone SCAP, which mediates processing and activation of SREBPs in the endoplasmic reticulum (ER), fail to engage glycolysis and OXPHOS and are susceptible to viral infection (Kidani et al., 2013). Modulation of T cell responses by cholesterol also occurs at the level of T cell receptor (TCR) signaling. TCRs accumulate in lipid rafts where they interact with cholesterol that keeps the TCRs in nanoclusters, enhancing avidity (Molnar et al., 2012). At the same time, cholesterol maintains the TCR in an inactive status that blocks downstream signal transduction (Swamy et al., 2016a). These seemingly contrasting outcomes may be reconciled by the fact that cholesterol enhances TCR nanoclustering and sensitivity to agonistic ligands, but restrains spontaneous signalling. Memory T cells have more lipid rafts than naïve T cells, and this feature may potentially underlie the increased sensitivity of memory T cells upon antigen re-encounter. The naturally occurring cholesterol sulphate instead represses TCR signaling by replacing cholesterol and disrupting TCR nanoclustering. The level of cholesterol sulfate varies during T cell development, suggesting that cholesterol metabolism may fine tune T cell development by modulating TCR signalling (Wang et al., 2016). Pharmacological manipulation of cholesterol metabolism can increase the anti-tumor potential of CD8 T cells. Inhibition of ACAT1 and cholesterol esterification promotes accumulation of cholesterol in the cell membrane, sustaining TCR clustering and signaling. In turn, CD8 T cells better polarize cytotoxic granules to the immunological synapse and show enhanced anti-tumor activity (Yang et al., 2016b). The importance of cholesterol metabolism in immune responses is highlighted by the fact that tumors themselves use sterols to hi-jack the immune response. Tumors secrete ligands of LXR that suppress dendritic cell migration to the lymph nodes and the anti-tumor response (Villablanca et al., 2010).
The many intermediates and derivatives of cholesterol biosynthesis have different structural features that allow them to have different functions. Intermediates of cholesterol biosynthesis bind to the transcription factor RORγt to control differentiation of IL-17-producing T helper cells (Th17) (Hu et al., 2015; Santori et al., 2015). In contrast, LXR activation inhibits Th17 development via a mechanism involving competition between SREBP1 and AhR, a transcription factor that is known to sustain Th17 differentiation (Cui et al., 2011). The development of Th17 is tightly linked to the development of regulatory T cells (Treg) and the engagement of metabolic pathways underlies this fate decision. mTOR activation supports glycolytic flux and together with de novo fatty acid synthesis promotes the development of Th17 (Berod et al. 2014). Although being a negative regulator of de novo Treg differentiation, mTOR signalling coordinates cholesterol and lipid metabolisms to sustain the suppressive function of Treg (Zeng et al. 2013). The cholesterol derivative 7α,25-hydroxycholesterol influences the spatial coordination of immune responses by binding to the G protein-coupled receptor EBI2 and directly positioning B cells close to cognate T cells, thus favoring their interaction and an effective response (Hannedouche et al., 2011) (Liu et al., 2011).
The antiviral response further underscores the relationship between cholesterol homeostasis and immune responses. Viral infection, as well as interferon stimulation, induces STAT1-dependent transcription of Ch25h, encoding cholesterol 25-hydroxylase that generates 25-hydroxycholestrol (25-HC) (Blanc et al., 2013; 2011). The antiviral activity of 25-HC ranges from the alteration of the membrane lipid composition to impair viral entry, to inhibition of SREBP2 processing and activity (Liu et al., 2013; Radhakrishnan et al., 2007). Since viruses must rely on cellular metabolism to meet their needs, it was thought that alteration of cholesterol biosynthesis and nutrient availability may affect viral life cycle. However recent work suggests that reduction of cholesterol biosynthesis directly drives the anti-viral response via the ER-bound signalling protein STING rather than decreasing nutrient availability to the virus (York et al., 2015). Upon viral infection, reduction of de novo cholesterol synthesis is indeed coupled to enhanced exogenous uptake, and total intracellular levels are unaffected. However, despite being chemically identical, de novo-synthesized and exogenous cholesterol follow different intracellular trafficking routes. Therefore, depletion of endogenous cholesterol in the ER may drive the antiviral response, while import from the extracellular space provides the cell with the cholesterol required for other functions. Interestingly, 25-HC also inhibits inflammasome activation and IL-1β secretion. Mice lacking Ch25h show an overall pro-inflammatory phenotype where they are more susceptible to LPS-induced sepsis and EAE, while controlling pathogen burden better (Reboldi et al., 2014). Could the concomitant dual role of 25-HC contribute to the occurrence of bacterial co-infection during viral invasion?
Signals through immune receptors such as Toll-like receptors (TLR) may interfere with the crosstalk between SREBP2/LXR signals in innate immune cells. TLR engagement blocks LXR signaling and reduces expression of the ABC transporters that control cholesterol efflux (Castrillo et al., 2003). In a model of systemic inflammation, TLR agonists impaired the reverse cholesterol transport, a system used to clear excess cholesterol from the tissues (McGillicuddy et al., 2009). Of note, cholesterol crystals found in atherosclerotic lesions and scavenged by macrophages activate the inflammasome and may underlie the ability of macrophages to effect the inflammatory response in atherosclerosis (Duewell et al., 2010). Furthermore, the cholesterol synthesis intermediate desmosterol accumulates in cholesterol-loaded macrophages, binds LXR and exerts anti-inflammatory effects (Spann et al., 2012). These data provide insight into the interplay between inflammation and cholesterol metabolism during the development of atherosclerosis, and highlight the requirement for exogenous cues to break homeostasis and drive the pro-inflammatory phenotype of macrophages.
