Abstract
Heptosyltransferase I (HepI) catalyzes the addition of L-glycero-β-D-manno-heptose onto Kdo2-Lipid A, as part of the biosynthesis of the core region of lipopolysaccharide (LPS). Gram-negative bacteria with gene knockouts of HepI have reduced virulence and enhanced susceptibility to hydrophobic antibiotics, making design of inhibitors against HepI of interest. Since HepI protein dynamics are partially rate-limiting, disruption of protein dynamics might provide a new strategy for inhibiting HepI. Discerning the global mechanism of HepI is anticipated to aid development of inhibitors of LPS biosynthesis. Herein, dynamic protein rearrangements involved in the HepI catalytic cycle were probed by combining mutagenesis with intrinsic tryptophan fluorescence and circular dichroism analyses. Using wild-type and mutant forms of HepI, multiple dynamic regions were identified via monitoring fluorescence wavelength changes. Interestingly, Trp residues (Trp199 and Trp217) in the C-terminal domain (which binds ADP-Heptose) are in a more hydrophobic environment upon ODLA binding to the N-terminal domain. These residues are adjacent to the ADP-Heptose binding site (with Trp217 in van der Waals contact with the Adenine ring of ADP-Heptose), suggesting that the two binding sites interact to report on the occupancy state of the enzyme. ODLA binding was also accompanied by a significant stabilization of HepI (heating to 95 °C fails to denature the protein when in the presence of ODLA). These results suggest that conformational rearrangements, from an induced fit model of substrate binding to HepI, are important for catalysis, and the disruption of these dynamics may serve as novel mechanism for inhibiting this and other glycosyltransferase enzymes.
Keywords: Heptosyltransferase, glycosyltransferase, GT-B, tryptophan fluorescence, structural rearrangement, circular dichroism, protein dynamics
Table of Contents Graphic
Introduction
Rapid increase of antibiotic resistance in Gram-negative bacteria has arisen due to overuse of many commonly used antibiotics, thus making them ineffective.1 Although antibiotic resistance is a global health concern and has the potential to reach pandemic proportions, new antibiotics are scarce. Glycosyltransferases (GTs) catalyze the addition of various sugars to other biomolecules, which are often essential to many biological processes (ranging from the synthesis of small molecules to enabling cell-lectin interactions). GTs encompass a large group of enzymes that are highly diverse, in both structure and mechanism.2, 3 Despite their differences, most GTs catalyze the formation of a glyosidic bond, where a high-energy nucleotide sugar donates a monosaccharide to an acceptor molecule. This acceptor can be a variety of molecules, such as oligosaccharides, monosaccharides, proteins and lipids.4
Escherichia coli Heptosyltransferase I (HepI) is a glycosyltransferase, containing two βαβ Rossmann-like folds connected by an inter-domain linker region, characteristic of the GT-B structural superfamily (Figure 1A). HepI catalyzes the addition of L-glycero-D-manno-heptose (heptose) from ADP-Heptose (ADPH) to the first 3-deoxy-α-D-manno-oct-2-ulopyranosonic acid (Kdo) of E. coli Kdo2-LipidA, generating heptose-Kdo2-LipidA.5–8 This is a crucial step for the formation of the Lipopolysaccharide (LPS), which is located on the outer membrane of Gram-negative bacteria and contributes to bacterial pathogenicity and virulence.8 LPS isolated from bacteria that are deficient in HepI, lack heptose and all additional sugars typically added afterwards.9 Although formation of heptose-Kdo2-LipidA is not necessary for cell viability, these HepI deficient mutants result in decreased bacterial pathogenicity in infected mice, have an increase in susceptibility to phagocytosis by macrophages, and lead to an increase in bacterial sensitivity to hydrophobic antibiotics.8, 10, 11 Efforts to design inhibitors of HepI have previously been reported, however all have resulted in inhibitors with IC50 greater than 1 µM.12, 13 Enhancing mechanistic understanding of HepI action would provide a molecular basis for development of inhibitors for HepI as potential therapeutic agents for the treatment of Gram-negative bacterial infections.
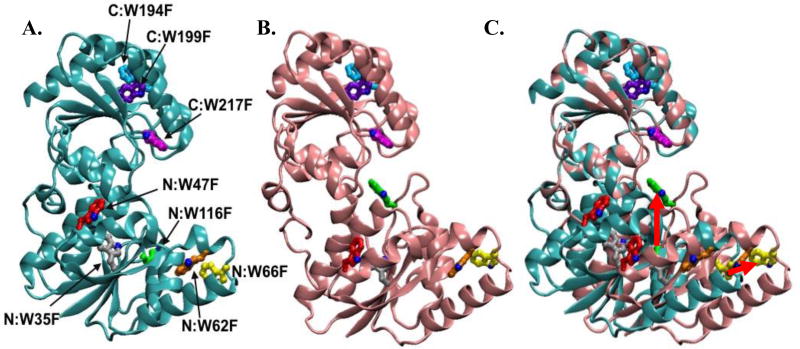
Figure 1.
A) HepI open structure colored cyan with Trp residues labeled (PDB ID: 2H1H:A). B) Structural model of HepI closed, colored pink (methods for closed model generation can be found in Reference 5). C) Superimposed HepI open with structural model of closed HepI. Movement of select Trp residues are shown with red arrows.
While the overall reaction of HepI was discerned in the 1970s, little is known about the detailed chemical and kinetic mechanisms by which HepI catalyzes the addition of heptose onto Kdo2-Lipid A. Recently multiple enzymes of the GT-B structural class, including GftA and MshA (the epivancosaminyl transferase from Amycolatopsis orientalis and the N-acetylglucosamine transferase from Corynebacterium glutamicum), have been crystalized in both open and closed configurations, dependent upon the ligation state of the proteins.6, 7, 13–17 Currently, structures are available for the apo HepI, and HepI complexed with ADP and an ADPH analogue, all of which adopt an open configuration.6 To date there are no crystal structures of HepI with Lipid A or both substrates bound; however a model of a hypothesized closed structure was previously generated.6 Despite the lack of structural evidence supporting the hypothesis that HepI adopts a closed conformation, it is thought that HepI, like other GT-Bs, also interconverts between an open and closed conformations to enable catalysis (Figure 1A–C). This hypothesis is supported by the observation from the apo crystal structure of HepI that the catalytic residue Asp13, which is needed to deprotonate Lipid A generating a nucleophile, is over 8 Å from the anomeric carbon of ADPH where deprotonated Lipid A nucleophile attacks.15 In addition, our previous transient kinetic analysis of the intrinsic protein fluorescence revealed substrate induced changes in fluorescence suggestive of multiple events occurring prior to chemistry, including conformational changes. Specifically, the fluorescence changes upon mixing of O-deacylated E. coli Kdo2-LipidA (ODLA) with HepI report on the ODLA binding event and subsequent one (or more) conformational change(s).6
In this work, the intrinsic tryptophan (Trp) fluorescence of wild-type HepI (HepI-WT) and mutants are used further to isolate the regions undergoing conformational changes to better understand the catalytically active structure of HepI. It has previously been shown that in the presence of ODLA, the fluorescence spectrum of HepI undergoes a blue shift. These results suggest that one or more Trp must move from a more solvated to a less solvated environment upon mixing with ODLA.6 To identify which Trp could be the source of this signal, each of the seven solvent exposed Trp residues (Figure 1) were individually mutated to phenylalanine (Phe) to investigate their roles in the previously observed transient kinetics experiments. Kinetic analyses were performed to assess the magnitude of catalytic alterations induced by the current set of mutations. Circular dichroism (CD) measurements were used to determine the effect of the mutations on substrate binding and thermostability. Furthermore, to distinguish which Trp residues are responsible for the previously observed substrate induced blue shift of HepI intrinsic fluorescence, WT and mutant HepI fluorescence spectra were obtained, revealing both N and C-terminal domains are altered upon binding ODLA. Ultimately, our results enhance understanding of HepI dynamics and the conformational changes that occur in GT-B enzymes.
