Abstract
The expression of liver-specific genes is regulated by unequivocally allocated transcription factors via proper responsible elements within their promoters. We identified a novel transcription factor, CREB-H, and found that its expression was restricted in the liver among 16 human tissues tested. A region of CREB-H exhibited significant homology to the basic leucine zipper (b-Zip) domain of members of the CREB/ATF family: mammalian LZIP and Drosophila BBF-2 that binds to box-B, a Drosophila enhancer modulating the fat-body-specific gene expression. CREB-H contained a hydrophobic region representing a putative transmembrane domain, like LZIP. Constructing a variety of CREB-H fusion proteins with the GAL4 DNA-binding domain disclosed that CREB-H functioned as a transcriptional activator and its N-terminal 149 amino acids accounted for the activation ability. Gel mobility sift assays revealed that CREB-H did not bind to the C/EBP, AP-1 and NF-κB elements but specifically bound to CRE and the box-B element. Luciferase reporter assays demonstrated that like BBF-2, CREB-H activated transcription via the box-B element and that a deletion of the putative transmembrane domain increased the activation of reporter expression significantly. Furthermore, a fusion protein of GFP and full-length CREB-H was localized in reticular structures surrounding the nucleus, whereas a fusion protein of GFP and a deletion mutant lacking the putative transmembrane domain was mainly in the nucleus. These findings suggest that CREB-H plays an important role in transcriptional regulation of genes specifically expressed in the liver, and that the putative transmembrane domain may be associated with modulation of its function as the transcriptional activator.
INTRODUCTION
Key determinants for tissue-specific gene expression involve unequivocally allocated transcription factors which bind to proper regulatory elements within the promoters for individual genes. Several transcription factors regulating the tissue-specific expression are known to be expressed in a tissue-specific manner. For example, MyoD, a basic helix–loop–helix transcription factor specifically expressed in the muscle, regulates the expression of muscle-specific genes such as muscle creatine kinase (1), myosin light chain 1/3 (2) and troponin C (3) via the E-box element within their promoters. On the other hand, C/EBP-α, a basic leucine zipper (b-Zip) protein, and HNF1, a POU domain protein, modulate gene expression of albumin, a typical liver-specific constituent (4–6) in a synergistic manner. Although these transcription factors are more abundantly expressed in the liver than other organs, their expressions are not restricted to the liver (7,8). Strictly tissue-specific transcription factors like MyoD have not been identified in the liver so far.
In Drosophila, a transcription factor regulating tissue-specific gene expression has been identified in the fat body, which is the functional counterpart of the mammalian liver. The transcription factor activates expression of the Drosophila alcohol dehydrogenase (ADH) gene via the box-B element in a fat-body-specific manner. Thus, this transcriptional activator was designated box-B binding factor-2 (BBF-2/dCREB-B) (9,10). BBF-2 belongs to the CREB/ATF family of the b-Zip protein and binds to not only the box-B element but also CRE (9) and the box-B-like element identified within the promoters for mammalian liver-specific genes, ADH and tyrosine aminotransferase. However, mammalian transcription factors binding to the box-B and box-B-like elements have not yet been identified.
A mammalian member of the CREB/ATF family, which is homologous to BBF-2 and expressed ubiquitously in human tissues, has been identified and designated LZIP (Luman/CREB3) (11–13). Although LZIP activates transcription through binding to CRE, it has not been assessed whether LZIP binds to the box-B-like element. Moreover, LZIP has been reported to associate with a liver-specific pathogen, hepatitis C virus (HCV); LZIP interacts with the HCV core protein and potentiates cellular transformation (13). LZIP contains a hydrophobic region representing a putative transmembrane domain following to the b-Zip domain. A deletion of the putative transmembrane domain of LZIP alters its subcellular localization from the endoplasmic reticulum (ER) membrane to the nucleus (14).
Here, we show a novel member of the CREB/ATF family, designated CREB-H, with a liver-specific expression. CREB-H contains a region highly related to the b-Zip domains of BBF-2 and LZIP and activates transcription through binding to the box-B element.
MATERIALS AND METHODS
Plasmids
A plasmid, pME-CREB-H, expressing the CREB-H protein was isolated from full-length-enriched cDNA libraries described previously (15,16) and contained 2586 bp of the full-length CREB-H cDNA in an SRα-driven expression vector, pME18S (GenBank accession no. AB009864). A plasmid, pME-CREB-HdelTM, that expresses CREB-H lacking the putative transmembrane domain was constructed by insertion of a PCR product (amino acids 1–320) between the EcoRI and XbaI sites of pME18S. A PCR product of CREB-H was produced by amplifying with two primers: 5′-Eco-P1, TTgaattcCATCTGCAGACAGAACTGGATGGAC; and TM-XBE, AAggatccgaattcTCTAGATCATGTCTGGGCTGACTTGCTGGTGGACTGC, with pfu TURBO (Stratagene). The nucleotide sequence of the construct was validated by a 377 autosequencer (PE Biosystems). To confirm the nucleotide sequence of the CREB-H ORF, a fragment containing the ORF was amplified by PCR from cDNA libraries of HepG2 cells and human liver tissue with KOD dash DNA polymerase (Toyobo) and subcloned into a TA cloning vector, pT7blue (Novagen). Five clones containing the ORF were validated by sequencing.