Hexosamine Biosynthesis
While most glucose that enters the cell is metabolized through glycolysis, this sugar is also used in an essential off-shoot of the glycolysis pathway known as the hexosamine biosynthesis pathway (HBP). The HBP diverges from glycolysis when fructose 6-phosphate is converted to glucosamine 6-phosphate by the HBP rate-limiting enzyme glutamine fructose-6-phophate amidotransferase (GFAT) (Ferrer et al., 2016). Following incorporation of nitrogen from glutamine, nucleotides and acetyl-CoA, the primary product of the HBP is the amino sugar uridine diphosphate N-acetylglucosamine (UDP-GlcNAc). UDP-GlcNAc is utilised by cells to mediate the reversible addition of O-linked β-N-acetyl glucosamine (O-GlcNAc) – a protein modification known as O-GlcNAcylation (Bond and Hanover, 2015) (Bond and Hanover, 2015) (Figure 4). As many as 4,000 proteins are thought to be targets for O-GlcNAcylation (Ma and Hart, 2014), but remarkably this modification is mediated by a single enzyme, O-GlcNAc transferase (OGT), that catalyzes O-GlcNAc addition to serine and threonine residues using UDP-GlcNAc as a substrate. In parallel, O-GlcNAc is removed by the action of O-GlcNAcase (OGA). While O-GlcNacylation occurs independently on many proteins, there is extensive cross-talk with protein phosphorylation. O-GlcNAcylation often occurs at the site of, or proximal to, the same serine and threonine residues modified by kinases (Hart et al., 2011) and this competition permits a dynamic interplay that can alter signaling and protein function (Bond and Hanover, 2015). Although the implications of O-GlcNAcylation in immunity are only just emerging, its biological significance is highlighted in the embryonic and perinatal lethality of OGT and OGA deletion in mice (Shafi et al., 2000; Yang et al., 2012). High expression levels of these enzymes observed in immune cells (Bond and Hanover, 2015) likely reflects their potential to rapidly modify protein activity during cellular activation and differentiation. One such example of this is the O-GlcNAcylation of NF-κB subunit c-Rel that is required for its DNA binding activity and subsequent cytokine expression in activated T cells (Ramakrishnan et al., 2013). O-GlcNAcylation inhibition in this context is sufficient to repress IL2 and IFNG expression following TCR stimulation. O-GlcNAcylation of STAT3 exerts an inhibitory effect on STAT3 phosphorylation resulting in loss of IL-10 production in macrophages (Li et al., 2017), showing how O-GlcNAc modifications can control important transcriptional programs in immune cells.
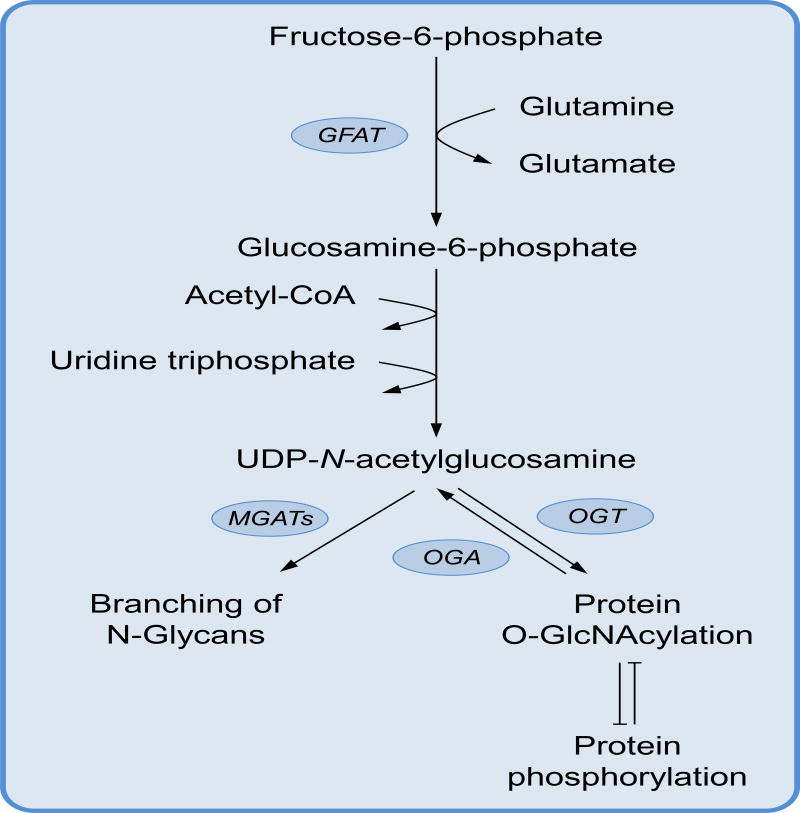
Figure 4. The hexosamines biosynthesis pathway.
The hexosamine biosynthesis pathway is an off-shoot of glycolysis that starts with the conversion of fructose-6-phosphate and the amino group donor glutamine to glucosamine-6-phosphate. Glucosamine-6-phosphate is then linked to the nucleotide UTP and N-acetylated to form the bioactive molecule UDP-N-acetylglucosamine. The enzymes O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) catalyse addition or removal of the post-translational modification O-GlcNAcylation. O-GlcNAcylation and phosphorylation often compete for the same amino acid residues and their competition influence protein stability and function. On the other hand, the enzymes MGATs use UDP-N-acetylglucosamine to modify the branching of N-glycans, a post-translational modification of surface proteins that regulates cell membrane dynamics.
In T cells, glucose and glutamine are essential for T cell activation. However, these metabolites also contribute to UDP-GlcNAc formation that is required throughout T cell development and activation. An increase in O-GlcNAc is an early event in T cell activation that supports proliferation and differentiation as genetic or pharmacological ablation of OGT blunts these processes (Lund et al., 2016; Swamy et al., 2016b). Interestingly, accumulation of OGT has been observed at the immunological synapse suggesting the O-GlcNAcylation machinery is polarised to the TCR during activation to augment signaling during priming (Lund et al., 2016). This is supported by a global analysis of the O-GlcNAc glycoproteome in activated human T cells that found key downstream components of the TCR, such as Lck and ZAP-70, O-GlcNAc-modified (Lund et al., 2016). O-GlycNAcylation in activated cells is controlled by Myc as Myc−/− T cells fail to increase protein O-GlycNAcylation. This regulation functions in a feedback loop as O-GlcNAcylation of Myc inhibits its phosphorylation thereby preventing proteolytic degradation (Swamy et al., 2016b). How O-GlycNAcylation impacts T cells at later stages of differentiation requires further exploration, although O-GlcNAcylation can modulate mTOR and autophagy (Ferrer et al., 2016), both of which regulate memory T cell formation (Araki et al., 2009; Puleston et al., 2014; Xu et al., 2014; Schlie et al., 2015).
