Abstract
Objective:
To describe the clinical features of a novel fused in sarcoma (FUS) mutation in a young adult female amyotrophic lateral sclerosis (ALS) patient with rapid progression of weakness and to experimentally validate the consequences of the P525R mutation in cellular neuronal models.
Methods:
We conducted sequencing of genomic DNA from the index patient and her family members. Immunocytochemistry was performed in various cellular models to determine whether the newly identified P525R mutant FUS protein accumulated in cytoplasmic inclusions. Clinical features of the index patient were compared with 19 other patients with ALS carrying the P525L mutation in the same amino acid position.
Results:
A novel mutation c.1574C>G (p.525P>R) in the FUS gene was identified in the index patient. The clinical symptoms are similar to those in familial ALS patients with the P525L mutation at the same position. The P525R mutant FUS protein showed cytoplasmic localization and formed large stress granule–like cytoplasmic inclusions in multiple cellular models.
Conclusions:
The clinical features of the patient and the cytoplasmic inclusions of the P525R mutant FUS protein strengthen the notion that mutations at position 525 of the FUS protein result in a coherent phenotype characterized by juvenile or young adult onset, rapid progression, variable positive family history, and female preponderance.
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease affecting the upper and lower motor neurons of the brain and spinal cord. It causes progressive paralysis and finally death due to respiratory failure typically within 3 years of symptom onset. Approximately 10%–15% of the ALS cases are familial ALS (fALS) and caused by mutations in several different genes. Recently, mutations in the fused in sarcoma (FUS) gene have been identified in a subset of patients with fALS.1,2 The FUS gene is located on chromosome 16p11.2 and encodes a multidomain protein with 526 amino acids. The FUS protein is predominantly localized in the nucleus in most tissues, whereas it also has a notable cytoplasmic presence in neurons.3 Most fALS mutations are located in the C-terminal nuclear localization sequence (NLS),4 causing accumulation of FUS-containing inclusions in the cytoplasm. The FUS protein binds to RNA/DNA and is involved in a variety of RNA metabolism pathways, including transcription,5–8 splicing,8–10 nucleocytoplasmic RNA shuttling11 and RNA transport,12 mitochondrial RNA biogenesis and function,13,14 and DNA repair.15,16
In this study, we report a novel mutation c.1574C>G (p.525P>R) in FUS in a 26-year-old woman with rapid onset and progression of weakness. The P525R mutant FUS protein showed cytoplasmic localization and formed large stress granule–like cytoplasmic inclusions in multiple cellular models. The phenotype is similar to fALS individuals with the p.525P>L mutation, suggesting that mutations at position 525 in FUS cause fulminant motor neuron disease (MND).
METHODS
Patient information.
The index patient was a 26-year-old woman who experienced subacute onset of proximal upper extremity weakness over 4–6 weeks followed by progression. By 5 months, she had evidence of acute and chronic denervation on EMG testing in the upper extremities, lower extremities, thoracic paraspinous, and sternocleidomastoid muscles. The forced vital capacity was 64% of predicted; the following were normal/negative: comprehensive metabolic panel, complete blood count, vitamin B12, hemoglobin A1C, heavy metals, creatine kinase, aldolase, Lyme, rapid plasma reagin, antinuclear antibody, rheumatoid factor, SSa/SSb, serum protein electrophoresis, and HIV. She was evaluated for a second opinion (M.B.) at 6 months when her ALSFRSr score was 37/48. She was started on riluzole 50 mg bid.
By 9 months, she was receiving total care in a nursing home. Her ALSFRSr score was 24/48. On examination, she had bifacial weakness, normal tongue contour and power, and extensive weakness of neck flexion and extension. An obese body habitus with extremity edema precluded evaluation of muscle atrophy or fasciculations. She did not have antigravity power in any upper extremity muscle and profound proximal lower extremity weakness. There was muscle hypotonia in the upper extremities and increased tone in the lower extremities with elevated tendon reflexes and bilateral Babinski signs. She died of respiratory weakness after a 12-month course.
Additional history revealed that her mother was evaluated 12 years back for upper motor neuron predominant ALS (E.J.K.). By 8 months after the onset of progressive weakness, she had grade 3/5 power in the upper extremities, grade 4−/5 power in the lower extremities, normal bulbar function, and corticospinal tract findings in extremities and in the bulbar region. Detailed clinical sensory testing was normal. EMG testing demonstrated fasciculations, few fibrillations, and large amplitude polyphasic motor unit potentials. She died at the age of 40 after a 6-year progressive course.
Standard protocol approvals, registrations, and patient consents.
The study was approved by the Institutional Review Board of the University of Kentucky. Written informed consent was obtained from all subjects who participated in the study.
Mutation analysis (genomic DNA sequencing).