A plasmid, pGST–CREB-H, expressing the GST–CREB-H fusion protein was constructed by an in-frame insertion of a fragment containing the CREB-H lacking the transmembrane domain between the BamHI and EcoRI sites of a GST-expressing plasmid, pGEX-3X (Pharmacia Biotech). A fragment containing the CREB-H lacking the transmembrane domain was amplified with the following primers: Bam5′-2, 5′-TTGGATCCCCATGAATACGGATTTAGCTGCTGG-3′; and TM-XBE with pfu TURBO. The nucleotide sequence of the constructs was checked by the 377 autosequencer.
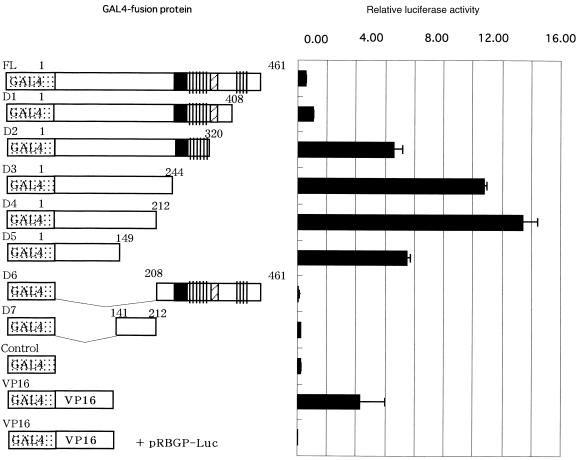
Plasmids expressing fusion proteins of the GAL4 DNA-binding domain and CREB-H deletion mutants were constructed by an in-frame insertion of DNA fragments encoding the full-length and the truncated CREB-H proteins corresponding to amino acids shown in Figure 6. The CREB-H products were inserted between the HindIII and BamHI sites of a plasmid pEF/Gal4-VP16 based on pSG5 (Promega) containing a HindIII–EcoRI fragment of the human elongation factor 1α cDNA promoter derived from pEF-BOS (17), and an EcoRI–BamHI fragment encoding a fusion protein of the DNA-binding domain of Gal4 (amino acids 1–147) and the acidic transactivator domain of VP16 (amino acids 413–490) linked by the AAGCTTAGATCT (HindIII–BglII, encoding Lys-Leu-Arg-Ser) sequence.
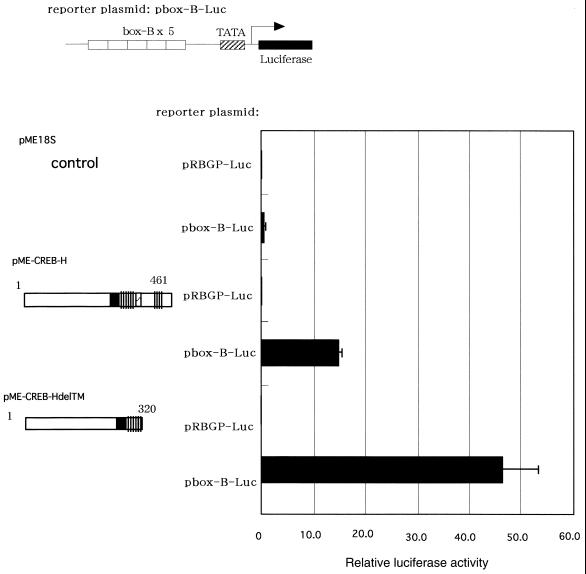
Figure 6.
CREB-H activates transcription of luciferase through the box-B element. The luciferase-reporter construct was transfected into COS7 cells with a CREB-H-expressing plasmid pME-CREB-H, or pME-CREB-HdelTM as well as a control vector plasmid, pME18S. The constructs expressing CREB-H are shown schematically on the left. The luciferase activity was quantified 24 h after transfection. Relative luciferase activities are shown as mean values with standard errors of three independent experiments.
Plasmids expressing GFP fusion proteins with the full-length CREB-H and a deletion mutant of the transmembrane domain were constructed by replacing an EcoRI–HindIII fragment encoding GFP with the GAL4 DNA binding domain of pEF/Gal4 plasmids containing CREB-H.
A luciferase reporter plasmid, pbox-B-Luc, was constructed by an insertion of oligonucleotides of the box-B element into an XhoI site of pRBGP-luc derived from pGL2-basic (Promega) and containing the rabbit β-globin TATA box (18). The oligonucleotide sequences for the box-B element of the Drosophila mulleri ADH-1 promoter (19) were as follows: box-B-sense, 5′-TCGAGCTCGGATGTACACGTAATCGTATTACTC-3′; and box-b-anti-sense, 5′-CGAGAGTAATACGATTACGTGTACATCCGAGCT-3′.
The plasmids were purified with a Qiagen Plasmid Kit (Qiagen) according to the manufacturer’s instruction and their nucleotide sequences were certified by the ABI 377 autosequencer.
Northern blot analysis
Northern blot analyses were performed with Multiple Tissue Northern Blots I and II (Clontech). A 708 bp probe was produced by PCR with primers P7 (5′-TGGGCCACCAGCTTGGAGCAGAGAC-3′) and Bam3 (5′-AAGGATCCTCACTCCTGACAGTGCCCAGCCCCAGGTC-3′). PCR products were purified by a QIAquick gel extraction kit (Qiagen) and labeled with [α-32P]dCTP by a Random Primer DNA Labeling Kit v.2 (Takara). Blots were prehybridized for 1 h and then hybridized for 18 h at 42°C in a hybridization buffer of 50% formamide, 5× SSC, 5× Denhardt’s solution, 0.5% SDS and 500 µg/ml denatured salmon sperm DNA. Hybridized blots were washed with 2× SSC/0.1% SDS at room temperature for 30 min and then with 0.1× SSC/0.1% SDS at 55°C for 60 min. The washed membranes were analyzed by a BAS-2500 bio-image analyzer (Fuji film).