HBP-derived UDP-GlcNAc can also provide a substrate for the branching of N-glycans - a post-translational modification found on surface glycoproteins. Most transmembrane receptors on mammalian cells are modified by N-glycosylation en route to the cell surface. GlcNAc-branching of N-glycans is mediated by the N-acetylglucosaminyltransferases I, II, IV and V (also known as Mgat1, 2, 4a/b, and 5) through the sequential transfer of GlcNAc from UDP-GlcNAc. Once at the cell surface, glycoproteins bind galectins through their N-glycan modifications to form a molecular lattice that negatively regulates lateral movement and endocytosis of surface receptors. The affinity of N-glycans on glycoproteins for surface galectins is directly proportional to the level of GlcNAc branching and the number of attached N-glycans (Chen et al., 2009).
In macrophages, high levels of UDP-GlcNAc are linked to upregulation of N-glycan synthesis enzymes during M2 polarisation (Jha et al., 2015). Although inhibition of N-glycosylation only mildly affects M1 polarization, it is sufficient to significantly impede the M2 program, further highlighting the functional importance of the HBP in macrophages. In T cells, large variations in N-glycan branching are observed throughout thymic development (Zhou et al., 2014). Deleting Mgat1, and thus N-glycan branching, was found to influence positive selection at both the lower and upper affinity boundaries through differential regulation of the TCR and CD4 and CD8 coreceptors. CD4 and CD8 N-glycan branching enhances their surface retention and recruitment of Lck to the TCR, increasing the sensitivity of the TCR to low-affinity peptide-MHC. In parallel, branching limits negative selection driven by Ca2+ flux in response to high affinity peptide-MHC (Zhou et al., 2014). In peripheral T cells, N-glycan branching is induced by signals from the TCR and regulates T cell activation and expansion (Chen et al., 2009). The TCR contains a relatively high number of N-glycan modifications that in resting cells promotes strong interactions with galectins that preclude spontaneous TCR oligomerization in the absence of antigen (Chen et al., 2007). During clonal expansion branching increases, enhancing the affinity of the inhibitory receptor CTLA-4 for galectins, thereby stabilising it at the cell surface and reducing receptor endocytosis (Lau et al., 2007). Thus, N-glycan branching regulates T cell responses by modulating the threshold for activation in resting cells and the activity of CTLA-4 in proliferating cells.
Induction of aerobic glycolysis and fatty acid oxidation (FAO) are needed for in vitro Th17 and Treg formation, respectively. During Th17 differentiation, T cells will polarise toward Tregs if glycolysis is inhibited, but inhibiting FAO blocks Treg induction and favors Th17 cells (Michalek et al., 2011; Shi et al., 2011). While the balance between Th17 and Treg cells is metabolically linked, a comprehensive understanding of this paradigm is lacking. A recent study by Demetriou and colleagues has implicated UDP-GlcNAc and N-glycan branching in this process (Araujo et al., 2017). During Th17 differentiation, the engagement of aerobic glycolysis and glutaminolysis starves the HBP of substrates resulting in a drop in GlcNAc branching of N-glycans. Reduced branching decreases CD25 expression and concomitant IL-2R signalling through STAT5 – an inhibitory event for Th17 bias, but one required for Treg formation. Remarkably, exogenous GlcNAc or overexpression of the branching enzyme Mgat5 biased polarisation toward FoxP3+ Treg formation even in the presence of Th17-differentiating cytokines. These observations place the HBP at the fulcrum of Th17/Treg balance and provide mechanistic insight into the metabolic relationship between Th17 and Treg cells.
The pentose phosphate pathway, NADPH and nucleotide biosynthesis
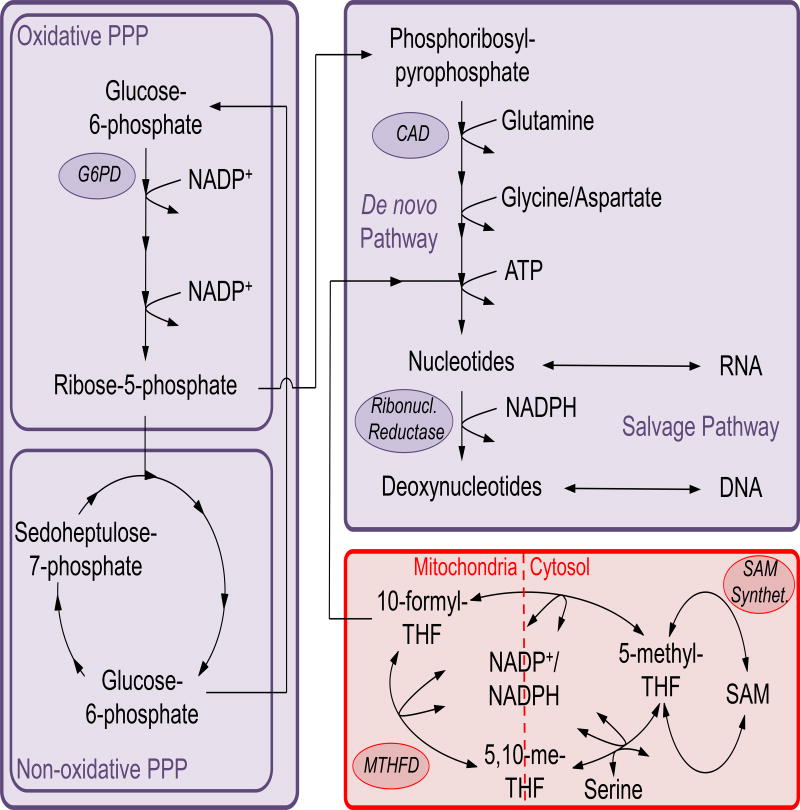
Immune cells rapidly activate in response to breakdown of tissue homeostasis. Cell activation results in proliferation and acquisition of effector functions that require nucleotides to synthesize RNA and DNA, or to coordinate intracellular and extracellular signaling. Biosynthesis of nucleotides requires ribose-5-phosphate, produced upon shunting of glucose from glycolysis to the pentose phosphate pathway (PPP). The PPP is divided into oxidative and non-oxidative arms (Stincone et al., 2015). Oxidative reactions produce ribose-5-phosphate, oxidized from glucose-6-phosphate generated during glycolysis. Ribose-5-phosphate can then be used in nucleotide biosynthesis or in the non-oxidative arm of the PPP. The oxidative PPP also reduces NADP+ to NADPH, which is critical for the biosynthesis of lipids and cholesterol as well as the regeneration of the antioxidant glutathione, and is also used as an electron carrier in folate metabolism that eventually feeds into nucleotide and polyamine biosynthesis. On the other hand, the non-oxidative PPP consists of a series of enzymatic reactions that convert the pentose phosphates produced by the oxidative reactions to hexose phosphates that can recycle back into the oxidative PPP, forming a loop that sustains NADPH production. Thus the PPP generates reducing equivalents in form of NADPH, in addition to generating ribose-5-phosphate moieties (Figure 5).