Genomic DNA was extracted from peripheral blood using the Gentra Puregene Blood Kit (158467; Qiagen, Hilden, Germany). Mutation analysis on FUS was performed by amplifying the exon encoding position P525 and its flanking regions, followed by PCR product sequencing.
Plasmids.
The enhanced green fluorescent protein (EGFP) tagged pEGFP-C3-FUS WT, R521G, P525L, and R495X plasmids were constructed in previous studies.17,18 The P525R FUS mutant was amplified using PCR and subcloned into the pEGFP-C3 (Clontech, Mountain View, CA) vector using the BglII and KpnI sites.
Cell culture and transfection (N2A, primary cortical neurons, and human skin fibroblasts).
Mouse neuroblastoma N2A cells were cultured in Dulbecco's Modified Eagle Medium (D5796; Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum, 100 unit/mL penicillin, and 100 μg/mL streptomycin in a humidified incubator at 37°C under 5% CO2/95% air. Transient transfection was performed using Lipofectamine 2000 (Invitrogen, Life Technologies, Grand Island, NY).
Mouse primary cortical neuron cultures were prepared as previously reported.18 Briefly, neonatal mice (strain C57BL/6; Jackson Laboratory, Bar Harbor, ME) were killed by decapitation within 24 hours of birth. After the incubation with trypsin, cells were dissociated using pipette lavage and cultured in Neurobasal Medium (21103049; Life Technologies) with B27 supplement (17504044; Life Technologies), l-glutamine, and penicillin/streptomycin. After the treatment of 5-fluoro-2′-deoxyuridine (F0503; Sigma-Aldrich) for 4 days to kill glial cells, the primary neurons were transfected with EGFP-tagged WT or mutant FUS expression plasmids using Lipofectamine 2000 (11668; Life Technologies). The primary neurons were fixed for immunofluorescence 48 hours after the transfection.
Human skin fibroblast cell cultures were established as previously described.19 Briefly, a 3-mm punch skin biopsy was obtained after informed consent from the patient with symptomatic ALS and family members who were free of neurologic disease. Skin biopsies were washed with phosphate-buffered saline (PBS), minced into small pieces, and incubated in a fibroblast growth medium (MEM [M5650; Sigma-Aldrich] supplemented with 20% fetal bovine serum, 2 mM l-glutamine, 100 unit/mL penicillin, and 100 μg/mL streptomycin) in tissue culture plates in a humidified incubator at 37°C under 5% CO2/95% air. Fibroblast cells grew from tissue fragments and were maintained under the same conditions as above.
Immunofluorescence microscopy.
N2A cells or primary cortical neurons were seeded on gelatin or poly-d-lysine hydrobromide-coated glass coverslips. Twenty-four hours after the transfection with EGFP-FUS, cells were rinsed with 1× PBS, fixed with 4% formaldehyde in 1× PBS, and permeabilized with 0.25% Triton X-100 in 1× PBS. Primary fibroblast cells were cultured, fixed, and permeabilized similarly as above. The primary antibodies used in this study were mouse anti-FUS (sc-47711; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-G3BP1 (13057-2-AP; Proteintech), and goat anti-TIA1 (sc-1751; Santa Cruz). The secondary antibodies were Alexa Fluor 488 donkey anti-mouse (A-21202; Life Technologies), Alexa Fluor 647 donkey anti-rabbit (A-31573; Life Technologies), and Alexa Fluor 568 donkey anti-goat (A-11057; Life Technologies). The samples were mounted using Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA). Images were acquired using a Nikon A1 confocal microscope with a 60× objective.
RESULTS
P525R mutation in the index patient.