Transfection and luciferase assay
COS7 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and seeded in six-well plates. At 48 h after seeding, cells were washed with OPTI-MEM I Reduced Serum Medium (Life Technologies) and transfected with Lipofectamine 2000 (Life Technologies) in OPTI-MEM I Reduced Serum Medium. In each transfection, 0.4 µg of the luciferase-reporter plasmid, 1 µg of the CREB-H-expressing plasmid, 1 ng of the pRL-CMV plasmid (Promega) and 10 µl of Lipofectamine 2000 were used. At 24 h after transfection, cells were lysed and assayed for the Firefly and the Renilla luciferase activities by a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. The results were normalized against the Renilla luciferase activities obtained from the pRL-TK plasmid as an internal control.
Gel mobility shift assay
The plasmids were transformed into Escherichia coli strain XL-1blue. Transformed bacteria were pre-cultured overnight, transferred to fresh medium and grown for 2 h. After induction with 1 mM IPTG for 2 h, bacteria were sonicated in a sonication buffer containing 50 mM Tris–HCl pH 8.0, 50 mM NaCl, 5 mM EDTA and 2 mM DDT. The GST–CREB-H fusion protein was purified by glutathione–Sepharose 4B in the sonication buffer, washed with the sonication buffer, and eluted by an elution buffer containing 50 mM Tris–HCl pH 8.0, 50 mM NaCl, 5 mM EDTA, 2 mM DDT and 20 mM glutathione. For each binding reaction, 0.1 µg of GST–CREB-H was used. Annealed oligonucleotides were labeled with [γ-32P]ATP with a Megalabel DNA 5′-end-labeling kit (Takara). GST–CREB-H and labeled oligonucleotides were incubated for 30 min at room temperature in 10 µl of a binding buffer containing 50 mM Tris–HCl pH 8.0, 5 mM EDTA, 1 mM DTT, 50 mM KCl, 0.1 µg/µl poly(dI–dC) and 20% glycerol. Electrophoresis was performed for 2 h at 10 mA with a mini-gel system, a 4.5% acrylamide gel and a 1× electrode buffer of a GelShift Assay Kit (Stratagene). The gels were dried down, and the autoradiogram was analyzed by the BAS-2500 bio-image analyzer (Fuji film). The nucleotide sequences of the probes were as follows: 5′-TCGAGCTCGGATGGCTGACGTCAGAGATTACTC-3′ and 5′-CGAGAGTAATCTCTGACGTCAGCCATCCGAGCT-3′ for CRE of the somatostatin promoter (20); 5′-TCGAGCTCGGATGATTTTGTAATGGGGTTACTC-3′ and 5′-CGAGAGTAACCCCATTACAAAATCATCCGAGCT-3′ for the C/EBP element of the albumin promoter (21); 5′-TCGAGCTCGGATCAAAGTTTAGTCAATTACTC-3′ and 5′-CGAGAGTAATTGACTAAACTTTGATCCGAGCT-3′ for the AP-1 element of the c-jun promoter (22); 5′-TCGAGCTCGGAGGGGAATCTCCCGGGTTACTC-3′ and 5′-CGAGAGTAACCCGGGAGATTCCCCTCCGAGCT-3′ for the NF-κB of the IL-2 receptor-α promoter (23).
For the box-B element, the same sequences for the luciferase construct, pbox-B-luc, were used.
Fluorescent microscopy
Transfected cells were seeded on a chamber slide (Lab-Tek). After incubation for 24 h, the cells were washed with phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde in PBS for 5 min, washed with PBS and permeabilized with 0.1% Triton X-100 in PBS for 5 min. The cells were incubated with 0.5 µg/ml of 4′,6-diamidino-2-phenylindole (DAPI; Polysciences, Inc.) for 30 min. The chamber slide was washed three times with PBS and placed with a drop of 90% glycerol in PBS. Microscopic examination was carried out under fluorescent light.
RESULTS
Cloning of a novel member of the CREB/ATF family
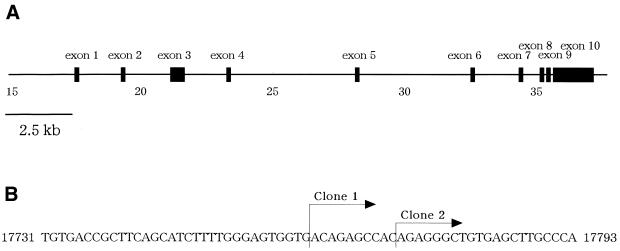
To identify transcription factors expressed in a liver-specific manner, we randomly sequenced clones of human cDNA libraries derived from a hepatoma cell line, HepG2. We chose novel genes after homology search against GenBank and investigated expression profiles for individual clones by RT–PCR, using total RNA extracted from human tissues. A clone showed the expression restricted in the liver among 16 human tissues tested (data not shown); at cycle 30 of RT–PCR a robust band of a PCR product was detected specifically in the liver, whereas at cycle 35 of RT–PCR, a faint band was observed in the intestine (data not shown). Next, we determined the entire nucleotide sequence of the clone. To obtain the full-length sequence of the cDNA, we screened a full-length enriched cDNA library and obtained four clones containing the sequence identical to that initially isolated. Among these clones, we selected the two longest cDNAs and determined their nucleotide sequence. These longest clones contained nearly identical sequences whose 5′ ends differed in 10 bases (Fig. 2B). Subsequently, we set primers for PCR, based on the nucleotide sequence of the longest clone, and directly analyzed PCR products amplified from a normal liver cDNA library. Eventually, we compiled all the nucleotide sequences determined and obtained the full-length cDNA sequence for this novel gene. The entire nucleotide sequence of the novel cDNA that we identified indicates a 2.5 kb sequence (GenBank accession no. AB050902) that contains an ORF of 463 amino acid residues and Kozak consensus around the first ATG (24). We designated the novel gene as CREB-H owing to the reason described below.