Materials and Methods
Primers were designed using BioMath Tm calculator (http://www.promega.com/a/apps/biomath/index.html?calc=tm). All primers, and the E. coli strains DH5α, and BL-21-AI were obtained from Invitrogen (Carlsbad, CA). Quikchange Lightning Site-Directed Mutagenesis Kit and E. coli strain XL10-gold were obtained from Stratagene (La Jolla, CA). Zyppy plasmid mini prep kit was attained from Zymo Research (Irvine, CA). IPTG was obtained from Gold Bio Technologies (St. Louis, MO). B-PER II Bacterial Protein Extraction Reagent, EDTA free protease inhibitor tablets, Sartorius Vivaspin 20 centrifugal concentrators 10,000 and 3000 Molecular Weight Cut Off (MWCO), tryptone and yeast extract were obtained from Fisher Thermo Scientific (Pittsburg, PA). Sodium chloride, sodium hydroxide, ampicillin (Amp), tetracycline (Tet), HEPES, imidazole, EDTA, cobalt sulfate, and L-arabinose were acquired from Sigma (St. Louis, MO). 30% acrylamide, Bio-Scale Mini Bio-Gel P-6 desalting cartridge, and Coomassie Brilliant Blue R-250 were obtained from Bio-Rad (Hercules, CA). GenAmp 9700 PCR thermocycler was obtained from Applied Biosystems (Foster City, CA). All UV-Vis measurements were taken using Cary 100 Bio UV-Vis from Agilent (Santa Clara, CA). Fluorescence spectra were measured using Fluoromax-4 from Horiba Scientific (Edison, NJ). All cells were lysed using a EmulsiFlex-C5 homogenizer manufactured by Avestin Inc. (Ottawa, ON). Toyopearl AF-Chelate-640 column was obtained from Tosoh (Grove City, OH) and used to purify HepI. All ESI-MS spectra were collected using a Thermo Scientific (Waltham, MA) ESI spectrometer. Previously, HepI assays were performed at 37 °C; however, HepI thermal instability observed as part of this study, led us to perform all biophysical experiments at lower temperatures. HepI kinetics assay performed at 25 °C revealed a 2-fold reduction in kcat at this lower temperature, supporting the validity of comparisons between these biophysical measurements (Table S1).
Substrate isolation and deacylation
Both ADPH and Lipid A were extracted from E. coli WBB06 cells (HepI and HepII knockout E. coli strain), as previously described.5, 7, 18, 19 In brief, from a 4 liter growth of fresh WBB06 cells ADPH was extracted with ethanol and placed over a DEAE column to purify (yield is ~10 mg). KDO2-Lipid A was extracted from 4 grams of frozen, dried down (cells were dried with acetone followed by diethyl ether) WBB06 cells using a solution of 2:5:8 phenol, chloroform, and petroleum ether (yield ~50 mg). O-deacylation of Lipid A was done by stirring 60 mg Lipid A with 6 mL hydrazine for 1 hr at 37 °C. The solution was then placed on ice and 60 mL of cold acetone was added to precipitate ODLA followed by centrifugation for 30 min at 11,000 rpm. The pellet was washed twice with cold acetone and once with diethyl ether, then dissolved in water and placed in −80 °C freezer for 20 minutes to freeze the solution, followed by lyophilization to dry. Deacylation was confirmed by ESI-mass spectroscopy in 50/50 acetonitrile and water by observation of the half mass (m/z 696 corresponding to M−2-H complex).
Mutagenesis and solubility expression optimization
E. coli pTOM HepI vector DNA was extracted from DH5α cells using Zyppy plasmid mini prep kit. Hep I tryptophan single mutants: Trp217Phe, Trp199Phe, Trp194Phe, Trp116Phe, Trp66Phe, Trp62Phe, Trp47Phe were prepared using QuikChange mutagenesis kit using primers shown in Supplemental Table S2. Protein expression and purification for all mutants except HepI Trp116Phe were the same as wild-type HepI previously described by Czyzyk et.al.6 The Trp116Phe was expressed in artic express cell at 10 °C and left to shake for 48 hours. The rest of the purification procedure for Trp116Phe was the same as wild-type. Protein was stored at 4 °C and assays were performed within two weeks of purification.
HepI activity assay
As previously reported,5, 6 an ADP/NADH coupled assay was used to monitor Hep I activity by monitoring the absorbance change at 340 nm at 37 °C on a Cary spectrometer. Under normal conditions, the assay buffer was composed of 50 mM HEPES, 50 mM KCl, 10 mM MgCl2, pH 7.5. High salt assays were performed with 50 mM HEPES, 2 M KCl, 10 mM MgCl2, pH 7.5. The coupled enzyme reaction additionally contained 100 µM phosphoenolpyruvate, 100 µM NADH, 100 µM ADP-Heptose, 100 µM ODLA, and 0.005 U/µL of both pyruvate kinase and lactate dehydrogenase. Once a stable baseline was established the reaction was initiated by addition of 50 nM enzyme and all reported reaction rates are after background subtraction.
Circular Dichroism (CD)
Protein thermal stability was assessed via CD melt analysis. Spectra were measured in triplicate at 20 °C or as a function of increasing temperature (5 – 95 °C, in 5 °C increments) using quartz cuvettes (0.2×1 cm) containing 5 μM HepI and 100 μM ODLA in 10 mM Tris-HCl and 100 mM KCl (pH = 7.5) buffer; melts were done in the presence and absence of ODLA. For salt titration experiments, the KCl concentration was increased to 250 mM, 500 mM, 1 M and 2 M while keeping other components constant. All measurements were taken using a Jasco J-810 Spectropolarimeter. Thermodynamic stability parameters of HepI-WT and mutants were calculated as previously reported by Mehl et.al, using ellipticities at 211 and 222 nm (Supplemental Table S3).20 Percent composition of secondary structure (α-helicity and β-strand percentages) was determined using the K2D3 CD spectral analysis tool (http://cbdm-01.zdv.uni-mainz.de/~andrade/k2d3//info/about.html).
Intrinsic tryptophan fluorescence spectra measurements
Fluorescence spectra were measured in triplicate at room temperature using 200 μL samples containing 1 μM HepI, in a 10 mM HEPES, 50 mM KCl, and 10 mM MgCl2, pH = 7.5 buffer, using a 0.3 × 0.3 cm cuvette. Substrate concentrations were 100 μM ODLA and 100 μM ADP-Hep. All measurements were taken using a Fluoromax-4 fluorimeter with an excitation slit bandpass of 2 nM and an emission slit bandpass of 4 nM (λex = 290 nm, λem = 310 – 450 nm). Spekwin32 (version 1.71.6.1) spectroscopy software was use to convert spectra to csv files, subtract spectra and normalize data. Peak emission wavelengths were determined by taking the first derivative of each fluorescence spectrum using MATLAB (R2012B) with the λmax wavelength being determined from a linear fit of 5 points about zero.
Molecular Dynamics (MD) Simulation
Apo-form HepI protein system (PDB: 2GT1, chain A) was prepared for MD using the Schrodinger modeling package (v. 2015.3, Schrodinger LLC, New York, NY) and related utilities. Unless noted otherwise, all procedures performed within Schrodinger’s Maestro GUI. Addition of unresolved missing atoms and hydrogens were modeled using Prime. Proper protonation of charged residues at pH 7.0 was determined via PROPKA, ensuring optimal hydrogen bonding network. System was solvated in an orthorhombic TIP3 explicit water box with a 15 Å buffer region, followed by addition of sodium and chloride counter-ions to ensure electrostatic neutrality. Final salt concentration of solvated, neutralized system was 0.15 M NaCl. Both system initialization (restrained coordinates) and subsequent MD production (100 ns, unrestrained coordinates) employed default settings and were carried out using the Desmond MD software (D.E. Shaw Research, New York, NY) with the OPLS-AA 2005 force field. Production stage dynamics were propagated using the RESPA integrator with a time step of 2 fs. The isothermal-isobaric (NPT) ensemble at 300K with Nose-Hoover chain thermostat and Martyna-Tobias-Klein barostat (isotropic) was used. Particle-Mesh Ewald accounted for long-range electrostatic interactions under periodic boundary conditions. Production run was performed on the Minnesota Supercomputing Institutes (MSI) Mesabi supercomputer resource with system coordinates collected every 4.8 ps.