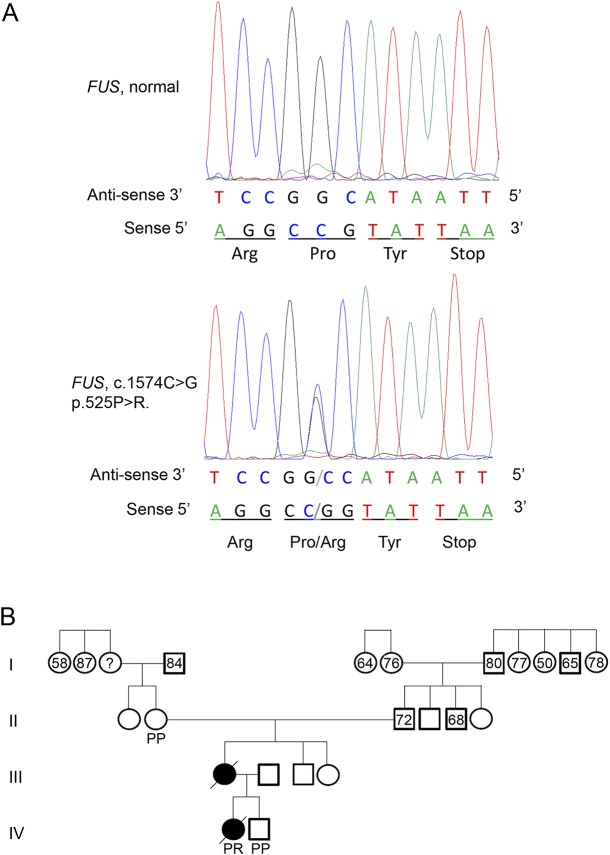
Commercial genetic testing of the ALS patient (Prevention Genetics, Marshfield, WI) revealed 2 normal alleles in C9orf72 with 5 and 14 GGGGCC repeats. The patient was heterozygous in the FUS gene at c.1574C>G (p.Pro525Arg) and heterozygous for a predicted benign variant in the SQSTM1 gene c.C350C>T (p.Ala117Val). Sequencing of 18 additional genes implicated in ALS, frontotemporal dementia (FTD), and ALS/FTD (i.e., ANG, ARHGEF28, CDH13, CHMP2B, GRN, HNRNPA1, HNRNPA2B1, MAPT, OPTN, PFN1, PSEN1, PSEN2, SOD1, TARDBP, TREM2, UBQLN2, VAPB, and VCP) did not reveal any pathologic variants. Sequencing results confirmed that the patient was heterozygous in the FUS gene at c.1574C>G and that her sibling and maternal grandmother were homozygous for normal alleles in FUS (figure 1A). DNA was not available from her mother who died prior to the study. Examination of the pedigree using publicly available records did not identify any other members with a diagnosis of ALS, MND, or neurologic impairment (figure 1B).
Figure 1. Identification of the P525R mutation and patient pedigree.
(A) Sequencing results of amplified patient genomic DNA. The FUS mutation, c.1574C>G (p.Pro525Arg) was identified in the patient. The antisense strand electropherogram is shown on the top, and the reading frame depicting the corresponding amino acid substitution is shown below. The normal genomic DNA was from the unaffected brother of the patient. (B) The family pedigree of the patient with ALS. Numbers indicate the age at death based on family history and public records. Predicted protein composition at position 525 of FUS based on DNA sequencing. ALS = amyotrophic lateral sclerosis; FUS = fused in sarcoma; P = proline; R = arginine.
FUS P525R mutation is mislocalized in cytoplasmic inclusions in N2A cells and primary cortical neurons.
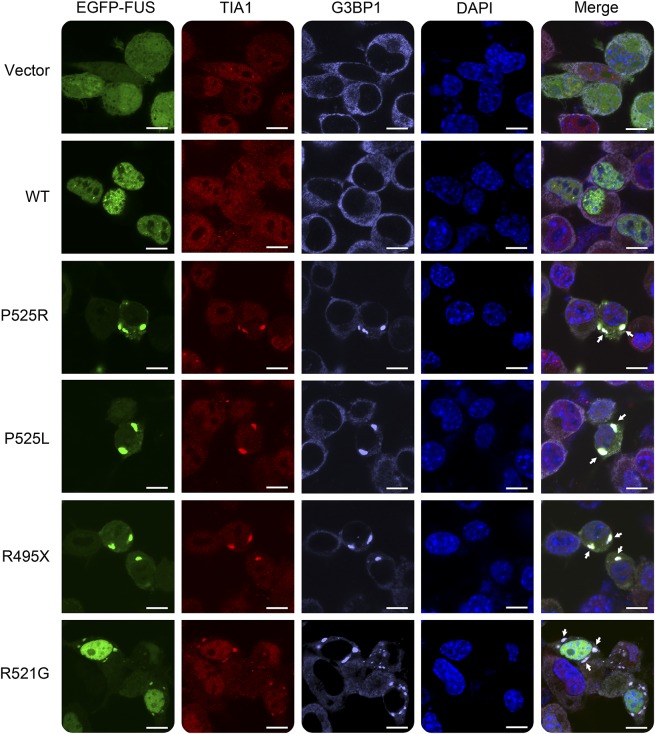
Because the P525R mutation is located in the NLS of FUS, we first tested whether this mutation caused FUS mislocalization in cultured N2A cells compared with other fALS FUS mutants. The wild-type (WT) FUS was located in the nucleus, whereas the P525R mutant FUS protein mislocalized from the nucleus to the cytoplasm and formed large inclusions (figure 2). The mislocalization and inclusions are similar to those observed in P525L and R495X mutations. In comparison, the R521G mutation also caused cytoplasmic localization of FUS, but the majority of the mutant protein was still retained in the nucleus.
Figure 2. The P525R mutant FUS is mislocalized in cytoplasmic inclusions in N2A cells.
N2A cells were transfected with EGFP-tagged WT or mutant FUS. Cytoplasmic inclusions of mutant FUS were colocalized with stress granule markers TIA1 and G3BP1 as indicated by arrows. Scale bar, 10 μm. FUS = fused in sarcoma; WT = wild type.