Figure 2.
(A) The genomic structure of CREB-H. CREB-H consisted of 10 exons distributing within a 19.4 kb region of the genome (cosmid R33590, accession no. AC005620; 40351 bp) mapped to human chromosome 19p13.3. (B) The mRNA start sites of CREB-H. The mRNA start site of two clones for full-length CREB-H were indicated. The nucleotide numbers in the cosmid R33590 sequence are shown.
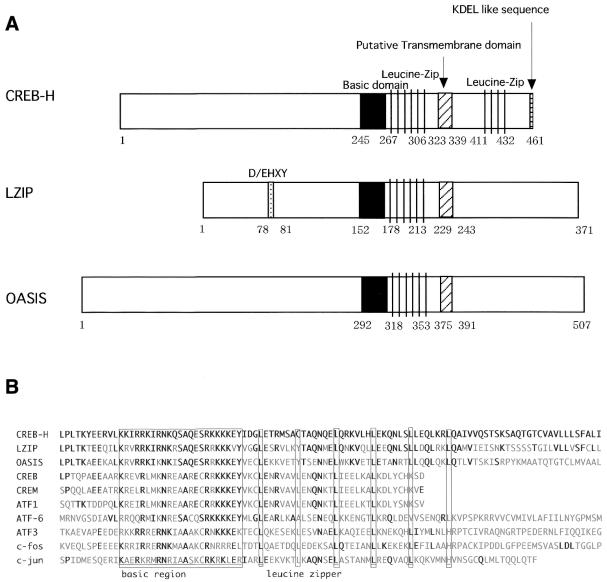
Homology search against protein databases revealed that the amino acid sequence of CREB-H contained a region extensively homologous to the b-Zip domain for three transcription factors belonging to the CREB/ATF family: Drosophila BBF-2, human LZIP and mouse OASIS (Fig. 1B). Amino acid similarities of the b-Zip domain between CREB-H and LZIP, OASIS and BBF-2 were 84, 69 and 71%, respectively. In addition to the b-Zip domain, we noticed moderate homologies within regions surrounding the b-Zip domain between CREB-H and these transcription factors. On the other hand, as a structural difference that was unique to CREB-H, we noticed another leucine zipper motif in the C-terminal region following the b-Zip domain (Fig. 1A), which consisted of three repeats of the ‘LXNXTXX’ sequence. Moreover, we found the third feature for CREB-H by analyzing the amino acid sequence of CREB-H with PSORTII program (K.Nakai and P.Horton, http://psort.ims.u-tokyo.ac.jp/form2.html; 25); CREB-H also contained a hydrophobic stretch of 17 amino acids between the b-Zip domain and the other leucine zipper, which may potentially constitute a transmembrane domain as found in LZIP (Fig. 1A and B). As the fourth feature for CREB-H, we perceived a KDEL-like sequence, ‘GDEL’ (amino acids 458–461), which can behave as an ER-retrieval sequence (26).
Figure 1.
(A) Schematic diagrams of CREB-H, LZIP and OASIS. The b-Zip, D/EHXY, putative transmembrane domain, KDEL-like sequence and leucine-zipper domains and their amino acid positions are indicated. (B) A multiple alignment of the b-Zip domain among CREB-H and other members of the CREB/ATF family. The basic and leucine zipper domains are indicated. Invariant residues are marked by bold letters.
To uncover the exon–intron organization of the novel gene that we identified, we searched GenBank genomic sequence database and retrieved the complete sequence of a cosmid (cosmid number, R33590; GenBank accession no. AC005620) which includes the entire CREB-H cDNA sequence. The CREB-H gene consisted of 10 exons spanning within a 19.4 kb region (Fig. 2A) mapped onto chromosome region 19p13.3 (GenBank accession no. NT_000904).
Live-specific expression of CREB-H
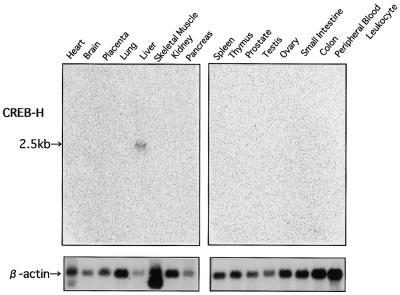
In the initial screening experiment with RT–PCR, we observed the liver-specific expression of CREB-H using 16 human tissues. To confirm this observation, we performed northern blotting analysis with the 16 human tissues (Fig. 3). As a probe, we chose a 708 bp fragment in the 5′ region of the initially isolated CREB-H cDNA, which did not include the b-Zip domain, to avoid cross hybridization with other members of the CREB/ATF family. A 2.5 kb band of the CREB-H mRNA was exclusively detectable in the liver. Though we observed a faint expression of CREB-H in the intestine by extended RT–PCR, we detected no hybridized signal from the intestine even after an extensive overexposure (data not shown). This tissue-specific expression of CREB-H substantially differs from those of LZIP and OASIS ubiquitously expressed in mammalian tissues (12,27).
