Abstract
Type I interferon (IFNα/β) induces antiviral and antiproliferative responses in cells through the induction of IFN-stimulated genes (ISGs). Although the roles of IFN-activated STAT1 and STAT2 in the IFN response are well described, the function of STAT3 is poorly characterized. We investigated the role of STAT3 in the biological response to IFNα/β in mouse embryonic fibroblasts (MEFs) with a germ line deletion of STAT3. These STAT3 knockout (STAT3-KO) MEFs were reconstituted with STAT3 or the F705-STAT3 mutant (unphosphorylated STAT3) where the canonical Y705 tyrosine phosphorylation site was mutated. We show that both STAT3 and unphosphorylated STAT3 expression enhance the sensitivity of MEFs to the antiviral, antiproliferative and gene-inducing actions of IFN. By chromatin immunoprecipitation assays, unphosphorylated STAT3 appears to bind, albeit weakly, to select gene promoters to enhance their expression. These results suggest that unphosphorylated STAT3 plays an important role in the IFN response pathway.
Keywords: interferon, gene expression, STAT, antiviral, antiproliferative
INTRODUCTION
Interferons (IFNs) are antiviral cytokines that have significant effects on cell proliferation, differentiation, apoptosis, and the immune system. Binding of the type I IFNs (IFNα, IFNβ, and IFNω) to their cognate cell surface receptor leads to the activation of the receptor-associated JAK1 and TYK2 tyrosine kinases. This process then results in the recruitment, tyrosine phosphorylation, dimerization, and nuclear translocation of the signal transducers and activators of transcription (STAT) proteins [1, 2]. In the classical JAK-STAT signaling pathway, STAT1 and STAT2 in a complex with IRF9 bind to the conserved IFN-stimulus response element (ISRE) within the promoters of the early response IFN-stimulated genes (ISGs), inducing their expression. However, IFNα/β may also activate additional signaling pathways that play important roles in the induction of IFN’s biological effects.
STAT3, which was originally identified as the transcription factor for acute phase response genes, is activated by a wide variety of cytokines suggesting that it may integrate diverse signals into common transcriptional responses [3–5]. Activation of STAT3 is defined by the phosphorylation of tyrosine 705 within the transactivation domain, which is required for STAT3 dimerization, nuclear translocation, and induction of gene transcription. However, Y705 phosphorylation may not be needed for all of STAT3 functions. For example, overexpression of a F705-STAT3 mutant, which cannot be phosphorylated on residue 705 and is denoted as “unphosphorylated” STAT3, enhanced the expression of a number of genes critical to tumorigenesis, suggesting that STAT3 also regulates transcription by “nontraditional” pathways [6]. A critical role in cellular physiology is also implicated by the finding that knockout of the STAT3 gene in mice leads to early embryonic lethality [7], presumably due in part to the finding that STAT3 plays an important role in mitochondrial respiration [8]. Moreover, STAT3 apparently plays an important role in tumorigenesis by regulating cell cycle progression, apoptosis, angiogenesis, invasion and metastasis, and evasion of immune surveillance [9–12]. Many of these processes have been identified as targets of IFN action. As a point of convergence for numerous oncogenic signaling pathways, STAT3 is constitutively activated at a frequency of 50 to 90% in diverse human cancers [13]. Thus, the activation of STAT3 by IFN would be expected to have important cellular consequences. In studies characterizing the IFN response pathway in an IFN-resistant subclone of human lymphoblastoid cells, we identified a defective STAT3-dependent IFN signaling pathway that can be rescued by stable expression of STAT3 [5, 14, 15]. Moreover, Cxcl11 is a STAT3-regulated gene induced by IFNα/β that apparently does not require STAT3 tyrosine phosphorylation for its induction [16].
The present study explored the involvement of “unphosphorylated” STAT3 in the biological response to IFNα/β in mouse embryonic fibroblasts (MEFs) with a germ line deletion of STAT3. These STAT3 knockout (STAT3-KO) MEFs were reconstituted with wild-type STAT3 as well as the F705-STAT3 mutant where the canonical Y705 tyrosine phosphorylation site was mutated to a phenylalanine, i.e. “unphosphorylated” STAT3.