Solvent Accessible Surface Area (SASA) calculations
SASA values (Å2) were computed for the HepI reference apo-form crystal structure (PDB: 2GT1, chain A) and for the 20,833 protein coordinates derived from the 100 ns HepI MD simulation using the Shrake-Rupley algorithm with a probe radius of 1.4 Å. Only the aromatic Trp side-chain atoms (heavy and hydrogen) were considered in the calculation.
Results and Discussion
Previously, our equilibrium and transient kinetic analyses of Trp fluorescence of HepI suggested that protein conformational changes were induced upon binding of ODLA. Biphasic pre-steady state kinetics were observed over a range of concentrations of ODLA, with the fast rate exhibiting a hyperbolic dependence on ODLA concentration (saturating to reveal a concentration independent process occurring at ~80 sec−1), and a concentration independent slow rate of ~5 sec−1. Since a catalytically impaired mutant (D13A) of HepI yielded the same pre-steady state kinetics, we hypothesized that ODLA induces two conformational changes prior to catalysis – a grasping of the substrate likely caused by loop movements, followed by a slower, larger conformational change.6 To better understand the protein configuration of the catalytic HepI Michaelis complex, we examined single Trp mutants using fluorescence and CD spectroscopy to identify the regions undergoing these conformational changes.
Secondary structural changes and thermal unfolding of HepI
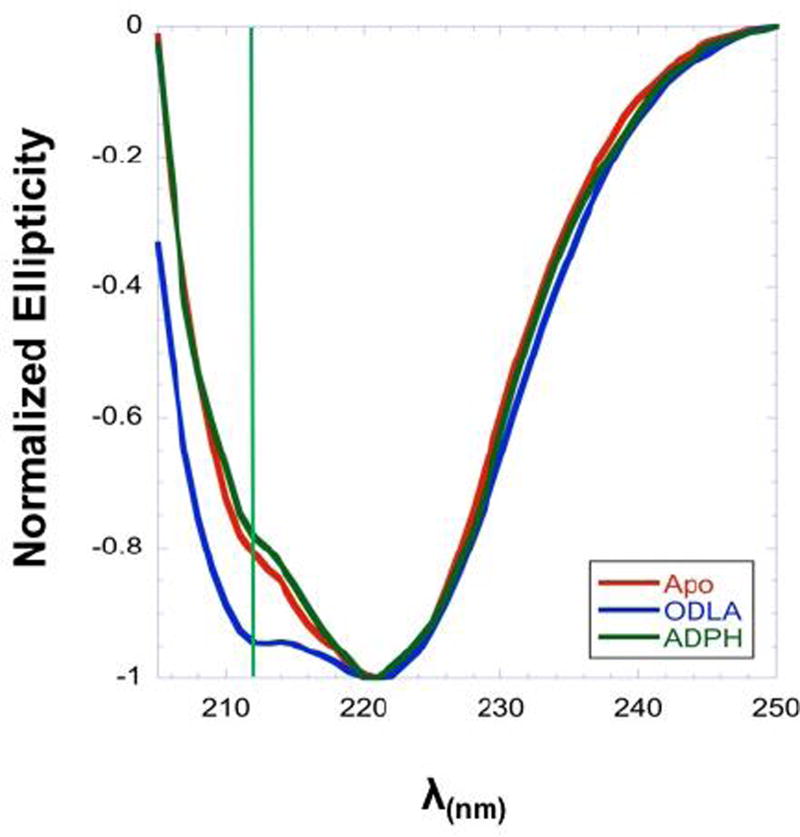
CD spectroscopy is a powerful tool for assessing the secondary structural elements of a protein under a variety of conditions.21–23 To assess the conformational states of HepI, CD spectra were obtained for apo HepI and HepI in the presence of either ODLA or ADPH (Figure 2). Apo HepI exhibits a double minimum with unequal intensity, with a sharp peak at about 222 nm and a small peak at 211 nm, characteristic of a protein with significant α-helical content. The CD spectra obtained for HepI with ADPH bound resembles that of apo HepI; however, the spectra for the HepI·ODLA complex has an approximately 12-fold increase in ellipticity at 211 nm. Analysis of the spectra of apo HepI and HepI·ADPH complex with the K2D3 CD analysis webserver shows little change in the overall calculated α-helical or β-strand percentages upon ADPH binding (with the apo protein having 43.4% α-helical and 12.5% β-strand, as compared to 39.4% and 18.5% for the ADPH bound enzyme, respectively).24 Consistent with the increase in the intensity of the peak minimum, the HepI·ODLA complex was calculated to have an approximately 14% increase in α-helicity as compared to apo HepI (with HepI·ODLA complex having 57.2% α-helical and 3.3% β-strand percentages; See Table S3).
Figure 2.
Far UV CD spectra of HepI apo (red), HepI with 100 µM ADPH (dark green), and HepI with 100 µM ODLA (blue) at 5 °C. Green line shows change in ellipticity at 211nm.
Since the crystal structure of apo HepI and HepI with a fluorinated ADPH analogue (PDB IDs 2GT1 and 2H1H, respectively) bound are virtually identical (the RMSD is 0.19 Å), no change in the CD spectra was expected. The similar CD spectra obtained for HepI and HepI·ADPH are consistent with this expectation and support an “open form” structure for both species and aligns with our earlier conclusion from fluorescence experiments that ADPH binding minimally changes HepI conformation.6 The increase in the ellipticity at 211 nm in the presence of ODLA is also in keeping with our previously observed changes in the intrinsic fluorescence of HepI upon binding of ODLA, which suggested that the protein conformation changes upon formation of the HepI·ODLA complex. The current CD results suggest that binding of ODLA induces folding of loop regions into an alpha helical structure to enhance binding interactions with ODLA. A similar substrate induced increase in α-helicity has been observed previously for the glycosyltransferase N-Acetylglucosaminyltransferase V, suggesting that an induced-fit mechanism might be a general phenomenon for substrate binding to a Rossman-like domain.25 The observed increases in helicity upon ODLA binding suggest that the active site only fully assembles upon substrate binding; therefore the use of current crystal structures to design inhibitors is limited as the structures are static and do not encapsulate the specific conformational changes that are occurring.
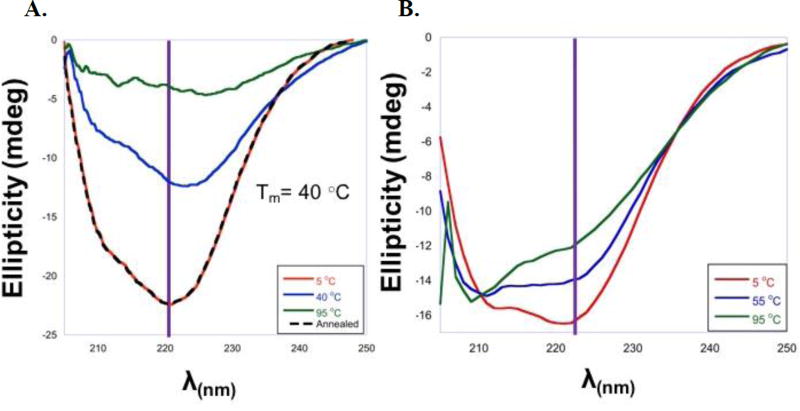
To explore the relative stability of the ligand bound forms, the thermodynamic stability of HepI in the presence and absence of substrates was assessed via a series of thermal denaturation CD experiments (performed from 5 °C – 95 °C, in 5 °C intervals; See Figure 3). As the protein unfolds a loss of ellipticity is observed signifying a loss in secondary structure; as the experiment is performed under equilibrium conditions determination of the melting temperature (Tm; Supplemental Figure S1)26 allows us to assess whether the stability of the protein is altered by the presence of ligand. From the CD melt experiment, Apo HepI was shown to have a Tm of 40 °C and HepI·ADPH has a Tm of 47 °C (Figure 3A, Supplemental Table S3). Interestingly, when ODLA was bound it appears that HepI remains mostly folded (with minimal change in ellipticity) even at 95 °C (Figure 3B). This suggests that ODLA is significantly stabilizing HepI, and due to the lack of unfolding a Tm cannot be calculated for the HepI·ODLA complex, and must be greater than 100 °C. Surprisingly, heating of HepI to 95 °C in the presence of ODLA also did not lead to a significant loss of activity, underscoring that binding of ODLA maintains the protein in an active conformation at temperatures >50 °C above the apo Tm. We hypothesize the stabilization of HepI is most likely caused by a conformational change upon ODLA binding which leads to the formation of hydrogen bonds and/or salt bridges (ionic interactions) within HepI and between HepI and ODLA. In contrast, ADPH alone does not have a major effect on protein stability. This finding is consistent with previously reported stop-flow experiments and current CD results where ADPH binding does not significantly alter HepI conformational.6 Since the two substrates bind to different Rossmann-like domains of HepI (with ODLA binding in the N-terminal domain while ADPH binds in the C-terminal domain), this is suggestive that HepI stabilization arises predominantly from conformational changes in the N-terminal domain.