Figure 3.
The expression profile of the CREB-H mRNA in multiple human tissues. Each lane contained ∼2 µg of poly(A)+ RNA. A probe for CREB-H was a 708 bp fragment of the 5′-portion of the CREB-H cDNA outside the b-Zip domain. A 2.5 kb band corresponding to the CREB-H mRNA is marked. The blot hybridized with a β-actin probe is shown in the lower panel.
Transcriptional activity of CREB-H fusion proteins with the GAL4 DNA-binding domain
Some members of the CREB/ATF family activate transcription and others repress expression of their target genes. To determine whether CREB-H is a transcription activator or not, we constructed plasmids expressing GAL4–CREB-H fusion proteins and co-transfected with a luciferase-reporter plasmid containing the GAL4 DNA-binding element into COS7 cells. We evaluated expression of these fusion proteins by western blotting and confirmed that they were expressed at similar levels (data not shown). The transfected full-length CREB-H cDNA fused to GAL4 activated the reporter expression (Fig. 4), indicating that the full-length CREB-H cDNA contains a transcriptional activation domain. To determine localization of the activation domain within CREB-H, we constructed a variety of deletion mutants of CREB-H fused to the GAL4 DNA-binding domain and co-transfected with the luciferase-reporter plasmid. GAL4–CREB-H fusion constructs lacking the C-terminal regions (designated D1, D2, D3, D4 and D5 in Fig. 4) could activate the reporter expression, whereas constructs lacking the N-terminal regions (D6 and D7) lost that ability. The minimum region responsible for the activation ability was localized within D5 (amino acids 1–141). In this region, the CREB-H protein contained a proline-rich stretch, consistent with the finding that transcriptional activation domains of many transcription factors contain proline-rich sequences (28). Interestingly, the deletion mutants lacking the putative transmembrane domain (D2, D3, D4 and D5) showed activation abilities greater than the full-length (FL) and D1 constructs containing the putative transmembrane domain. These results suggest that the putative transmembrane domain may be associated with modulation of the CREB-H function as the transcriptional activator.
Figure 4.
Mapping of the transcriptional activation domain in CREB-H. The GAL4-DNA binding domain and various deletion mutants of CREB-H were fused, and expression constructs of various GAL4–CREB-H fusion proteins were transfected with a reporter plasmid, pGAL-Luc, into COS7 cells. The luciferase activity was quantified 24 h after transfection. Relative luciferase activities are shown as mean values with standard errors of three independent experiments.
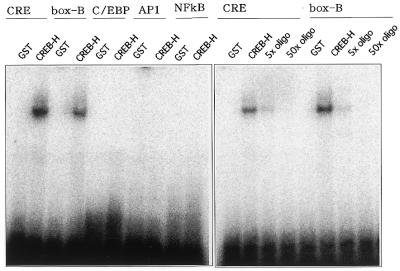
DNA-binding ability of CREB-H
The CREB/ATF family protein specifically binds to the CRE consensus sequence, TGACGTCA (20). To determine whether CREB-H can bind to the CRE sequence, we performed gel-shift assays. We constructed a fusion protein of GST and CREB-H lacking the putative transmembrane domain. The GST–CREB-H fusion protein was produced in bacteria and purified by glutathione–Sepharose 4B. Using the fusion protein, we examined binding ability of CREB-H to CRE and other major transcription factor-binding elements, including AP1, C/EBP and NF-κB. The GST–CREB-H fusion protein formed a complex with the CRE sequence but did not develop any detectable complex with the C/EBP, AP1 and NF-κB elements (Fig. 5). BBF-2, which is highly homologous to CREB-H around the b-Zip region, bound to the box-B element previously identified within the promoter for genes specifically expressed in the Drosophila fat-body. We assessed whether CREB-H binds to the box-B element preferentially. Figure 5 shows that like BBF-2, CREB-H also bound to the box-B element specifically. The shifted bands with the CRE and the box-B element were competed out by 5- and 50-fold excesses of non-labeled oligonucleotides, respectively. We also constructed a fusion protein with full-length CREB-H and eventually obtained results essentially comparable with those with the mutant CREB-H lacking the putative transmembrane domain (data not shown).
Figure 5.
DNA-binding ability of CREB-H analyzed by gel-shift assays. A fusion protein of GST and a CREB-H mutant lacking the putative transmembrane domain was produced in bacteria and purified by glutathione–Sepharose 4B. Oligonucleotides representing CRE, C/EBP, AP1, NF-κB and box-B elements were annealed and labeled with [γ-32P]ATP and incubated with the GST–CREB-H fusion protein. Purified non-fused GST was used in a control lane. Competitive assays for CRE and the box-B element were performed with 5- and 50-fold excesses of unlabeled oligonucleotides for CRE and the box-B element, respectively.