In addition, cytoplasmic inclusions of the FUS P525R mutant colocalized with stress granule markers G3BP1 and TIA-1 in N2A cells (figure 2). Similarly, other ALS FUS mutants also showed colocalization of cytoplasmic inclusions and stress granule markers. The results are consistent with previous studies that cytoplasmic inclusions of ALS FUS mutants are colocalized with stress granule markers.17,20
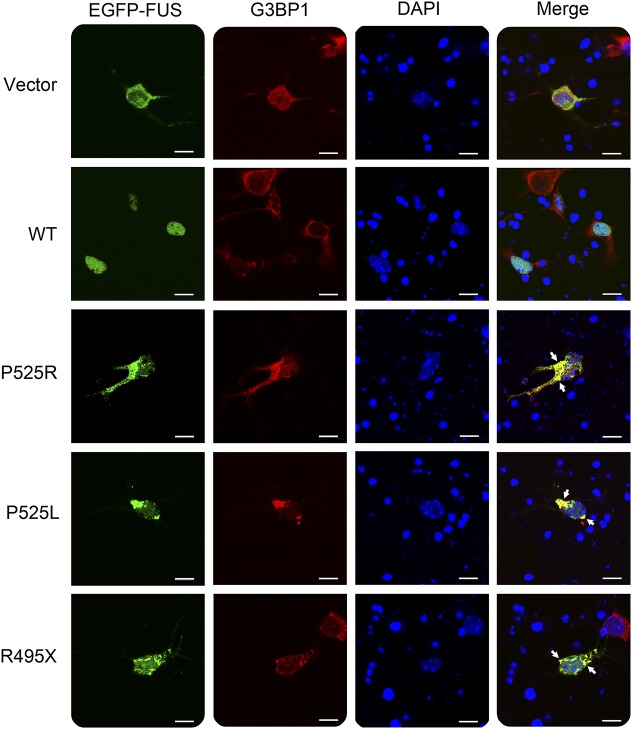
We further examined the subcellular localization of P525R mutant FUS in mouse primary cortical neurons (figure 3). The P525R mutant FUS protein was largely localized in the cytoplasm of primary neurons and formed inclusions positive for stress granule marker G3BP1 in a similar fashion as P525L and R495X mutants. By contrast, the WT FUS protein was predominantly in the nucleus. The results from both N2A and primary neurons consistently indicate that P525R mutant FUS forms cytoplasmic inclusions reminiscent of stress granules.
Figure 3. The P525R mutant FUS is mislocalized in cytoplasmic inclusions in mouse primary cortical neurons.
Mouse primary cortical neurons were transfected with EGFP-tagged WT or mutant FUS. Cytoplasmic inclusions of mutant FUS were colocalized with the stress granule marker G3BP1 as indicated by arrows. Scale bar, 10 µm. FUS = fused in sarcoma; WT = wild type.
Cytoplasmic inclusions of the P525R mutant FUS protein in human fibroblasts.
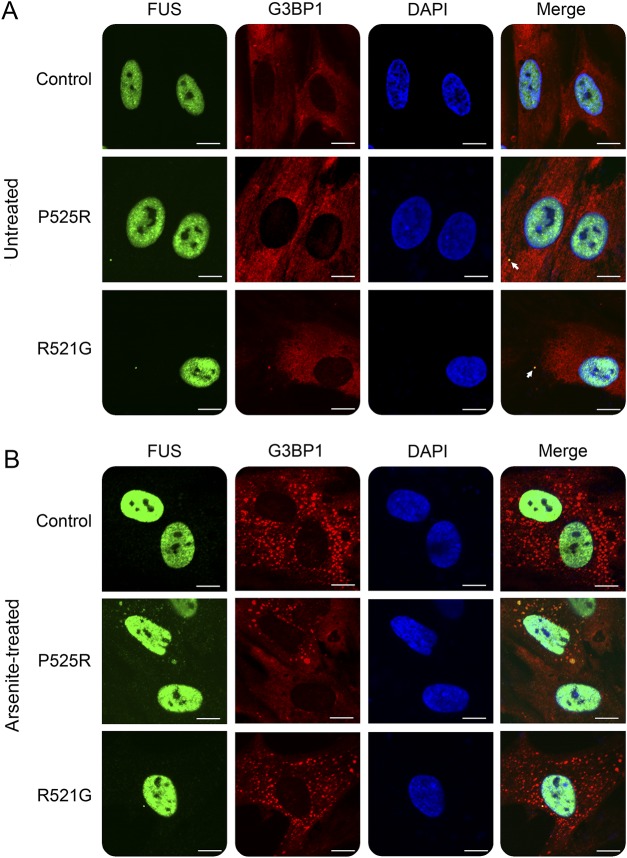
Skin fibroblast cells derived from this patient were examined along with fibroblast cells from patients with fALS carrying the R521G mutation or healthy controls. Under normal cell culture conditions, the majority of the mutant FUS protein was still localized in the nucleus with visible cytoplasmic distribution. Several cytoplasmic puncta of mutant FUS were observed colocalizing with the stress granule marker G3BP1 (figure 4A). The partial mislocalization in patient-derived primary fibroblasts is similar to the previously published study.19
Figure 4. Mislocalization of the P525R mutant FUS in skin fibroblast cells derived from the patient with ALS.