Figure 5. The pentose phosphate pathway, nucleotide biosynthesis and one-carbon metabolism.
The pentose phosphate pathway (PPP) is divided into an oxidative and a non-oxidative arm. In the former, the glycolytic intermediate glucose-6-phosphate is oxidized to ribose-5-phosphate that can then feed the non-oxidative PPP. The non-oxidative PPP converts ribose-5-phosphate to glucose-6-phosphate that re-enters the oxidative PPP and sustains a cycle aimed at regenerating NADP+ to NADPH. Ribose-5-phosphate can otherwise fuel the de novo pathway of nucleotide biosynthesis to generate the building blocks for DNA replication and RNA transcription. Quiescent cells mostly use the salvage pathway of nucleotide regeneration that recycles senescent DNA and RNA to feed the nucleotide pool. The one-carbon metabolism is compartmentalized between cytosol and mitochondria and provided essential carbon equivalents for purine and pyrimidine biosynthesis. The THF cycle is the entry point of the essential amino acid serine to sustain proliferation of activate T cells.
T cells proliferate in response to activating stimuli to expand their effector potential. T cell expansion requires de novo generation of biomass and, indeed, activated T cells shunt carbon equivalents toward the PPP. Inhibition of glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the PPP, blocks T cell proliferation (Wang et al., 2011). The transcription factor Myc is partially responsible of this effect as Myc deletion reduces carbon flux toward the PPP, reduces accumulation of nucleotides and impairs up-regulation of the PPP enzymes G6PD and transketolase. mTORC1 contributes to the metabolic reprogramming driving PPP engagement since it activates the transcription factors SREBP1 and 2 that control expression of G6PD (Düvel et al., 2010). Of note, mTORC1 links the activation of the PPP to cholesterol biosynthesis via the induction of SREBPs, while the PPP provides reducing equivalents in the form of NADPH for cholesterol biosynthesis. The reduced form of glutathione (GSH), regenerated during the oxidative PPP, has recently emerged to be essential for T cell effector function. Deficiency of GSH does not affect early T cell activation. However, GSH buffers the reactive oxygen species produced during T cell activation, supports mTOR and NFAT activity and regulates Myc-dependent metabolic reprogramming of activated T cells (Mak et al. 2017).
A study investigating the metabolic reprogramming of macrophage polarization found that CARKL, the sedoheptulose kinase, is a rheostat modulating carbon flux into the PPP (Haschemi et al., 2012). CARKL phosphorylates sedoheptulose to sedoheptulose-7-phosphate (an intermediate of the non-oxidative PPP) and by a simple mass-action limits the carbon flux through the PPP, favoring its utilization toward pyruvate oxidation and entry into the TCA cycle. LPS-driven M1 activation, known to promote glycolysis rather than oxidative phosphorylation, blunts CARKL expression and induces carbon flux toward the PPP. PPP engagement was also highlighted by integrative analysis of RNA and metabolic profiles in LPS-polarized macrophages, further supporting the role of the PPP in defining the M1 polarization (Jha et al., 2015; Tannahill et al., 2013). Similarly to macrophages, LPS-activated DCs engage the PPP to regenerate NADPH and support fatty acid synthesis that in turn sustains the anabolic demands of cell activation (Everts et al., 2014).
Pyruvate kinase M2 (PKM2) is important in the metabolic control of immune cell differentiation, by regulating the carbon flux toward the PPP. The highly active tetrameric form of PKM2 converts phosphoenolpyruvate (PEP) to pyruvate and directs carbons to the TCA cycle. Instead, LPS-activated macrophages and proliferating cells, amongst which include activated lymphocytes and cancer cells, present PKM2 in a dimeric form with low affinity for PEP. The lower rate of conversion of PEP to pyruvate leads to accumulation of glycolytic and PPP metabolites, thus favoring PPP engagement and biosynthetic processes. Moreover, dimeric PKM2 can also translocate to the nucleus to control, in concert with HIF-1α, the expression of glycolysis-related enzymes (Luo et al., 2011; Palsson-McDermott et al., 2015).
Instead of being recycled between the non-oxidative and the oxidative PPP to sustain NADPH production, Ribose-5-phosphate can also exit the PPP and funnel into nucleotide biosynthesis (Figure 5). Ribose-5-phosphate is phosphorylated to phosphoribosyl-pyrophosphate that is used together with amino groups (NH4+) from glutamine, and amino acids to generate nucleotides. This de novo pathway is complex and energy-expensive and used by actively proliferating cells, such as T cells; resting cells instead use the salvage pathway that recycles free bases and nucleosides released from nucleic acid breakdown (Fairbanks et al., 1995). Nucleotide biosynthesis inhibition has obvious effects on cell proliferation and effector functions (Turka et al., 1991), however the underlying mechanisms are often unexpected. For instance, pyrimidine nucleoside deoxythymidine triphosphate (dTTP) levels, recycled via the salvage pathway, must be tightly controlled to allow development of haematological lineages in the bone marrow. Elevated levels of dTTP arrest cell cycle progression by allosteric inhibition of ribonucleotide reductase that impairs generation of deoxyribonucleotides and eventually DNA synthesis (Austin et al., 2012). Glutaminolysis catabolizes glutamine to feed the TCA cycle and support several biosynthetic pathways, among which are polyamines and hexosamines. Of note, glutaminolysis provides amino groups to de novo nucleotide biosynthesis, and is coupled to Myc transcriptional activity. Myc deficiency affects carbon flux through glutaminolysis and polyamine and nucleotide accumulation upon T cell activation (Wang and Green, 2012).