Figure 3.
A) Far UV CD Spectra HepI apo at 5 °C (red), 40 °C (blue), 95 °C (green), and Annealed at 5 °C (black). B) Far UV CD Spectra HepI with 100 µM ODLA at 5 °C (red), 40 °C (blue), 95 °C (green).
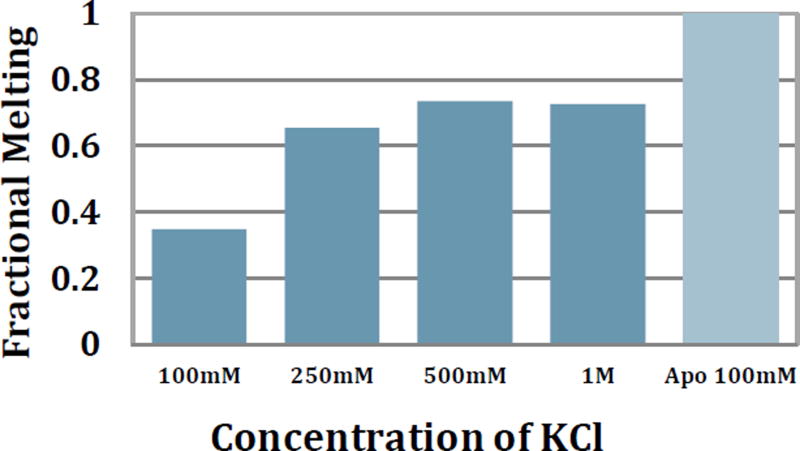
Without a priori information about where ODLA binds, examination of ODLA structure suggests phosphates and carboxylate moieties in ODLA might coordinate to positively charged residues within HepI. To test for the presence of these hypothetical electrostatic binding interactions between HepI and ODLA, CD melt experiments were performed in the presence of increasing salt concentrations (Figures 4 and S2).27 If an increase of salt concentration disrupts these interactions formed between ODLA and HepI, we would anticipate an enhancement of melting of the HepI·ODLA complex. It was observed that at high salt (1 M KCl) there is a 70% change in the overall ellipticity at 222 nm over the temperature range monitored, as compared to the 30% change at 100 mM KCl. At 1 M KCl there is sufficient melting of HepI to allow Tm determination, giving a value of 64.7±0.5 °C. To ensure that higher salt concentration was not causing the protein to adopt a catalytically impaired or non-native conformation, reaction kinetics were determined at both 100 mM and 2 M KCl (the extremes described herein). Under the high salt conditions, there was only a 10-fold decrease in the ODLA dependent Km and a small increase in kcat relative to the lower salt conditions (Supplemental Table S1). A slight increase in HepI stability was also observed in CD melts done with no substrate present at high salt concentrations (Supplemental Table S3). We suggest therefore that HepI is stabilized by ionic interactions (either from ODLA or salt) and ODLA affinity is impacted by the high salt. The nearly 2-fold increase in observable melting at 250 mM KCl suggests that a high salt environment reduces HepI·ODLA stability and facilitates unfolding, which suggests that ionic interactions between ODLA and HepI must have been at least partially disrupted. Additionally, it has previously been shown that salts can have a stabilizing effect on α-helices and proteins.28 We therefore examined the α-helicity under high salt (1M KCl), and observed that it was relatively unchanged from the 100 mM salt concentration described above for the apo protein (40.2% α-helical and 17.0% β-strand). This suggested that the changes in behavior are the result of substrate-induced changes rather than from a change in protein conformation induced by salt. Furthermore, the HepI stability (Tm) did not revert to what was observed for the apo protein at high salt concentrations, as HepI coordination of ODLA probably also involves other interactions, such as hydrogen bonds and hydrophobic interactions, in addition to the electrostatic interactions (based upon its structure). Consequently, the high salt concentration can only partially disrupt the stabilization effects caused by ODLA binding (Table S3).
Figure 4.
Far UV CD spectra monitoring the change in ellipticity at 222 nm with varying salt concentrations in the presence of ODLA as temperature increases from 5 °C to 95 °C. Fractional Melt was determined to be [(max – min)/min]/ (max WT–min WT)/min WT.
Isolating the protein conformational changes causing HepI stabilization
Building upon an extensive literature describing the use of tryptophan (Trp) residues to report on conformational changes upon substrate binding,29–38 we saw HepI, with its 8 Trp residues, as an ideal candidate to explore the protein dynamics of a GT-B protein. Previously, we observed a blue shift in the intrinsic tryptophan (Trp) fluorescence spectrum of HepI upon binding ODLA, which suggests that one or more Trp residue moves from a more polar to a more non-polar or hydrophobic environment upon complex formation. While there are many factors that can affect tryptophan fluorescence such as solvent polarity, conformational changes, hydrogen bonding, excited state reactions and tryptophan-tryptophan interactions,39 we hypothesized that this spectral change could enable us to understand the HepI protein dynamics that occur prior to catalysis. We noted that HepI contains 8 Trp residues at positions 35, 47, 62, 66, 116, 194, 199 and 217, and elected to study how each Trp residue contributes to the fluorescence and by inference, protein dynamics (Figure 1A). In advance of mutagenesis, we performed a solvent accessibility surface area (SASA) analysis on the apo crystal structure, as well as from the ensemble of spatial orientations adopted throughout a 100 ns MD simulation to identify the Trp residues most likely to contribute to the observed fluorescence change (Table S4, Figure 5). Figure 5 shows that most Trp residues have small deviations in their geometries as compared to the crystal structure with the exception of Trp66 and Trp194, both of which have large changes and therefore are either very dynamic and/or are more solvent accessible than would be predicted by the crystal structure (Table S4). Trp35 was observed to have zero or near zero solvent accessibility either in the crystal structure or in the simulation. Since Trp35 is already fully in a hydrophobic environment, we judged it unlikely to contribute to the observed changes in intrinsic HepI fluorescence and therefore we chose not to mutate this residue.
Figure 5.
Three Dimensional Spatial Distribution of Trp Side-chains. Colored regions indicate the predicted, aggregate occupied volume of HepI Trp side-chain heavy atoms as reported by a 100ns molecular dynamics trajectory of the wild-type HepI apo form structure (PDB: 2GT1) with > 20,000 atomic coordinate frames. The description of this figure in the text and this description do not quite match. In the text, it suggests that we are looking at the difference in SASA – but here it is just the volume that the Trp residue is occupying. The 3-D spatial distribution is more about how much space the Trp residue is occupying and how dynamic it is. Perhaps you can report the SASA numbers in a table to highlight the differences. Also a color code for the Trp residues would be useful here.