(A) Relatively a small amount of mutant FUS was mislocalized in the cytoplasm and formed puncta colocalized with G3BP1 (as indicated by arrows) under normal cell culture conditions. (B) Skin fibroblast cells were treated with 0.5 mM sodium arsenite for 1 hour before fixation. Under arsenite (oxidative stress) treatment, a notable amount of FUS P525R mutant mislocalized in the cytoplasm and formed inclusions colocalized with G3BP1. Scale bar, 10 µm. ALS = amyotrophic lateral sclerosis; FUS = fused in sarcoma.
To determine the response of WT and different mutant FUS proteins to stress, we treated fibroblasts with sodium arsenite that induces oxidative stress. All fibroblasts responded to arsenite treatment and formed numerous stress granules in the cytoplasm as marked by the stress granule marker G3BP1 (figure 4B). However, the P525R mutant FUS protein responded differently from the WT or R521G mutant. A substantial portion of P525R mutant FUS was mislocalized into the cytoplasm and colocalized with G3BP1 in stress granules (figure 4B). By contrast, WT FUS in healthy control cells and the R521G mutant FUS in fibroblast cells derived from another patient with ALS showed little response to the arsenite treatment. The result suggests that fibroblasts carrying the P525R mutation are more vulnerable to oxidative stress compared with either the WT or the R521G mutation.
DISCUSSION
Here, we report a patient with ALS carrying a novel heterozygous FUS mutation P525R (figure 1). Based on the family history, her mother also died of ALS, and we speculate that she carried the FUS P525R mutation. The index patient's grandmother is healthy in her 70s and has 2 WT FUS alleles; thus, we speculate that her mother likely gained a spontaneous mutation at c.1574C>T. We identified 19 ALS cases carrying the P525L mutation in the FUS gene in the literature and summarized them in table. The literature indicates that approximately 70% of ALS subjects with the documented P525L mutation have no family history of ALS (table), suggesting that either spontaneous mutation event at this locus is not infrequent and/or that individuals with ALS do not survive long enough to reproduce, thus appearing to be sporadic ALS. Previous studies determined that the C-terminal 32 amino acids of FUS functions as an NLS and is critical for its nuclear import mediated by transportin (Trn1).21–23 Indeed, many ALS FUS mutations are located in the NLS. Mutations on the very end of the C-terminus appear to cause the most rapidly progressive phenotype with basophilic cytoplasmic inclusions in motor neurons at autopsy. At the position of amino acid 525 of FUS, P525L mutation has been linked to familial ALS and apparently sporadic cases24 (table). Patients with a P525L mutation were found to have an early disease onset and rapidly progressive disease with short survival when compared with patients with other FUS mutations.21,24 In this study, the patient with the P525R mutation also had an early onset in her mid-20s and a rapid progression over a 12-month course to death. The clinical features are similar to those carrying the truncated R495X mutation lacking the entire NLS (mean age at onset: 35 ± 16 years; average survival: 16.4 ± 10 months from disease onset).25 The novel P525R mutation in this case strengthens the notion that mutations at position 525 of FUS are more fulminant as compared to other FUS mutations. There is a female preponderance in the reported patients with P525L FUS occurring in 75% of recorded cases (table). We currently have no explanation for this phenomenon.