Intermediates of the one-carbon metabolism are major carbon sources that support the biosynthesis of purines and pyrimidines (Ducker and Rabinowitz, 2017). The core of the one-carbon metabolism consists of two interconnected cycles, the tetrahydrofolate (THF) cycle and the methionine cycle. The THF cycle is centred around three active molecules with distinct redox states and bioactivities. 5,10-methylene-THF regulates the balance between the pyrimidines deoxyuridine monophosphate (dUMP) and deoxythymidine monophosphate (dTMP) and it is the entry point for the non-essential amino acid serine, a major carbon donor to the pathway. 10-formyl-THF is the most oxidized form of folate and is required for de novo synthesis of purines. In proliferating cells, the demand of purines pushes the THF cycle to funnel carbon equivalent into 10-formyl-THF that feeds the purinosome, an enzymatic complex containing all the enzymes required to support purine synthesis. 5-methyl-THF is the most reduced form of folate and links the THF cycle to the methionine cycle. 5-methyl-THF is used to methylate homocysteine to methionine that is the substrate of S-adenosyl-methionine (SAM) synthetase. SAM is the most important methyl carrier in mammalian cells and has a role in DNA methylation, phospholipid biogenesis and polyamine synthesis. The efficiency of the THF cycle is strongly coupled to cellular redox balance. The THF cycle runs both in the cytosol and in the mitochondria where, respectively, NADP+/NADPH and NAD+/NADH ratios are used to balance the amount of 5,10-methylene-THF, 10-formyl-THF and 5-methyl-THF to drive distinct biosynthetic processes (Figure 5).
Recent work highlights the central role of one-carbon metabolism for nucleotide biosynthesis in T cells. Activated T cells increase serine uptake that is metabolized through the THF cycle to generate purines. Tracing experiments show that the THF and the methionine cycles are not interconnected in activated T cells as serine-derived carbons do not enter the methionine cycle but they are only used to feed the purine pool. T cell proliferation is reduced upon serine starvation, whereas cytokine production and the overall bioenergetic output are unaffected. Bypassing serine deficiency by feeding cells with formate and glycine, by-products of serine metabolism through the THF cycle, rescues the proliferation defect (Ma et al., 2017). Furthermore, the work of Ron-Harel et al. points out that the mitochondrial branch of the THF cycle modulates T cell activation. Activated T cells rewire the mitochondrial proteome to augment the flux through the THF cycle. Failure to engage the mitochondrial THF cycle impairs the purine pool and mitochondrial redox balance. Failure to regenerate NADP+ to NADPH depletes the reserve of reduced glutathione and results in DNA damage and increased cell death that is rescued by providing cells with formate and the anti-oxidant N-acetyl cysteine (Ron-Harel et al., 2016). Cell activation licenses mTORC1 to drive accumulation of both pyrimidines and purines. mTORC1 controls pyrimidine synthesis via the S6K1-dependent phosphorylation of the rate limiting multi-domain enzyme of pyrimidine biosynthesis, CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase) (Ben-Sahra et al., 2013). On the other hand mTORC1 modulates the purine pool by directly affecting the flux through the THF cycle; indeed, mTORC1 controls expression of MTHFD2 that keeps the balance between 10-formyl-THF and 5,10-methylene-THF (Ben-Sahra et al., 2016).
Metabolic plasticity allows cells to adapt to the surrounding environment and the available nutrients. Carmeliet and colleagues showed that fatty acid oxidation (FAO), a core metabolic pathway involved in energy production, provides carbon equivalents to nucleotide biosynthesis. The rate-limiting enzyme of FAO is carnitine palmitoyltransferase 1 (encoded by CPT1a), whose deficiency in endothelial cells impairs levels of TCA intermediates and TCA cycle-derived amino acids. Among these, aspartate and glutamate are involved in purine and pyrimidine synthesis, and CPT1a-deficient cells show a reduced pool of nucleotides and defective proliferation (Schoors et al., 2015). Although FAO has not been shown to feed nucleotide biosynthesis in T cells in vitro, it is feasible that immune cells may remodel their metabolism in response to the changing environment and use fatty acids as a rich carbon source to synthetize nucleotides in vivo.
Conclusion
The field of immunometabolism has elucidated many of the ways in which specific metabolic pathways are linked to fate and function in immune subsets. Understandably, much of the focus has been on the major pathways that drive energy production in cells, such as glycolysis and fatty acid oxidation. The aim of this review is to bring attention to the wider metabolic networks that operate in the immune system. Although the metabolic demand of these pathways may be small in terms of energy supply, they have a significant impact on cell function. However, many questions remain on the importance of these ‘peripheral’ networks in immunity (Box 1). There are many ancillary pathways, such as the urea cycle, branched chain amino acids catabolism, and ammonia metabolism, which we have not described here and have yet to be fully explored in immune cells. Finally, elucidating precisely why cells engage particular metabolic pathways, be it for energy, redox balance, biosynthesis, or protein modification, will be a focus of ongoing work.
Box 1. Outstanding questions in ancillary immunometabolism.
How are polyamines integrated into the metabolic and gene expression network of immune cells?
The spermidine-dependent translation factor eIF5a is one of the most expressed proteins in activated T cells (Hukelmann et al., 2015). What is its role in immunity?
Intermediates of cholesterol biosynthesis can bind nuclear receptors like RORγt, can other metabolites moonlight as ligands or modulators for other immunologically relevant receptors and transcription factors?
Key metabolic regulators such as AMPK and Myc can be O-GlcNAcylated but how O-GlcNAc modifications facilitate dynamic changes in immune cell metabolism is yet to be fully explored.
How do pathogens hi-jack immune cell metabolism to evade the immune response?
In this review article, Puleston et al. highlight the role of auxiliary metabolic pathways in immune cell function. The biosynthesis of polyamines, cholesterol, hexosamines, and nucleotides not only supports core metabolism, but also critically influences the immune response.