Transcriptional activity of CREB-H through the box-B element
BBF-2 can activate transcription of a CAT reporter directly through the box-B element (9). To assess whether CREB-H activates transcription through the box-B element directly, we co-transfected a full-length CREB-H-expressing plasmid and a luciferase-reporter construct containing the box-B element into COS7 cells. The co-transfection of the full-length CREB-H-expressing construct with box-B-containing reporter plasmid activated the reporter expression by 39-fold relative to a control plasmid (Fig. 6).
As mentioned above, the GAL4 fusion construct lacking the putative transmembrane domain activated the reporter expression significantly when compared to the full-length CREB-H construct (Fig. 4), indicating that a deletion of the putative transmembrane domain of CREB-H enhanced its transcriptional activity in a transient assay in vitro. To confirm the results of the GAL4 fusion experiment, we evaluated the effect of the deletion of the putative transmembrane domain using a box-B-containing reporter plasmid. The co-transfection of the CREB-H construct lacking the putative transmembrane domain with the reporter plasmid containing the box-B element activated the luciferase expression by 124-fold (Fig. 6), confirming the enhancing effect of the deletion on the transcriptional activation ability of CREB-H.
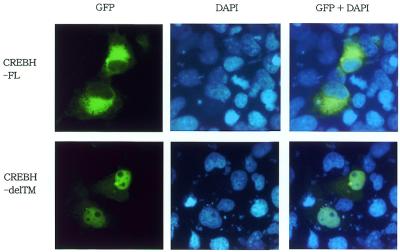
Subcellular localization of GFP–CREB-H fusion proteins
To investigate the subcellular localization of CREB-H, we constructed fusion proteins of GFP and of full-length CREB-H or a deletion mutant lacking the transmembrane domain. We transfected the constructs to COS7 cells and investigated the subcellular localization of fluorescent signal of GFP. The cells were stained by DAPI for visualization of the nucleus 24 h after transfection. The signal of the fusion protein of GFP and the full-length construct of CREB-H was detected in reticular structures surrounding the nucleus, which probably corresponded to the ER (CREBH-FL in Fig. 7). On the other hand, the fusion protein of GFP and the CREB-H mutant lacking the putative transmembrane domain was detected mainly in the nucleus (CREBH-delTM in Fig. 7). In cells transfected with a control vector expressing only GFP, the signal was observed all over the cell, including the nucleus and the cytoplasm (data not shown).
Figure 7.
Subcellular localization of fusion proteins of GFP and CREB-H. The full-length (CREBH-FL) or a deletion mutant of the transmembrane domain (CREBH-delTM) of CREB-H was fused to GFP and transfected to COS7 cells. The cells were stained by DAPI for visualization of the nucleus 24 h after transfection.
DISCUSSION
Identification of a novel transcription factor
In this study, we have identified a novel gene whose product functioned as a transcriptional activator in transient transfection assays in vitro. A homology search disclosed that the novel gene product contained a characteristic region that exhibited extensive similarity to transcription factors belonging to the CREB/ATF family. An in vitro binding analysis with transcription regulatory elements revealed that the novel gene product bound to CRE, consistent with the activity that the novel gene exhibited towards the CREB/ATF family genes. Moreover, we have demonstrated that the novel gene was expressed in a liver-specific manner among the 16 tissues tested. Based on these structural and functional characteristics, we propose that the novel gene encodes a transcription factor belonging to the CREB/ATF family with a liver-specific expression and designated CREB-H (CRE-binding and hepatocyte-specific factor).
CREB-H and its homologs, BBF-2, LZIP and OASIS
Although all members of the CREB/ATF family share structural similarity in their b-Zip domains, some members exhibit additional similarity within other regions and can be grouped into several subfamilies. For example, ATF1, CREB and CREM are grouped into an individual subfamily (29–31). Our results demonstrate that CREB-H exhibits homology in the b-Zip domain especially to LZIP and OASIS and that between CREB-H and these transcription factors, regions surrounding the b-Zip domain also manifest homology to one another. This suggests that they may be classified into the identical subfamily of the CREB/ATF family.
The b-Zip transcription factor forms a homo- or heterodimer via the leucine zipper (5). The heterodimer formation in a different combination between distinct b-Zip factors results in yielding a distinguishable function in the transcriptional regulation. The affinity of the heterodimer formation depends on the structural similarity of the b-Zip domain. For example, CREB and CREM can form a heterodimer, and their leucine zipper domains display significant similarity to each other (31). Since CREB-H, LZIP and OASIS showed the extensive similarity in the b-Zip domain (Fig. 1B), they may potentially form heterodimers between them and function cooperatively.
Though CREB-H, LZIP and OASIS demonstrated the sequence similarity of the b-Zip domain, they also exhibited structural differences outside the b-Zip domain. We noticed another leucine zipper motif in the C-terminal region following the b-Zip domain (Fig. 1A) in CREB-H, which consisted of three repeats of the ‘LXNXTXX’ sequence. We did not find this feature in LZIP and other members of the CREB/ATF family. As the leucine zipper is frequently present within functional domains associated with protein–protein interactions (32), the second leucine zipper domain of CREB-H might interact with other proteins. In contrast, LZIP possesses the D/EHXY motif to associate with host cell factor which is known to interact with herpes simplex virus transactivator, VP16 (33). CREB-H did not include the D/EHXY motif.