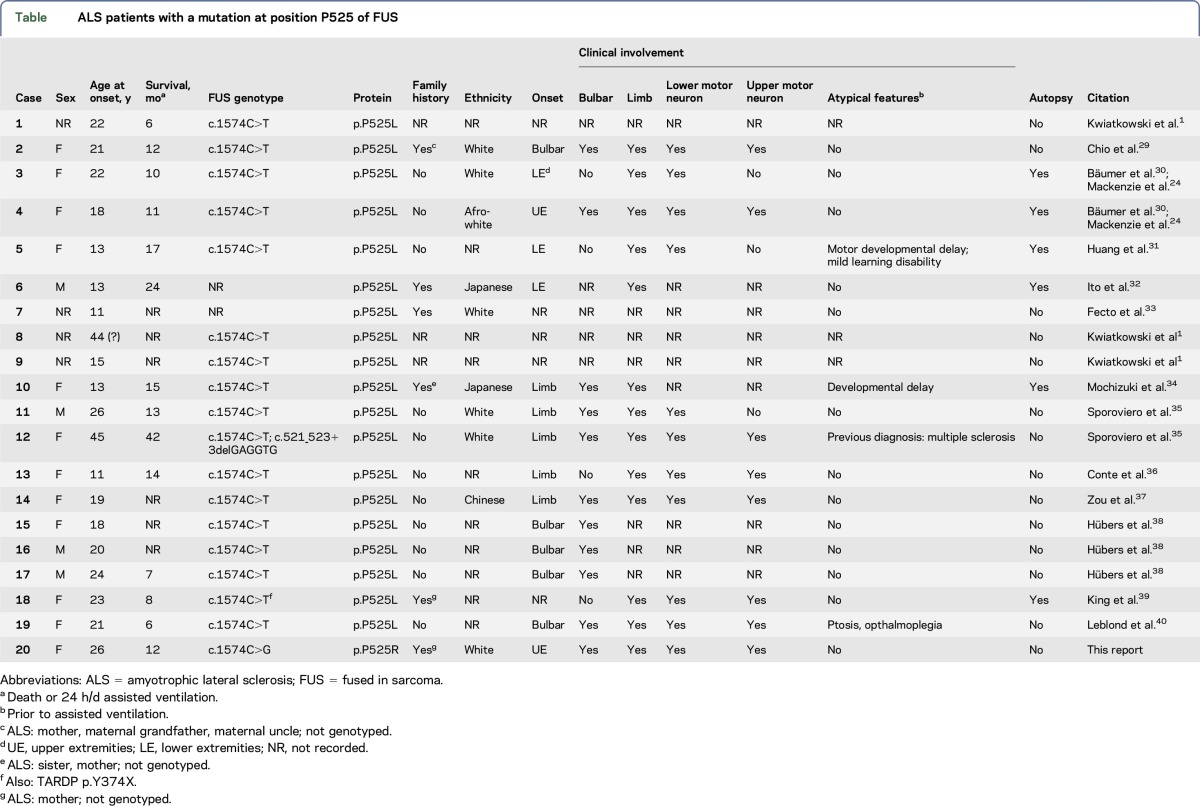
Table.
ALS patients with a mutation at position P525 of FUS
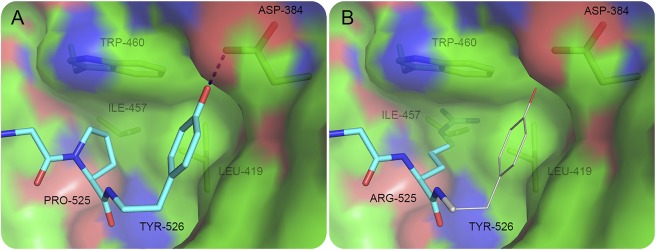
The importance of the C-terminal proline residue was reported for other NLS in which Pro525 and Tyr526 are strictly conserved as the signature PY-NLS.26 Figure 5, which is based on our previously published structural analysis, shows that P525 plays a critical role in the binding of FUS-NLS to Trn1 through the hydrophobic interaction with L419, I457, and W460 of Trn1.21 In addition, P525 puts a C-terminal bend in the peptide and positions Y526 to form a hydrogen bond with D384 of Trn1. In the modeling structure of P525R, the strong charge of Arg does not match the hydrophobicity of the cave formed by L419, I457, and W460 of Trn1. Moreover, the side chain of Arg is predicted to be too large to fit in the small cavity (figure 5). Thus, not only the hydrophobic interaction formed by P525 would be destroyed but also the relocation of residue 525 would push Y526 away and break the hydrogen bond between Y526 and D384. The modeling analysis is consistent with previous reports that the P525L mutation caused dramatic decrease in the binding affinity of FUS-NLS to Trn1,21,22 leading to substantial cytoplasmic accumulation.
Figure 5. The binding details of the C-terminus of FUS and Trn1.
P525 and Y526 of FUS (cyan) and L419, I457, W460 and D384 of Trn1 are shown in sticks. Trn1 is shown in static electric surface mode, in which the red, blue, and green represent negative charge, positive charge, and hydrophobicity, respectively. The hydrogen bond between Y526 and D384 is illustrated. (A) WT FUS binding is based on the structure of the FUS-NLS/Trn1 complex (protein data bank entry 4FQ3). (B) An illustration of the P525R mutant structure. If R525 should remain in the same position as P525, the side chain of R525 would insert into the surface of Trn1. The steric constrains will force R525 to adapt a different conformation. Consequently, the Y526 residue (shown in thin sticks) in the P525R mutant should not keep the same conformation as in the WT structure. FUS = fused in sarcoma; NLS = nuclear localization sequence; WT = wild type.