Driven by concern about protein alterations that might result from removing all of the Trp residues simultaneously and building upon precedent in the literature for more conservative approaches for studying proteins with many Trp residues,30, 38, 40–44 each of the remaining seven Trp residues were individually mutated to phenylalanine (Phe) residues. All of the point mutants were tested for alterations in kinetics, thermal stability and fluorescence spectra relative to HepI-WT. Reaction kinetics for each of the Trp mutants showed little change, with only slightly diminished kcat and slightly increased Km (Table S1). Given that none of the Trp residues are near the active site, it was anticipated that catalysis would not be dramatically impaired. Additionally, CD spectra for all mutants with and without ODLA were obtained and resulted in similar spectra as wild-type, implying that all mutants are folded and bind substrate normally. Interestingly, the CD melts revealed that the thermal stability of all apo mutants, except W47F and W194F, were increased relative to wild-type (Table S3), perhaps suggesting that the remaining Trp residues are dynamic in the protein. Due to the minimal impact of mutagenesis on kinetics and thermodynamics, we reasoned therefore that any changes observed in the fluorescence of these mutants would be due specifically to the removal of that tryptophan and not because of changes in the general behavior or folding of the mutant enzymes.
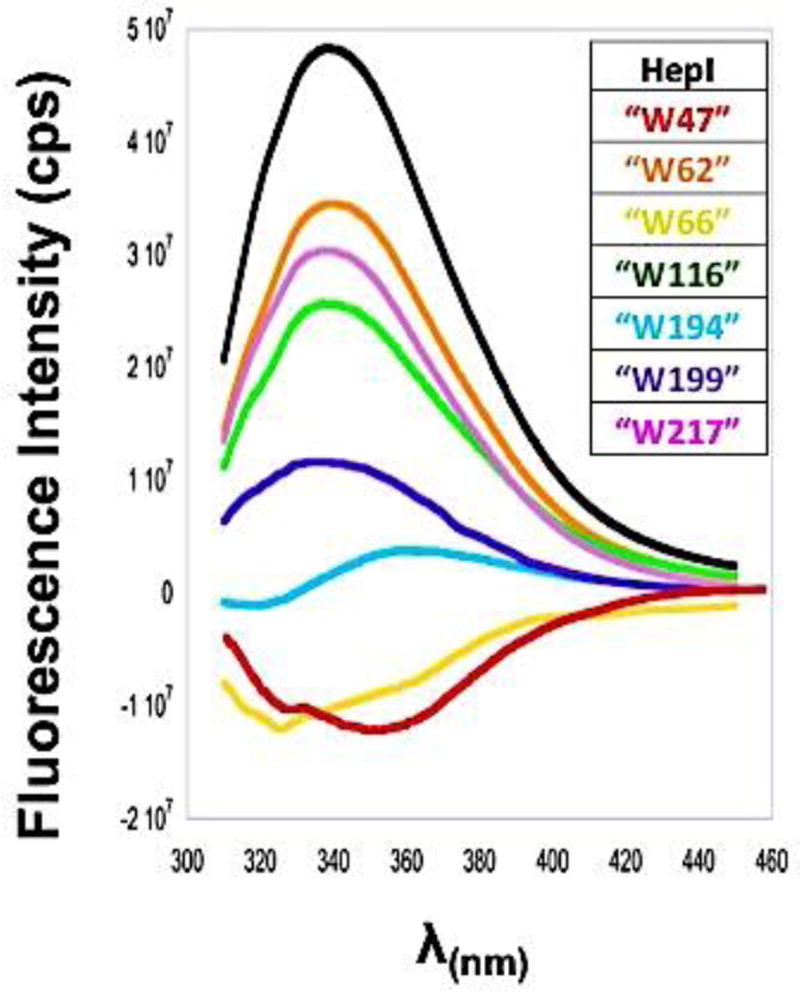
To assess the contribution of each Trp to the overall fluorescence spectrum of HepI, individual Trp florescence spectra were generated by subtracting those obtained for each mutant protein from that of HepI-WT (Figure 6). This analysis of Trp fluorescence has widely been used to approximate the contribution of a single Trp to the spectra of a protein. 40, 41, 45, 46 However, it is important to note that this analysis may not account for any of the complex interactions that may happen between each Trp residue and other residues within the protein, including Trp residues being quenched and/or having spectral shifts caused by neighboring phenylalanine, tryptophan or charge residues.47, 48 As shown in the case of the fluorescence spectra of the bacterial ribonuclease Barnase, energy transfer between proximal tryptophan residues was observed; additionally, a Trp residue was quenched by a nearby charged His residue.43, 45 Since there are multiple Trp residues proximal to each other in HepI, as well as lysine and arginine residues adjacent to some Trps, similar complex interactions could impact our analysis. As can be seen in Figure 6, Trp62, Trp116 and Trp217 contribute most significantly to HepI’s fluorescence,48 with Trp199 having a smaller but non-zero contribution. The remaining three residues (Trp47, Trp66, and Trp194) all have either very slight contribution or a negative contribution to the fluorescnce. Trp66 and Trp194 are both within close proximity to other Trp residues (Trp62 and Trp199, respectively), which could result in some fluorescence quenching could result from the interactions with these neighboring tryptophans.45
Figure 6.
Spectra of individual Trp residues and WT HepI, calculated by subtracting the mutant spectrum from WT spectrum using a subtraction factor of 1 in Spekwin32-optical spectroscopy software.
To examine how each Trp mutation contributed to the overall ODLA-induced fluorescence spectral changes of HepI, λmax were determined for all mutants in the Apo form and with ODLA, ADPH and both substrates bound (Table 1 and Figure S3). Based upon our prior results and those of others, Trp located in environments that are more non-polar are blue shifted relative to those in more polar/solvated environment.39, 49 Observing the changes in fluorescence wavelengths (or lack of changes) for a given mutant would allow us to determine the specific impact of substrate binding on the solvation state of that Trp residue. While the fitting of the data allowed for determination of λmax with high precision, all values were rounded to the nearest integer value to account for the intrinsic experimental error introduced by our acquisition parameters. For all mutants, when ADPH was bound, the fluorescence spectra were virtually identical to apo enzyme, as was observed for HepI-WT. In contrast, the binding of ODLA resulted in much larger changes to the peak emission wavelengths (Figure 7, S3, and Table 1). Removal of Trp47 resulted in nearly identical λmax values as wild-type under all conditions. The mutation of this residue resulted in an increase in the overall fluorescence intensity relative to HepI-WT, and the wavelengths for Trp47Phe λmax (for the apo or the ODLA complex) are also virtually unchanged (any change in λmax of ≤ 1 nm relative to HepI-WT was considered identical). This result suggests that Trp47 has little or no role in the overall ODLA induced blue shift.
Table 1.
Trp fluorescence emission maxima (λmax) for HepI-WT and Trp mutants. Spectra were obtained with 1 uM HepI at 25 °C and samples were excited at 290 nm. Excitation slit width = 2 nm and Emission slit width = 4 nm. All values have an associated error ± 1 nm.