Materials and Methods
Biological reagents and cell culture
The biological activity of recombinant rat IFNβ [17] was expressed in terms of international reference units/mL using the NIH reference mouse IFNβ standard [14]. STAT3-knockout (STAT3-KO) mouse embryo fibroblasts (MEFs) [18] were reconstituted with empty vector (EV), WT-STAT3 or a STAT3 mutant with the canonical phosphorylation site at Y705 mutated to a phenylalanine (F705-STAT3). The various constructs were cloned in the pcEF expression vector [5]. Cells (107) were transfected by electroporation (capacitance 300 μF, 250 V) with 50 μg of salmon sperm DNA and 20 μg of plasmid DNA for each sample. Stable transfectants were selected for neomycin resistance (0.4 mg/ml G418). MEFs were plated at 1 × 104 cells/cm2 every 3 days in DMEM supplemented with 10% defined calf serum (DCS), 100 U/mL penicillin G and 100 μg/ml streptomycin.
Antiviral and antiproliferative assays
To determine antiviral activity, cells were incubated overnight with rat IFNβ, followed by infection with vesicular stomatitis virus (VSV) for 1 h at 0.1 plaque-forming units (pfu) per cell. At 24 h post-infection, the virus yield in the medium was assayed by plaque formation on Vero cells [19]. To determine antiproliferative activity, MEF cultures were plated at 1 × 105 cells in 25-cm2 flasks, treated with rat IFNβ, and at 2 days after IFN addition cells were harvested by trypsinization and enumerated in a Coulter Counter [20].
RNA Preparation and Microarray Analysis
Total cellular RNA was prepared from three individual 75cm2 flasks of control and rat IFNβ-treated (1,000 units/ml for 5 h) STAT3-KO MEFs transfected with empty vector, STAT3 or the F705-STAT3 mutant using the RNeasy Mini kit (QIAGEN). RNA (~10 μg) from two separate experiments was submitted to the UTHSC Center of Genomics and Bioinformatics (Memphis, TN) for labeling and hybridization to Mouse-6 BeadChips (Illumina Inc.). Microarray data analysis was then carried out using IlluminaGUI (http://illuminagui.dnsalias.org). ISGs were defined as the genes whose expression was increased by IFN treatment by more than 2-fold (P<0.05). Unsupervised hierarchal clustering was carried out using standard correlation coefficients on the ISGs.
Quantitative Real time-PCR
Total RNA was isolated using RNeasy Mini kit. Quantitative real time-PCR (qPCR) was carried out on the iCyclerIQ detection system (BioRad, Hercules, CA) using iScript One-Step RT-PCR Kit with SYBR Green (BioRad). Reaction parameters were as follows: cDNA synthesis at 50°C for 20 min, iScript reverse transcriptase inactivation at 95°C for 5 min, PCR cycling at 95°C for 10 sec and 60°C for 30 sec for 40 cycles. Gene expression data was normalized to the expression of the β-actin housekeeping gene. The following forward and reverse primers were used for each gene: β-Actin (5′-AGTGTGACGTTGACATCCGTA-3′; 5′-GCCAGAGCAGTAATCTCCTTCT-3′); Adar (5′-TGAGCATAGCAAGTGGAGATACC-3′; 5′-GCCGCCCTTTGAGAAACTCT-3′); Cxcl10 (5′-CGTGTTGAGATCATTGCCAC-3′; 5′-TTAAGGAGCCCTTTTAGACC-3′); Cxcl11 (5′-GGCTTCCTTATGTTCAAACAGGG-3′; 5′-GCCGTTACTCGGGTAAATTACA-3′); Gadd45γ (5′-GGGAAAGCACTGCACGAACT-3′; 5′-AGCACGCAAAAGGTCACATTG-3′); Gbp1 (5′-GCAGCAAATAGAGCATTGGC; 5′-CCATTAACTCTTTCCGTTCC-3′); Ifi47 (5′-AAGTTCCCCCTTGATGTCTG-3′; 5′-TTGCCTGAACAAGAACAGTG-3′); Irf1 (5′-ATGCCAATCACTCGAATGCG-3′; 5′-TTGTATCGGCCTGTGTGAATG-3′); Myd88 (5′-AGGACAAACGCCGGAACTTTT-3′;5′-GCCGATAGTCTGTCTGTTCTAGT-3′); Osmr (5′-CATCCCGAAGCGAAGTCTTGG-3′; 5′-GGCTGGGACAGTCCATTCTAAA-3′); Stat2 (5′-TCCTGCCAATGGACGTTCG-3′; 5′-GTCCCACTGGTTCAGTTGGT-3′); T2bp (5′-GAACGGAGCTGTTGAACTGTT-3′; 5′-GACGCTGATACAGAGGAGACG-3′); Tnfsf10 (5′-GCTTGCAGGTTAAGAGGCAAC-3′; 5′-TCTCCGAGTGATCCCAGTAATG-3′).