ALS-linked FUS mutants form cytoplasmic inclusions that are colocalized with stress granule markers in various cell models.25,27 Using induced pluripotent stem cell–derived motor neurons, Lenzi et al.28 also showed that ALS mutant FUS was recruited into stress granules. In this study, we showed that cytoplasmic inclusions of FUS P525R were dominant in N2A cells (figure 2) and primary cortical neurons (figure 3). These inclusions were colocalized with stress granule markers TIA1 and G3BP1, suggesting that this novel mutant P525R was also recruited into stress granules. However, the mislocalization and inclusion formation were less prominent in fibroblast cells derived from patients with ALS under normal experimental conditions (figure 4A). It is likely that the heterozygous mutation and the endogenous expression levels of the mutant FUS protein contribute to the observation that the majority of the FUS protein remained in the nucleus. It is also likely that cell type difference between fibroblast cells and neurons contributes to the less prominent mislocalization of mutant FUS in fibroblast cells. Nevertheless, when exposed to oxidative stress by sodium arsenite treatment, FUS P525R mutant fibroblasts showed stronger response with substantial mislocalization and inclusion formation in cytoplasm as compared to WT and R521G mutants (figure 4B). This supports the hypothesis that the P525R mutant is more susceptible to stress conditions such as arsenite-induced oxidative stress.
We identified a novel mutation in the FUS gene that is associated with an early onset and rapid rate of disease progression. Mechanistically, the P525R mutation causes a cytoplasmic localization of the FUS protein that is prominently colocalized with stress granules, suggesting that a substantial cytoplasmic mislocalization and a strong association with stress granules could be an indicator of the degree of severity in FUS fALS.
ACKNOWLEDGMENT
The authors thank Meghann Bruno, RN, for assistance.
GLOSSARY
- ALS
amyotrophic lateral sclerosis
- EGFP
enhanced green fluorescent protein
- fALS
familial amyotrophic lateral sclerosis
- FTD
frontotemporal dementia
- FUS
fused in sarcoma
- MND
motor neuron disease
- NLS
nuclear localization sequence
- PBS
phosphate-buffered saline
- WT
wild type
AUTHOR CONTRIBUTIONS
Lisha Kuang performed experiments, analyzed data, and wrote the manuscript. Marisa Kamelgarn performed the experiments. Alexandra Arenas performed the experiments. Jozsef Gal performed the experiments. Deborah Taylor performed the pedigree analysis. Weiming Gong performed the structural analysis. Martin Brown reported the phenotype. Daret St. Clair conceptualized the study and wrote the manuscript. Edward J. Kasarskis conceptualized the study, designed experiments, analyzed data, and wrote the manuscript. Haining Zhu conceptualized the study, designed experiments, analyzed data, and wrote the manuscript.
STUDY FUNDING
This study was in part supported by the National Institutes of Neurological Disorder and Stroke grant R01NS077284, MDA grant MDA352743, ALS Association grant 6SE340 and VA MERIT award I01 BX002149 (to H.Z.), and Crispen and Heidrich endowments (to E.J.K.). The support from the Multidisciplinary Value Program (MVP) initiative in the University of Kentucky College of Medicine is appreciated. M.K. and A.A. are supported by the National Institute of Environmental Health Sciences training grant T32ES007266.
DISCLOSURE
L. Kuang reports no disclosures. M. Kamelgarn has received research support from the Muscular Dystrophy Association and the Amyotrophic Lateral Sclerosis Association. A. Arenas reports no disclosures. J. Gal has received research support from NIH/NINDS, the University of Kentucky, and the American Cancer Society. D. Taylor and W. Gong report no disclosures. M. Brown has received travel funding from NEALS (Northeast ALS Consortium). D. St. Clair reports no disclosures. E.J. Kasarskis has served on the scientific board of DSMB for Cytokinetics; has served on the editorial boards of Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration; has been a consultant for Neuraltus Pharmaceuticals; and has received research support from Neuraltus Pharmaceuticals, NINDS, NIEHS, and the ALS Association. H. Zhu has served on the editorial boards of the Journal of Biological Chemistry and Frontiers in Biology and has received research support from the Muscular Dystrophy Association and the Amyotrophic Lateral Sclerosis Association. Go to Neurology.org/ng for full disclosure forms.