A model for modulation of the CREB-H function
ATF6, a member of the CREB/ATF family, contains a transmembrane domain following to the b-Zip domain and is anchored in the ER membrane (34). The ER stress response caused by exogenous stimuli such as tunicamycin treatment induces proteolysis in the anchored ATF6 protein, resulting in translocation of ATF6 released from the ER into the nucleus. An ATF6 construct lacking the transmembrane domain enhances the activation ability significantly compared to the full-length ATF6 (35). LZIP also contains a putative transmembrane domain following the b-Zip domain and exhibits similar translocation from the ER to the nucleus after a deletion of the putative transmembrane domain, although the effect of the deletion on the transcriptional activation of LZIP has not been assessed yet (14). We showed that CREB-H contained a putative transmembrane domain and that a deletion of the putative transmembrane domain of CREB-H enhanced the activation ability in transient assays as shown in Figures 4 and 6. Furthermore, we also showed that a fusion protein of GFP and CREB-H was localized in reticular structures surrounding the nucleus and that a deletion of the putative transmembrane domain altered the subcellular localization to the nucleus. Based on these findings, we propose the following model for modulation of the transcriptional activation of CREB-H: first, CREB-H would localize in the ER by anchoring through the putative transmembrane domain; second, a proteolytic reaction between the b-Zip domain and the putative transmembrane domain would proceed and release the anchored CREB-H protein; and third, the released protein should be translocated to the nucleus and involved in the transcriptional activation. Finally, our finding that CREB-H contained the ER-retrieval signal in the C-terminus might support the idea that CREB-H can be localized to the ER in its full-length form.
CREB-H is a liver-specific transcription activator
We have shown that CREB-H directly activated the expression of the luciferase reporter gene through the box-B element in transient assays. The box-B element was initially identified within the promoter for the Drosophila ADH gene and acts as an enhancer specific to the Drosophila fat-body, which is the counterpart of the mammalian liver (19). The box-B-like element was also identified within the promoter for the human ADH gene expressed in the liver (9). Though Drosophila BBF-2 binds to the box-B-like element within the human ADH promoter, a mammalian factor binding to this element has not yet been identified. Thus, we would nominate CREB-H as the first candidate for a transcription factor binding to the box-B and box-B-like elements with a liver-specific expression. It is likely to assume that CREB-H may regulate the liver-dominant expression of ADH through the box-B-like element and that CREB-H may be involved in regulation of expression of other liver-specific genes via the box-B-like or unknown elements within their promoters.
Recently, LZIP was reported to interact with the HCV core protein and be involved in cellular transformation (13). Besides, the HCV core protein can induce hepatocellular carcinoma in transgenic mice, whereas other components of HCV cannot (36). Because of the structural similarity between LZIP and CREB-H, it is predictable that CREB-H potentially interacts with the HCV core protein. Moreover, it may be possible to infer that CREB-H with the liver-specific expression plays a role in pathogenesis of hepatitis C and subsequent hepatocellular carcinoma by interacting with the HCV core protein rather than LZIP with the ubiquitous expression.
Acknowledgments
ACKNOWLEDGEMENTS
We thank K.Nakagawa, H.Hata, Y.Shirai and Y.Takahashi for helpful suggestions on experimental procedures. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan and Special Coordination Founds for Promoting Science and Technology from Science and Technology Agency of Japan.
DDBJ/EMBL/GenBank accession no. AB050902
References
- 1.Lassar A.B., Buskin,J.N., Lockshon,D., Davis,R.L., Apone,S., Hauschka,S.D. and Weintraub,H. (1989) MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell, 58, 823–831. [DOI] [PubMed] [Google Scholar]
- 2.Donoghue M., Ernst,H., Wentworth,B., Nadal-Ginard,B. and Rosenthal,N. (1988) A muscle-specific enhancer is located at the 3′ end of the myosin light-chain 1/3 gene locus. Genes Dev., 2, 1779–1790. [DOI] [PubMed] [Google Scholar]
- 3.Christensen T.H., Prentice,H., Gahlmann,R. and Kedes,L. (1993) Regulation of the human cardiac/slow-twitch troponin C gene by multiple, cooperative, cell-type-specific, and MyoD-responsive elements. Mol. Cell. Biol., 13, 6752–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courtois G., Baumhueter,S. and Crabtree,G.R. (1988) Purified hepatocyte nuclear factor 1 interacts with a family of hepatocyte-specific promoters. Proc. Natl Acad. Sci. USA, 85, 7937–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landschulz W.H., Johnson,P.F. and McKnight,S.L. (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science, 240, 1759–1764. [DOI] [PubMed] [Google Scholar]
- 6.Maire P., Wuarin,J. and Schibler,U. (1989) The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science, 244, 343–346. [DOI] [PubMed] [Google Scholar]
- 7.Baumhueter S., Mendel,D.B., Conley,P.B., Kuo,C.J., Turk,C., Graves,M.K., Edwards,C.A., Courtois,G. and Crabtree,G.R. (1990) HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev., 4, 372–379. [DOI] [PubMed] [Google Scholar]