| Fluorescence λmax | ||||
|---|---|---|---|---|
| Apo (nm) | ADPH (nm) |
ODLA (nm) |
Both (nm) | |
| HepI | 340 | 340 | 334 | 334 |
| W47F | 340 | 340 | 334 | 334 |
| W62F | 337 | 337 | 336 | 335 |
| W66F | 337 | 337 | 334 | 334 |
| W116F | 339 | 339 | 336 | 337 |
| W194F | 337 | 337 | 334 | 333 |
| W199F | 341 | 338 | 336 | 336 |
| W217F | 340 | 340 | 341 | 340 |
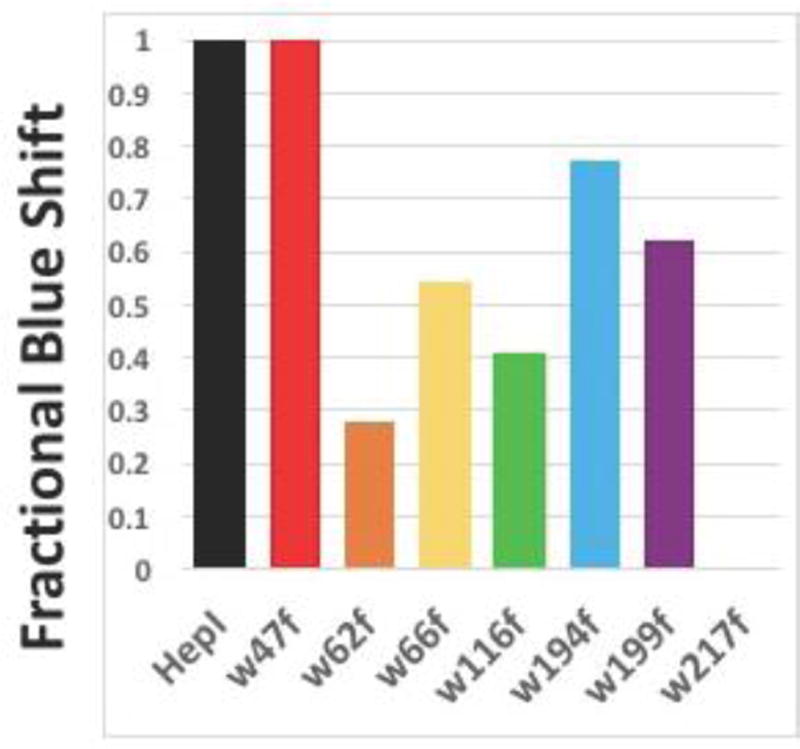
Figure 7.
Degree of blue shift upon ODLA binding by calculating the change between Apo and ODLA spectra. Fractional blue shift was determined to be [(Apo mutant – ODLA mutant)]/(Apo WT-ODLA WT). Note: Trp217Phe has a red shift, and therefore no blue shift is indicated.
Trp66Phe and Trp194Phe exhibited λmax changes in the apo form relative to HepI-WT; even though the HepI·ODLA complex peak wavelengths were unchanged relative to HepI-WT. Both of these mutants exhibit a 3 nm blue-shift in λmax compared to wild-type, which suggests these Trp residues reside in a highly polar environment in the apo HepI-WT protein and contribute to the overall emission maximum of the WT protein. For the Trp116Phe and Trp199Phe mutants, both exhibit peak wavelengths similar to apo HepI-WT in their apo forms, however neither of these proteins demonstrate wild-type-like blue shifts upon ODLA binding. The partial blue shifts exhibited by these two mutants suggest that these Trp residues are minor contributors of the ODLA-induced conformational changes. In the apo forms Trp62Phe, Trp66Phe and Trp194Phe exhibit a blue shift in peak emission resulting in a smaller wavelength shift relative to HepI-WT, like the Trp116Phe and Trp199Phe mutants. Interestingly, Trp62Phe, Trp116Phe and Trp199Phe all exhibit peak maxima that are red-shifted relative to WT HepI upon ODLA binding, suggesting that all three residues contribute to the observed blue shift. These results indicate that apo HepI adopts a conformation where Trp62 is in a polar environment, and moves to a more hydrophobic environment upon ODLA binding, making Trp62 a contributor to the overall fluorescence change. The remaining mutant, Trp217Phe, has a λmax value that is unchanged relative to HepI-WT as the apo protein, and surprisingly, upon ODLA binding exhibits a red shift (+7 nm) relative to HepI-WT. This is the most significant effect observed from mutagenesis and suggests that Trp217 is the major contributor to the ODLA-induced blue shift since its removal eliminates the ability to observe ODLA binding (Figures 7 and S3).
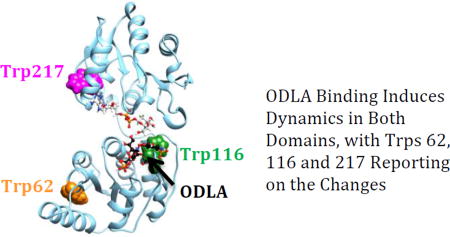
The ODLA binding induced changes in the behavior of Tryptophan residues in the N-terminus (Trp62, Trp66 and Trp116) were anticipated a priori since the sugar acceptor substrates have been shown in other systems to bind to the N-terminal domain.15, 50, 51 Mutation of Trp62 and Trp116 had no change from wild-type in their apo fluorescence spectra but exhibit a red-shift in λmax relative to HepI-WT upon binding of ODLA; thus, we consider Trp62 and Trp116 to be local reporters of ODLA; thus, we consider Trp62 and Trp116 to be local reporters of ODLA binding. These residues are adjacent to positively charged amino acids which may be interacting with the phosphates and carboxylates of the ODLA. This would then likely result in an internalization of these residues upon binding. Taken together, these spectral changes provide insights into the regions that may drive the increase in α-helicity observed in our CD experiments triggered by HepI·ODLA complex formation. Further investigations to discern the residues important for binding and the associated conformational changes are ongoing.
What is most unexpected is that both Trp199 and Trp 217 are in a more non-polar environment upon binding of ODLA, as determined from the peak emission of the point mutants. Mutagenesis of Trp217 (and Trp199) was not expected to impact the fluorescence spectra on the basis of their location in the C-terminal domain since ODLA binds to the N-terminal domain of HepI. Interestingly, these residues in the C-terminal domain are located proximal to the ADPH binding site. While it is possible that the mutation of Trp217 to a hydrophobic residue (Phe) resulted in small structural changes to the adjacent amino acids ultimately altering the intrinsic fluorescence, since the apo protein behaved like HepI-WT, this seems unlikely. The observation that mutagenesis of Trp217, which is located directly adjacent to ADPH but is over 21 Å from the catalytic Asp13 residue that deprotonates the nucleophile, has the most pronounced impact on the blue shift shows that the C-terminal domain residues are altered by the binding of a substrate to the N-terminal domain. This indicates that these two binding sites might be communicating their occupancy to each other via conformational changes. Perhaps binding of ODLA leads to the closing of HepI, ultimately by burying Trp217 through loop/loop interactions between the two domains as they become proximal. Dramatic changes observed in Trp217Phe mutant suggest that the true closed structure may even be more 'closed' then our model suggests (Figure 1B). Since crystal structures of GT-B structural enzymes that are unliganded (apo enzymes) uniformly adopt an open structural conformation, while the few closed GT-B structures are of liganded complexes, it is clear that the occupancy of the ligand binding sites impacts the overall conformational state of the enzyme.14–17 These results provide the first evidence of GT-B enzymes having inter-domain alterations related to the occupancy of a substrate binding domain. Further investigations examining the conformational dynamics (both in silico and in vitro) are ongoing and we hope to elucidate the origin of the scarcity of GT-B protein structures that adopt the closed conformation.
Conclusions
In order to effectively inhibit an enzyme, understanding not only its reaction, but also its dynamics is essential.51 Often crystal structures allow us to deduce static changes in an enzyme upon substrate binding. Unfortunately, these structures are not always available. Here, we were able to use circular dichroism and intrinsic tryptophan fluorescence spectra to observe conformational changes in HepI. CD experiments in the presence and absence of ODLA suggest that the HepI active site might not be fully formed in the apo protein, and that conformational changes which increase the overall α-helicity of the protein are necessary for formation of the Michaelis complex. This is an important insight which can help rationalize the dearth of potent inhibitors for glycosyltransferase enzymes, despite prior in silico drug design efforts.50, 52, 53
Additionally, 7 Trp mutants were examined to identify the regions of HepI that are altered upon ligand binding. W62F, and W116F, which are located in the N-terminal ODLA binding domain exhibited smaller changes in peak fluorescence upon ODLA binding, as compared to WT-HepI. These residues are located on dynamic loops (N-3 and N-7) and it is hypothesized that binding of ODLA results in a conformational change stabilizing these loops, which potentially changes the local environment of these residues. We hypothesize based upon the investigations of the HepI stability changes in conjunction with the fluorescence behavior of these mutants, that the dynamic N-3 and N-7 loops bring positively charged amino acid residues proximal to the negatively charged regions of ODLA (phosphates and carboxylates), to form salt-bridges that can be destabilized with high salt. The fluorescence and CD melt experiments perhaps suggest loop movements that are consistent with our previously observed ~80 sec−1, concentration independent, blue shift from our pre-steady state kinetics experiments.6 Additionally, these experiments suggest that HepI functions either through an ordered mechanism (ODLA must bind second for chemistry to occur) or a dynamic mechanism where enzyme is converting between open and closed states, where binding of ODLA shifts the thermodynamic landscape to favor the closed conformational state.