REFERENCES
- 1.Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009;323:1205–1208. [DOI] [PubMed] [Google Scholar]
- 2.Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009;323:1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson MK, Stahlberg A, Arvidsson Y, et al. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol 2008;9:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci 2012;15:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008;454:126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan AY, Riley TR, Coady T, Bussemaker HJ, Manley JL. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc Natl Acad Sci USA 2012;109:6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhar SK, Zhang J, Gal J, et al. FUsed in sarcoma is a novel regulator of manganese superoxide dismutase gene transcription. Antioxid Redox Signal 2014;20:1550–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Gal J, Chen J, Zhu H. Self-assembled FUS binds active chromatin and regulates gene transcription. Proc Natl Acad Sci USA 2014;111:17809–17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dichmann DS, Harland RM. fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes Dev 2012;26:1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem 1998;273:27761–27764. [DOI] [PubMed] [Google Scholar]
- 11.Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci 1997;110:1741–1750. [DOI] [PubMed] [Google Scholar]
- 12.Sephton CF, Tang AA, Kulkarni A, et al. Activity-dependent FUS dysregulation disrupts synaptic homeostasis. Proc Natl Acad Sci USA 2014;111:E4769–E4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morlando M, Dini Modigliani S, Torrelli G, et al. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J 2012;31:4502–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dini Modigliani S, Morlando M, Errichelli L, Sabatelli M, Bozzoni I. An ALS-associated mutation in the FUS 3'-UTR disrupts a microRNA-FUS regulatory circuitry. Nat Commun 2014;5:4335. [DOI] [PubMed] [Google Scholar]
- 15.Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem 2013;288:24731–24741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang WY, Pan L, Su SC, et al. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 2013;16:1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gal J, Zhang J, Kwinter DM, et al. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging 2011;32:2323.e27–2323.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamelgarn M, Chen J, Kuang L, et al. Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim Biophys Acta 2016;1862:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gal J, Kuang L, Barnett KR, et al. ALS mutant SOD1 interacts with G3BP1 and affects stress granule dynamics. Acta Neuropathol 2016;132:563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Zhang J, Kamelgarn M, et al. Subcellular localization and RNAs determine FUS architecture in different cellular compartments. Hum Mol Genet 2015;24:5174–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu C, Zhang J, Gao F, et al. FUS-NLS/Transportin 1 complex structure provides insights into the nuclear targeting mechanism of FUS and the implications in ALS. PLoS One 2012;7:e47056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang ZC, Chook YM. Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the fused in sarcoma protein (FUS). Proc Natl Acad Sci USA 2012;109:12017–12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dormann D, Madl T, Valori CF, et al. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J 2012;31:4258–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackenzie IR, Ansorge O, Strong M, et al. Pathological heterogeneity in amyotrophic lateral sclerosis with FUS mutations: two distinct patterns correlating with disease severity and mutation. Acta Neuropathol 2011;122:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosco DA, Lemay N, Ko HK, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 2010;19:4160–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006;126:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gal J, Strom AL, Kwinter DM, et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem 2009;111:1062–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenzi J, De Santis R, de Turris V, et al. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis Model Mech 2015;8:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chio A, Restagno G, Brunetti M, et al. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol Aging 2009;30:1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumer D, Hilton D, Paine SM, et al. Juvenile ALS with basophilic inclusions is a FUS proteinopathy with FUS mutations. Neurology 2010;75:611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang EJ, Zhang J, Geser F, et al. Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol 2010;20:1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito H, Fujita K, Nakamura M, et al. Optineurin is co-localized with FUS in basophilic inclusions of ALS with FUS mutation and in basophilic inclusion body disease. Acta Neuropathol 2011;121:555–557. [DOI] [PubMed] [Google Scholar]
- 33.Fecto F, Siddique T. Making connections: pathology and genetics link amyotrophic lateral sclerosis with frontotemporal lobe dementia. J Mol Neurosci 2011;45:663–675. [DOI] [PubMed] [Google Scholar]
- 34.Mochizuki Y, Isozaki E, Takao M, et al. Familial ALS with FUS P525L mutation: two Japanese sisters with multiple systems involvement. J Neurol Sci 2012;323:85–92. [DOI] [PubMed] [Google Scholar]
- 35.Sproviero W, La Bella V, Mazzei R, et al. FUS mutations in sporadic amyotrophic lateral sclerosis: clinical and genetic analysis. Neurobiol Aging 2012;33:837.e1–837.e5. [DOI] [PubMed] [Google Scholar]
- 36.Conte A, Lattante S, Zollino M, et al. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul Disord 2012;22:73–75. [DOI] [PubMed] [Google Scholar]
- 37.Zou ZY, Cui LY, Sun Q, et al. De novo FUS gene mutations are associated with juvenile-onset sporadic amyotrophic lateral sclerosis in China. Neurobiol Aging 2013;34:1312.e1–1312.e8. [DOI] [PubMed] [Google Scholar]
- 38.Hübers A, Just W, Rosenbohm A, et al. De novo FUS mutations are the most frequent genetic cause in early-onset German ALS patients. Neurobiol Aging 2015;36:3117.e1–3117.e6. [DOI] [PubMed] [Google Scholar]
- 39.King A, Troakes C, Smith B, et al. ALS-FUS pathology revisited: singleton FUS mutations and an unusual case with both a FUS and TARDBP mutation. Acta Neuropathol Commun 2015;3:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leblond CS, Webber A, Gan-Or Z, et al. De novo FUS P525L mutation in Juvenile amyotrophic lateral sclerosis with dysphonia and diplopia. Neurol Genet 2016;2:e63 doi: 10.1212/NXG.0000000000000063. [DOI] [PMC free article] [PubMed] [Google Scholar]