- 8.Birkenmeier E.H., Gwynn,B., Howard,S., Jerry,J., Gordon,J.I., Landschulz,W.H. and McKnight,S.L. (1989) Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev., 3, 1146–1156. [DOI] [PubMed] [Google Scholar]
- 9.Abel T., Bhatt,R. and Maniatis,T. (1992) A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev., 6, 466–480. [DOI] [PubMed] [Google Scholar]
- 10.Smolik S.M., Rose,R.E. and Goodman,R.H.(1992) A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol. Cell. Biol., 12, 4123–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burbelo P.D., Gabriel,G.C., Kibbey,M.C., Yamada,Y., Kleinman,H.K. and Weeks,B.S. (1994) LZIP-1 and LZIP-2: two novel members of the bZIP family. Gene, 25, 241–245. [DOI] [PubMed] [Google Scholar]
- 12.Lu R., Yang,P., O’Hare,P. and Misra,V. (1997) Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol., 17, 5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin D.Y., Wang,H.L., Zhou,Y., Chun,A.C., Kibler,K.V., Hou,Y.D., Kung,H. and Jeang,K.T. (2000) Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J., 19, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R. and Misra,V.(2000) Potential role for Luman, the cellular homologue of herpes simplex virus VP16 (α gene trans-inducing factor), in herpes virus latency. J. Virol., 74, 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki Y., Yoshitomo-Nakagawa,K., Maruyama,K., Suyama,A. and Sugano,S. (1997) Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene, 200, 149–156. [DOI] [PubMed] [Google Scholar]
- 16.Marra M., Hillier,L., Kucaba,T., Allen,M., Barstead,R., Beck,C., Blistain,A., Bonaldo,M., Bowers,Y., Bowles,L. et al. (1999) An encyclopedia of mouse genes. Nat. Genet., 21, 191–194. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima S. and Nagata,S. (1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res., 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller M.M., Ruppert,S., Schaffner,W. and Matthias,P. (1988) A cloned octamer transcription factor stimulates transcription from lymphoid-specific promoters in non-B cells. Nature, 336, 544–551. [DOI] [PubMed] [Google Scholar]
- 19.Fischer J.A. and Maniatis,T. (1988) Drosophila Adh: a promoter element expands the tissue specificity of an enhancer. Cell, 53, 451–461. [DOI] [PubMed] [Google Scholar]
- 20.Montminy M.R., Sevarino,K.A., Wagner,J.A., Mandel,G. and Goodman,R.H. (1986) Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc. Natl Acad. Sci. USA, 83, 6682–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams S.C., Cantwell,C.A. and Johnson,P.F. (1991) A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev., 5, 1553–1567. [DOI] [PubMed] [Google Scholar]
- 22.Angel P., Hattori,K., Smeal,T. and Karin,M. (1988) The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell, 55, 875–885. [DOI] [PubMed] [Google Scholar]
- 23.Bohnlein E., Ballard,D.W., Bogerd,H., Peffer,N.J., Lowenthal,J.W. and Greene,W.C. (1989) Induction of interleukin-2 receptor-α gene expression is regulated by post-translational activation of κB specific DNA binding proteins. J. Biol. Chem., 264, 8475–8478. [PubMed] [Google Scholar]
- 24.Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horton P. and Nakai,K. (1997) Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol., 5, 147–152. [PubMed] [Google Scholar]
- 26.Pelham H.R. (1991) Recycling of proteins between the endoplasmic reticulum and Golgi complex. Curr. Opin. Cell Biol., 3, 585–591. [DOI] [PubMed] [Google Scholar]
- 27.Honma Y., Kanazawa,K., Mori,T., Tanno,Y., Tojo,M., Kiyosawa,H., Takeda,J., Nikaido,T., Tsukamoto,T., Yokoya,S. and Wanaka A. (1999) Identification of a novel gene, OASIS, which encodes for a putative CREB/ATF family transcription factor in the long-term cultured astrocytes and gliotic tissue. Brain Res. Mol. Brain Res., 69, 93–103. [DOI] [PubMed] [Google Scholar]
- 28.Chen J.L., Attardi,L.D., Verrijzer,C.P., Yokomori,K. and Tjian,R.(1994) Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell, 79, 93–105. [DOI] [PubMed] [Google Scholar]
- 29.Hai T.W., Liu,F., Coukos,W.J. and Green,M.R. (1989) Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev., 3, 2083–2090. [DOI] [PubMed] [Google Scholar]
- 30.Hoeffler J.P., Meyer,T.E., Yun,Y., Jameson,J.L. and Habener,J.F. (1988) Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science, 242, 1430–1433. [DOI] [PubMed] [Google Scholar]
- 31.Foulkes N.S., Borrelli,E. and Sassone-Corsi,P. (1991) CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell, 64, 739–749. [DOI] [PubMed] [Google Scholar]
- 32.Rabindran S.K., Haroun,R.I., Clos,J., Wisniewski,J. and Wu,C. (1993) Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science, 259, 230–234. [DOI] [PubMed] [Google Scholar]
- 33.Freiman R.N. and Herr,W.(1997) Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev., 11, 3122–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haze K., Yoshida,H., Yanagi,H., Yura,T. and Mori,K.(1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell, 10, 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Shen,J., Arenzana,N., Tirasophon,W., Kaufman,R.J. and Prywes,R. (2000) Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem., 275, 27013–27020. [DOI] [PubMed] [Google Scholar]
- 36.Moriya K., Fujie,H., Shintani,Y., Yotsuyanagi,H., Tsutsumi,T., Ishibashi,K., Matsuura,Y., Kimura,S., Miyamura,T. and Koike,K.(1998) The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med., 4, 1065–1067. [DOI] [PubMed] [Google Scholar]