Surprisingly, mutation of Trp217 resulted in complete loss of the blue shift upon substrate binding. We hypothesize that Trp217 could be making contact with the N-terminal loops directly below it (where both Trp62 and Trp66 are located) or could be experiencing changes in its dynamic motions (propagated throughout the protein) as a result of ODLA binding; additional studies would be necessary to clearly understand how ODLA binding is impacting Trp217. While the available crystal structures suggest that the C-terminal domain is relatively static whether or not the sugar donor substrate is bound, this data suggests that ODLA binding leads to changes in the protein that are widespread and not solely confined to the N-terminal domain. This study has helped to isolate regions important for formation of the enzyme substrate complex. Further, there is now evidence that Heptosyltransferase I utilizes an induced fit model for ligand binding that involves rearrangements of both N and C terminal domains and may shed light on regions of the protein to target for development of an inhibitor of Heptosyltransferase I.
Supplementary Material
Acknowledgments
We would like to thank Candice Etson for advice on performing calculations with MATLAB.
Funding
E. A. T., J. M. C. and C. A. R. M. thank the National Institutes of Health for grant support (1R15AI119907-01). J. M. C. is grateful for National Institutes of Health Doctoral Studies in Molecular Biophysics Training Grant (2T32GM008271-24).
Abbreviations
- GT
glycosyltransferase
- HepI
heptosyltransferase I
- Heptose
L-glycero-D-manno-heptose
- ADP-Heptose or ADPH
ADP- L-glycero-D-manno-heptose
- LPS
lipopolysaccharide
- ODLA
O-deacylated E. coli Kdo2-LipidA
- Trp
tryptophan
- Phe
phenylalanine
- CD
circular dichroism
- MD
molecular dynamics
- SASA
solvent accessible surface area
Footnotes
This work was supported by NIH grants 1R15AI119907-01 (E. A. T.) and 2T32GM008271-24 (J. M. C.).
Supporting Information
Supporting information contains primer sequences used for site-directed mutagenesis, and additional kinetic, circular dichroism and fluorescence data.
References
- 1.Xu Z-QQ, Flavin MT, Flavin J. Combating multidrug-resistant Gram-negative bacterial infections. Expert opinion on investigational drugs. 2014;23:163–182. doi: 10.1517/13543784.2014.848853. [DOI] [PubMed] [Google Scholar]
- 2.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An Evolving Hierarchical Family Classification for Glycosyltransferases. Journal of Molecular Biology. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 3.Pedro MC, Emeline D, Gideon JD, Bernard H. An Evolving Hierarchical Family Classification for Glycosyltransferases. Journal of Molecular Biology. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 4.Ajit Varki RC, Jeffrey Esko, Hudson Freeze, Gerald Hart, Jamey Marth. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1999. [Google Scholar]
- 5.Czyzyk D, Liu C, Taylor E. Lipopolysaccharide biosynthesis without the lipids: recognition promiscuity of Escherichia coli heptosyltransferase I. Biochemistry. 2011;50:10570–10572. doi: 10.1021/bi201581b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czyzyk D, Sawant S, Ramirez-Mondragon C, Hingorani M, Taylor E. Escherichia coli Heptosyltransferase I: Investigation of Protein Dynamics of a GT-B Structural Enzyme. Biochemistry. 2013:5158–5160. doi: 10.1021/bi400807r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grizot S, Salem M, Vongsouthi V, Durand L, Moreau F, Dohi H, Vincent S, Escaich S, Ducruix A. Structure of the Escherichia coli heptosyltransferase WaaC: binary complexes with ADP and ADP-2-deoxy-2-fluoro heptose. Journal of molecular biology. 2006;363:383–394. doi: 10.1016/j.jmb.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 8.Coleman WG, Leive L. Two mutations which affect the barrier function of the Escherichia coli K-12 outer membrane. Journal of bacteriology. 1979;139:899–910. doi: 10.1128/jb.139.3.899-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadrmas JL, Raetz CRH. Enzymatic Synthesis of Lipopolysaccharide in Escherichia coli : Purification and Properties of Heptosyltransferase I. Journal of Biological Chemistry. 1998;273:2799–2807. doi: 10.1074/jbc.273.5.2799. [DOI] [PubMed] [Google Scholar]
- 10.Gronow S, Brade H. Invited review: Lipopolysaccharide biosynthesis: which steps do bacteria need to survive? Journal of Endotoxin Research. 2001;7:3–23. [PubMed] [Google Scholar]
- 11.Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zähringer U, Seydel U, Di Padova F. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 12.Maxime D, Kevin B, Julien I, Michel H, Jean-François N, Stéphane PV. The Inhibition of Liposaccharide Heptosyltransferase WaaC with Multivalent Glycosylated Fullerenes: A New Mode of Glycosyltransferase Inhibition. Chemistry - A European Journal. 2012;18:641–651. doi: 10.1002/chem.201102052. [DOI] [PubMed] [Google Scholar]
- 13.Moreau F, Desroy N, Genevard JM, Vongsouthi V, Gerusz V, Le Fralliec G, Oliveira C, Floquet S, Denis A, Escaich S, Wolf K, Busemann M, Aschenbrenner A. Discovery of new Gram-negative antivirulence drugs: structure and properties of novel E. coli WaaC inhibitors. Bioorganic & medicinal chemistry letters. 2008;18:4022–4026. doi: 10.1016/j.bmcl.2008.05.117. [DOI] [PubMed] [Google Scholar]
- 14.Albesa-Jové D, Giganti D, Jackson M, Alzari P, Guerin M. Structure-function relationships of membrane-associated GT-B glycosyltransferases. Glycobiology. 2014;24:108–124. doi: 10.1093/glycob/cwt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lairson L, Henrissat B, Davies G, Withers S. Glycosyltransferases: structures, functions, and mechanisms. Annual review of biochemistry. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 16.Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vetting MW, Frantom PA, Blanchard JS. Structural and Enzymatic Analysis of MshA from Corynebacterium glutamicum: SUBSTRATE-ASSISTED CATALYSIS. Journal of Biological Chemistry. 2008;283:15834–15844. doi: 10.1074/jbc.M801017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brabetz W, Muller-Loennies S, Holst O, Brade H. Deletion of the Heptosyltransferase Genes rfaC and rfaF in Escherichia coli K-12 Results in an Re-Type Lipopolysaccharide with a High Degree of 2-Aminoethanol Phosphate Substitution. European Journal of Biochemistry. 1997;247:716–724. doi: 10.1111/j.1432-1033.1997.00716.x. [DOI] [PubMed] [Google Scholar]
- 19.Mudapaka J, Taylor EA. Cloning and characterization of the Escherichia coli Heptosyltransferase III: Exploring substrate specificity in lipopolysaccharide core biosynthesis. FEBS letters. 2015;589:1423–1429. doi: 10.1016/j.febslet.2015.04.051. [DOI] [PubMed] [Google Scholar]
- 20.Mehl AF, Crawford MA, Zhang L. Determination of Myoglobin Stability by Circular Dichroism Spectroscopy: Classic and Modern Data Analysis. Journal of Chemical Education. 2009;86:600. [Google Scholar]
- 21.Greenfield NJ. Determination of the folding of proteins as a function of denaturants, osmolytes or ligands using circular dichroism. Nature Protocols. 2007;1:2733–2741. doi: 10.1038/nprot.2006.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Venyaminov S, Yang J. Determination of Protein Secondary Structure. Springer; 1996. [Google Scholar]