Acknowledgments
The work was supported by a Sir Henry Wellcome Fellowship awarded by The Wellcome Trust to D.J.P, by NIH grant NCI CA18125 to E.L.P, and by the Max Plank Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdulhussein AA, Wallace HM. Polyamines and membrane transporters. Amino Acids. 2014;46:655–660. doi: 10.1007/s00726-013-1553-6. [DOI] [PubMed] [Google Scholar]
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo L, Khim P, Mkhikian H, Mortales C-L, Demetriou M. Glycolysis and glutaminolysis cooperatively control T cell function by limiting metabolite supply to N-glycosylation. Elife. 2017;6:e21330. doi: 10.7554/eLife.21330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin WR, Armijo AL, Campbell DO, Singh AS, Hsieh T, Nathanson D, Herschman HR, Phelps ME, Witte ON, Czernin J, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. Journal of Experimental Medicine. 2012;209:2215–2228. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de Novo Pyrimidine Synthesis by Growth Signaling Through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, Sandouk A, Hesse C, Castro CN, Bähre H, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nature Medicine. 2014;20:1237–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, Shui G, Lacaze P, Watterson S, Griffiths SJ, Spann NJ, et al. The Transcription Factor STAT-1 Couples Macrophage Synthesis of 25-Hydroxycholesterol to the Interferon Antiviral Response. Immunity. 2013;38:106–118. doi: 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M, Hsieh WY, Robertson KA, Watterson S, Shui G, Lacaze P, Khondoker M, Dickinson P, Sing G, Rodriguez-Martin S, et al. Host Defense against Viral Infection Involves Interferon Mediated Down-Regulation of Sterol Biosynthesis. Plos Biol. 2011;9 doi: 10.1371/journal.pbio.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond MR, Hanover JA. A little sugar goes a long way: The cell biology of O-GlcNAc. The Journal of Cell Biology. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowlin TL, McKown BJ, Babcock GF, Sunkara PS. Intracellular Polyamine Biosynthesis Is Required for Interleukin-2 Responsiveness During Lymphocyte Mitogenesis. Cell. Immunol. 1987;106:420–427. doi: 10.1016/0008-8749(87)90184-5. [DOI] [PubMed] [Google Scholar]
- Bussiere FI, Chaturvedi R, Cheng YL, Gobert AP, Asim M, Blumberg DR, Xu HX, Kim PY, Hacker A, Casero RA, et al. Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. Journal of Biological Chemistry. 2005;280:2409–2412. doi: 10.1074/jbc.C400498200. [DOI] [PubMed] [Google Scholar]
- Casero RA, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Castrillo A, Joseph SB, Vaidya SA, Haberland M, Fogelman AM, Cheng GH, Tontonoz P. Crosstalk between LXR and Toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Molecular Cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- Celano P, Baylin SB, Casero RA. Polyamines Differentially Modulate the Transcription of Growth-Associated Genes in Human-Colon Carcinoma-Cells. Journal of Biological Chemistry. 1989;264:8922–8927. [PubMed] [Google Scholar]
- Chaturvedi R, Asim M, Hoge S, Lewis ND, Singh K, Barry DP, de Sablet T, Piazuelo MB, Sarvaria AR, Cheng Y, et al. Polyamines Impair Immunity to Helicobacter pylori by Inhibiting L-Arginine Uptake Required for Nitric Oxide Production. Gastroenterology. 2010;139 doi: 10.1053/j.gastro.2010.06.060. 1686–98–1698.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HL, Li CF, Grigorian A, Tian W, Demetriou M. T Cell Receptor Signaling Co-regulates Multiple Golgi Genes to Enhance N-Glycan Branching. Journal of Biological Chemistry. 2009;284:32454–32461. doi: 10.1074/jbc.M109.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IJ, Chen HL, Demetriou M. Lateral compartmentalization of T cell receptor versus CD45 by galectin-N-glycan binding and Microfilaments coordinate basal and activation signaling. Journal of Biological Chemistry. 2007;282:35361–35372. doi: 10.1074/jbc.M706923200. [DOI] [PubMed] [Google Scholar]
- Childs AC, Mehta DJ, Gerner EW. Polyamine-dependent gene expression. Cell. Mol. Life Sci. 2003;60:1394–1406. doi: 10.1007/s00018-003-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Qin X, Wu L, Zhang Y, Sheng X, Yu Q, Sheng H, Xi B, Zhang JZ, Zang YQ. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J. Clin. Invest. 2011;121:658–670. doi: 10.1172/JCI42974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nunez G, Schnurr M, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–U1357. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, Redmann V, Freitas TC, Blagih J, van der Windt GJ. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKε supports the anabolic demands of dendritic cell activation. Nat. Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. Importance of Ribonucleotide Availability to Proliferating T-Lymphocytes From Healthy Humans - Disproportionate Expansion of Pyrimidine Pools and Contrasting Effects of De-Novo Synthesis Inhibitors. J. Biol. Chem. 1995;270:29682–29689. [PubMed] [Google Scholar]
- Ferrer CM, Sodi VL, Reginato MJ. O-GlcNAcylation in Cancer Biology: Linking Metabolism and Signaling. J. Mol. Biol. 2016;428:3282–3294. doi: 10.1016/j.jmb.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidelus RK, Laughter AH, Twomey JJ. The role of mitogens and lymphokines in the induction of ornithine decarboxylase (ODC) in T lymphocytes. The Journal of Immunology. 1984;132:1462–1465. [PubMed] [Google Scholar]
- Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, Guerini D, Baumgarten BU, Roggo S, Wen B, et al. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475:524–527. doi: 10.1038/nature10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardbower DM, Asim M, Luis PB, Singh K, Barry DP, Yang C, Steeves MA, Cleveland JL, Schneider C, Piazuelo MB, et al. Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proc. Natl. Acad. Sci. U.S.a. 2017;114:E751–E760. doi: 10.1073/pnas.1614958114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschemi A, Kosma P, Gille L, Evans CR, Burant CF, Starkl P, Knapp B, Haas R, Schmid JA, Jandl C, et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012;15:813–826. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasko G, Kuhel DG, Marton A, Nemeth ZH, Deitch EA, Szabo C. Spermine differentially regulates the production of interleukin-12 p40 and interleukin-10 and suppresses the release of the T helper 1 cytokine interferon-gamma. Shock. 2000;14:144–149. doi: 10.1097/00024382-200014020-00012. [DOI] [PubMed] [Google Scholar]