- 24.Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins: Structure, Function, and Bioinformatics. 2012;80:374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 25.Zhang N, Peng K-C, Chen L, Puett D, Pierce M. Circular Dichroic Spectroscopy of N-Acetylglucosaminyltransferase V and Its Substrate Interactions. Journal of Biological Chemistry. 1997;272:4225–4229. doi: 10.1074/jbc.272.7.4225. [DOI] [PubMed] [Google Scholar]
- 26.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nature protocols. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson SW, Jonas A, Clayton DF, George JM. Stabilization of α-Synuclein Secondary Structure upon Binding to Synthetic Membranes. Journal of Biological Chemistry. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 28.Kumar A, Rani A, Venkatesu P. A comparative study of the effects of the Hofmeister series anions of the ionic salts and ionic liquids on the stability of α-chymotrypsin. New Journal of Chemistry. 2014;39:938–952. [Google Scholar]
- 29.Yao X, Bleile DW, Yuan Y, Chao J, Sarathy KP, Sanders DAR, Pinto BM, O'Neill MA. Substrate directs enzyme dynamics by bridging distal sites: UDP-galactopyranose mutase. Proteins: Structure, Function, and Bioinformatics. 2009;74:972–979. doi: 10.1002/prot.22206. [DOI] [PubMed] [Google Scholar]
- 30.Garrity JD, Pauff JM, Crowder MW. Probing the Dynamics of a Mobile Loop above the Active Site of L1, a Metallo-β-lactamase from Stenotrophomonas maltophilia via Site-directed Mutagenesis and Stopped-flow Fluorescence Spectroscopy. Journal of Biological Chemistry. 2004;279:39663–39670. doi: 10.1074/jbc.M406826200. [DOI] [PubMed] [Google Scholar]
- 31.Dumitrașcu L, Stănciuc N, Bahrim GE, Ciumac A, Aprodu I. pH and heat-dependent behaviour of glucose oxidase down to single molecule level by combined fluorescence spectroscopy and molecular modelling. Journal of the Science of Food and Agriculture. 2016;96:1906–1914. doi: 10.1002/jsfa.7296. [DOI] [PubMed] [Google Scholar]
- 32.Hogue CWV, Doublié S, Xue H, Wong JT, Carter CW, Jr, Szabo AG. A Concerted Tryptophanyl-adenylate-dependent Conformational Change in Bacillus subtilis Tryptophanyl-tRNA Synthetase Revealed by the Fluorescence of Trp92. Journal of Molecular Biology. 1996;260:446–466. doi: 10.1006/jmbi.1996.0413. [DOI] [PubMed] [Google Scholar]
- 33.Cavatorta P, Sartor G, Neyroz P, Farruggia C, Franzoni L, Szabo AG, Spisni A. Fluorescence and CD studies on the conformation of the gastrin releasing peptide in solution and in the presence of model membranes. Biopolymers. 1991;31:653–661. doi: 10.1002/bip.360310610. [DOI] [PubMed] [Google Scholar]
- 34.Cavatorta P, Spisni A, Szabo AG, Farruggia G, Franzoni L, Masotti L. Conformation of bombesin in buffer and in the presence of lysolecithin micelles: Nmr, CD and fluorescence studies. Biopolymers. 1989;28:441–463. doi: 10.1002/bip.360280140. [DOI] [PubMed] [Google Scholar]
- 35.Szabo AG, Rayner DM. Fluorescence decay of tryptophan conformers in aqueous solution. Journal of the American Chemical Society. 1980;102:554–563. [Google Scholar]
- 36.Jang DJ, el-Sayed MA. Tryptophan fluorescence quenching as a monitor for the protein conformation changes occurring during the photocycle of bacteriorhodopsin under different perturbations. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:5815–5819. doi: 10.1073/pnas.86.15.5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson ID, Hudson BS. Environmental modulation of M13 coat protein tryptophan fluorescence dynamics. Biochemistry. 1989;28:6392–6400. doi: 10.1021/bi00441a035. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Barkley MD. Toward Understanding Tryptophan Fluorescence in Proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 39.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer; 2006. [Google Scholar]
- 40.Bhanu MK, Kendall DA. Fluorescence spectroscopy of soluble E. coli SPase I Δ2-75 reveals conformational changes in response to ligand binding. Proteins. 2014;82:596–606. doi: 10.1002/prot.24429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haiyuan D, Ishita M, Donald O. Lipid and Signal Peptide-Induced Conformational Changes within the C-Domain of Escherichia coli SecA Protein. Biochemistry. 2001;40:1835–1843. doi: 10.1021/bi002058w. [DOI] [PubMed] [Google Scholar]
- 42.Imhof N, Kuhn A, Gerken U. Substrate-dependent conformational dynamics of the Escherichia coli membrane insertase YidC. Biochemistry. 2011;50:3229–3239. doi: 10.1021/bi1020293. [DOI] [PubMed] [Google Scholar]
- 43.Loewenthal R, Sancho J, Fersht AR. Fluorescence spectrum of barnase: contributions of three tryptophan residues and a histidine-related pH dependence. Biochemistry. 1991;30:6775–6779. doi: 10.1021/bi00241a021. [DOI] [PubMed] [Google Scholar]
- 44.Biesso A, Xu J, Muíño PL, Callis PR, Knutson JR. Charge invariant protein-water relaxation in GB1 via ultrafast tryptophan fluorescence. Journal of the American Chemical Society. 2014;136:2739–2747. doi: 10.1021/ja406126a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engelborghs Y, Fersht A. Barnase: Fluorescence Analysis of A Three Tryptophan Protein. In: Lakowicz JR, editor. Topics in Fluorescence Spectroscopy: Volume 6: Protein Fluorescence. Springer US; Boston, MA: 2000. pp. 83–101. [Google Scholar]
- 46.Eftink MR. Intrinsic Fluorescence of Proteins. In: Lakowicz JR, editor. Topics in Fluorescence Spectroscopy: Volume 6: Protein Fluorescence. Springer US; Boston, MA: 2000. pp. 1–15. [Google Scholar]
- 47.Rouviere N, Vincent M, Craescu CT, Gallay J. Immunosuppressor Binding to the Immunophilin FKBP59 Affects the Local Structural Dynamics of a Surface β-Strand: Time-Resolved Fluorescence Study†. Biochemistry. 1997;36:7339–7352. doi: 10.1021/bi962289w. [DOI] [PubMed] [Google Scholar]
- 48.Vivian JT, Callis PR. Mechanisms of Tryptophan Fluorescence Shifts in Proteins. Biophysical Journal. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakowicz JR. Protein fluorescence. Kluwer Academic; New York: 2002. [Google Scholar]
- 50.De Leon GP, Elowe NH, Koteva KP, Valvano MA, Wright GD. An In Vitro Screen of Bacterial Lipopolysaccharide Biosynthetic Enzymes Identifies an Inhibitor of ADP-Heptose Biosynthesis. Chemistry & Biology. 2006;13:437–441. doi: 10.1016/j.chembiol.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 51.Wong CF, McCammon AJ. Dynamics and design of enzymes and inhibitors. Journal of the American Chemical Society. 1986;108:3830–3832. [Google Scholar]
- 52.Liu X, Cao Y-F, Ran R-X, Dong P-P, Gonzalez FJ, Wu X, Huang T, Chen J-X, Fu Z-W, Li R-S, Liu Y-Z, Sun H-Z, Fang Z-Z. New insights into the risk of phthalates: Inhibition of UDP-glucuronosyltransferases. Chemosphere. 2016;144:1966–1972. doi: 10.1016/j.chemosphere.2015.10.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dkhar HK, Gopalsamy A, Loharch S, Kaur A, Bhutani I, Saminathan K, Bhagyaraj E, Chandra V, Swaminathan K, Agrawal P, Parkesh R, Gupta P. Discovery of Mycobacterium tuberculosis α-1,4-Glucan Branching Enzyme (GlgB) Inhibitors by Structure-and Ligand-based Virtual Screening. Journal of Biological Chemistry. 2015;290:76–89. doi: 10.1074/jbc.M114.589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.