- Hu X, Wang Y, Hao L-Y, Liu X, Lesch CA, Sanchez BM, Wendling JM, Morgan RW, Aicher TD, Carter LL, et al. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORγ agonists. Nat. Chem. Biol. 2015;11:141–147. doi: 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- Hukelmann JL, Anderson KE, Sinclair LV, Grzes KM, Murillo AB, Hawkins PT, Stephens LR, Lamond AI, Cantrell DA. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat Immunol. 2015 doi: 10.1038/ni.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AK, Huang SC-C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Kay JE, Lindsay VJ. Control of Ornithine Decarboxylase Activity in Stimulated Human Lymphocytes by Putrescine and Spermidine. Biochem. J. 1973;132:791–796. doi: 10.1042/bj1320791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAY JE, PEGG AE. Effect of Inhibition of Spermidine Formation on Protein and Nucleic-Acid Synthesis During Lymphocyte Activation. FEBS Letters. 1973;29:301–304. doi: 10.1016/0014-5793(73)80044-4. [DOI] [PubMed] [Google Scholar]
- Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, et al. Sterol regulatory element–binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Y, Swaisgood MH, Ramos BV, Steck TL. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J. Biol. Chem. 1989;264:3786–3793. [PubMed] [Google Scholar]
- Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Z, Li L, Gong W, Lazenby AJ, Swanson BJ, Herring LE, Asara JM, Singer JD, Wen H. Myeloid-derived cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. J. Exp. Med. 2017;214:1093–1109. doi: 10.1084/jem.20161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yang XV, Wu J, Kuei C, Mani NS, Zhang L, Yu J, Sutton SW, Qin N, Banie H, et al. Oxysterols direct B-cell migration through EBI2. Nature. 2011;475:519–U121. doi: 10.1038/nature10226. [DOI] [PubMed] [Google Scholar]
- Liu S-Y, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, et al. Interferon-Inducible Cholesterol-25-Hydroxylase Broadly Inhibits Viral Entry by Production of 25-Hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund PJ, Elias JE, Davis MM. Global Analysis of O-GlcNAc Glycoproteins in Activated Human T Cells. The Journal of Immunology. 2016;197:3086–3098. doi: 10.4049/jimmunol.1502031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer ML, et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab. 2017;25:345–357. doi: 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Ma J, Hart GW. O-GlcNAc profiling: from proteins to proteomes. Clin Proteomics. 2014;11 doi: 10.1186/1559-0275-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak TW, Grusdat M, Duncan GS, Dostert C, Nonnenmacher Y, Cox M, Binsfeld C, Hao Z, Brüstle A, Itsumi M, et al. Glutathione Primes T Cell Metabolism for Inflammation. Immunity. 2017;46(4):675–689. doi: 10.1016/j.immuni.2017.03.019. [DOI] [PubMed] [Google Scholar]
- McGillicuddy FC, Moya M, de LL, Hinkle CC, Joshi MR, Chiquoine EH, Billheimer JT, Rothblat GH, Reilly MP. Inflammation Impairs Reverse Cholesterol Transport In Vivo. Circulation. 2009;119:1135–U121. doi: 10.1161/CIRCULATIONAHA.108.810721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4(+) T Cell Subsets. The Journal of Immunology. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar E, Swamy M, Holzer M, Beck-Garcia K, Worch R, Thiele C, Guigas G, Boye K, Luescher IF, Schwille P, et al. Cholesterol and Sphingomyelin Drive Ligand-independent T-cell Antigen Receptor Nanoclustering. J. Biol. Chem. 2012;287:42664–42674. doi: 10.1074/jbc.M112.386045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondanelli G, Bianchi R, Pallotta MT, Orabona C, Albini E, Iacono A, Belladonna ML, Vacca C, Fallarino F, Macchiarulo A, et al. A Relay Pathway between Arginine and Tryptophan Metabolism Confers Immunosuppressive Properties on Dendritic Cells. Immunity. 2017;46:233–244. doi: 10.1016/j.immuni.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli LA, Buck MD, Flamar A-L, Saenz SA, Wojno EDT, Yudanin NA, Osborne LC, Hepworth MR, Tran SV, Rodewald HR, et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol. 2016;17:656. doi: 10.1038/ni.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounce BC, Poirier EZ, Passoni G, Simon-Loriere E, Cesaro T, Prot M, Stapleford KA, Moratorio G, Sakuntabhai A, Levraud J-P, et al. Interferon-Induced Spermidine-Spermine Acetyltransferase and Polyamine Depletion Restrict Zika and Chikungunya Viruses. Cell Host Microbe. 2016;20:167–177. doi: 10.1016/j.chom.2016.06.011. [DOI] [PubMed] [Google Scholar]
- O'Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packham G, Cleveland JL. Induction of ornithine decarboxylase by IL-3 is mediated by sequential c-Myc-independent and c-Myc-dependent pathways. Oncogene. 1997;15:1219–1232. doi: 10.1038/sj.onc.1201273. [DOI] [PubMed] [Google Scholar]
- Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MAR, Sheedy FJ, Gleeson LE, van den Bosch MWM, Quinn SR, Domingo-Fernandez R, Johnston DGW, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AR, Wang JY. Polyamines modulate transcription but not posttranscription of c-myc and c-jun in IEC-6 cells. Am. J. Physiol. 1997;273:C1020–C1029. doi: 10.1152/ajpcell.1997.273.3.C1020. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Pearce EJ. Metabolic Pathways in Immune Cell Activation and Quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. Functions of Polyamines in Mammals. J. Biol. Chem. 2016;291:14904–14912. doi: 10.1074/jbc.R116.731661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, Watson AS, Cerundolo V, Townsend AR, Klenerman P, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife. 2014;3 doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proceedings of the National Academy of Sciences. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan P, Clark PM, Mason DE, Peters EC, Hsieh-Wilson LC, Baltimore D. Activation of the Transcriptional Function of the NF-kappa B Protein c-Rel by O-GlcNAc Glycosylation. Science Signaling. 2013;6:ra75. doi: 10.1126/scisignal.2004097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Dang EV, McDonald JG, Liang G, Russell DW, Cyster JG. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 2014;345:679–684. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Spinelli JB, et al. Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell Metab. 2016;24:104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy UKB, Rial NS, Kachel KL, Gerner EW. Activated K-RAS increases polyamine uptake in human colon cancer cells through modulation of caveolar endocytosis. Mol. Carcinog. 2008;47:538–553. doi: 10.1002/mc.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santori FR, Huang P, van de Pavert SA, Douglass EF, Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, et al. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab. 2015;21:286–297. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schall RP, Sekar J, Tandon PM, Susskind BM. Difluoromethylornithine (Dfmo) Arrests Murine Ctl Development in the Late, Pre-Effector Stage. Immunopharmacology. 1991;21:129–143. doi: 10.1016/0162-3109(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Scott IG, Poso H, Akerman K, Andersson LC. Rapid Activation of Ornithine Decarboxylase by Mitogenic (but Not by Nonmitogenic) Ligands in Human Lymphocytes-T. Eur. J. Immunol. 1985a;15:783–787. doi: 10.1002/eji.1830150808. [DOI] [PubMed] [Google Scholar]
- Scott IG, Poso H, AK K, Andersson LC. Mitogens Cause a Rapid Induction of Ornithine Decarboxylase Activity in Human Lymphocytes-T. Biochem. Soc. Trans. 1985b;13:934–935. [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proceedings of the National Academy of Sciences. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlie K, Westerback A, DeVorkin L, Hughson LR, Brandon JM, MacPherson S, Gadawski I, Townsend KN, Poon VI, Elrick MA, et al. Survival of effector CD8+ T cells during infleunza infection is dependent on autophagy. Journal of Immunology. 2015;194:1–10. doi: 10.4049/jimmunol.1402571. [DOI] [PubMed] [Google Scholar]
- Soulet D, Gagnon B, Rivest S, Audette M, Poulin R. A fluorescent probe of polyamine transport accumulates into intracellular acidic vesicles via a two-step mechanism. Journal of Biological Chemistry. 2004;279:49355–49366. doi: 10.1074/jbc.M401287200. [DOI] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, et al. Regulated Accumulation of Desmosterol Integrates Macrophage Lipid Metabolism and Inflammatory Responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy M, Beck-Garcia K, Beck-Garcia E, Hartl FA, Morath A, Yousefi OS, Dopfer EP, Molnar E, Schulze AK, Blanco R, et al. A Cholesterol-Based Allostery Model of T Cell Receptor Phosphorylation. Immunity. 2016a;44:1091–1101. doi: 10.1016/j.immuni.2016.04.011. [DOI] [PubMed] [Google Scholar]
- Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DMF, Cantrell DA. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat Immunol. 2016b;17:712. doi: 10.1038/ni.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Southan GJ, Wood E, Thiemermann C, Vane JR. Inhibition by Spermine of the Induction of Nitric-Oxide Synthase in J774.2 Macrophages - Requirement of a Serum Factor. British Journal of Pharmacology. 1994;112:355–356. doi: 10.1111/j.1476-5381.1994.tb13078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turka LA, Dayton J, Sinclair G, Thompson CB, Mitchell BS. Guanine Ribonucleotide Depletion Inhibits T-Cell Activation - Mechanism of Action of the Immunosuppressive Drug Mizoribine. J. Clin. Invest. 1991;87:940–948. doi: 10.1172/JCI115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bossche J, Lamers WH, Koehler ES, Geuns JMC, Alhonen L, Uimari A, Pirnes-Karhu S, Van Overmeire E, Morias Y, Brys L, et al. Pivotal Advance: Arginase-1-independent polyamine production stimulates the expression of IL-4-induced alternatively activated macrophage markers while inhibiting LPS-induced expression of inflammatory genes. J. Leukoc. Biol. 2012;91:685–699. doi: 10.1189/jlb.0911453. [DOI] [PubMed] [Google Scholar]
- Villablanca EJ, Raccosta L, Zhou D, Fontana R, Maggioni D, Negro A, Sanvito F, Ponzoni M, Valentinis B, Bregni M, et al. Tumor-mediated liver X receptor-alpha activation inhibits CC chemokine receptor-7 expression on dendritic cells and dampens antitumor responses. Nature Medicine. 2010;16:98–U137. doi: 10.1038/nm.2074. [DOI] [PubMed] [Google Scholar]
- Wagner AJ, Meyers C, Laimins LA, Hay N. c-Myc induces the expression and activity of ornithine decarboxylase. Cell Growth Differ. 1993;4:879–883. [PubMed] [Google Scholar]
- Wang F, Beck-Garcia K, Zorzin C, Schamel WWA, Davis MM. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat Immunol. 2016;17:844. doi: 10.1038/ni.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Araki K, Li S, Han J-H, Ye L, Tan WG, Konieczny BT, Bruinsma MW, Martinez J, Pearce EL, et al. Autophagy is essential for effector CD8+ T cell survival and memory formation. Nat Immunol. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Zheng C, Cao J, Cao G, Shou P, Lin L, Velletri T, Jiang M, Chen Q, Han Y, et al. Spermidine alleviates experimental autoimmune encephalomyelitis through inducing inhibitory macrophages. Cell Death Differ. 2016a;23:1850–1861. doi: 10.1038/cdd.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Bai Y, Xiong Y, Zhang J, Chen S, Zheng X, Meng X, Li L, Wang J, Xu C, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016b;531:651. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YR, Song M, Lee H, Jeon Y, Choi E-J, Jang H-J, Moon HY, Byun H-Y, Kim E-K, Kim DH, et al. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell. 2012;11:439–448. doi: 10.1111/j.1474-9726.2012.00801.x. [DOI] [PubMed] [Google Scholar]
- York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, Gray EE, Zhen A, Wu NC, Yamada DH, et al. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou RW, Mkhikian H, Grigorian A, Hong A, Chen D, Arakelyan A, Demetriou M. N-glycosylation bidirectionally extends the boundaries of thymocyte positive selection by decoupling Lck from Ca2+ signaling. Nat Immunol. 2014;15:1038–1045. doi: 10.1038/ni.3007. [DOI] [PubMed] [Google Scholar]