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) assays were carried out using the ChIP-IT™ Express Enzymatic kit (Active Motif, Carlsbad, CA) according to the manufacturer’s instructions. To improve the sensitivity of the ChIP assay, cells were treated with the cross-linking agent Ethylene glycolbis(succinimidylsuccinate) (EGS, 1mM) for 30 min at 22°C prior to cross-linking with 1% formaldehyde (10 min at 22°C) [21]. Chromatin was sheared to an average size of 200 bp by enzymatic digestion, and then immunoprecipitated with anti-pY701-STAT1(A-2, Santa Cruz) or STAT3 (C-20, Santa Cruz). The ChIP-PCR primers for each gene were designed to amplify a proximal promoter region that contains putative STAT3 binding sites. The promoter sequences of ISGs (1kb upstream of the transcription start site) were retrieved using UCSC Genome Browser (http://genome.ucsc.edu) and putative STAT3 binding sites were identified by TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html). The following forward and reverse primers were used for ChIP-PCR:
Cxcl11, TTCCTGAGTTGGTGGGACTC; AAGCCACTGGAAGGTGAAAG; Gadd45γ, TCGCACAATGACTCTGGAAG; CCCACCCTGCAAACTTCTATC; Myd88, CTTTTAGGCTGGGTAGTTCTGG; TCGCCTGTGGAGTTTCCTAC; Oas1γ, TCGATGGGATCCTCAGAAAG; CCTGGCTGAAATGGGAAGTAG; Osmr, ACCAGGAGCAAATTCCTGTG; AATCAACTACGGGGCAAGTG; T2bp, ATTTCCATCCCAACCTCCTT; CAGAAGCCTCCTCACCTGTC; Tnfsf10, CATTGTTGCCTTCAGCAGTC; TCCTGGACAAAGGACAAAGG.
Data analysis
All quantitative data represent at least three independent experiments performed in duplicate, at least, and presented as mean ± S.D.
RESULTS
The role of STAT3 in the antiproliferative and antiviral actions of IFN
To define the role of STAT3 in IFN action, we compared the IFN sensitivity of WT-MEFs to that of STAT-KO MEFs. As shown in Fig 1A, STAT3-KO MEFs were relatively resistant to the antiviral and antiproliferative actions of IFN when compared to WT-MEFs. To investigate the role of unphosphorylated STAT3 in IFN action, STAT3-KO MEFs were reconstituted with empty vector (KO+EV), STAT3 (KO+STAT3) or the F705 mutant form of STAT3 (KO+F705−STAT3). To verify that the STAT3 transfectants expressed STAT3 and responded to IFN, transfectants were treated with IFN (1,000 IU/mL, 30 min), lysed and immunoblotted for STAT3 and Y705-STAT3. As shown in Figure 1A, STAT3 was expressed in transfectants expressing STAT3 or F705-STAT3, and in untransfected wild-type MEFs (WT-MEFs). STAT3-KO cells transfected with empty vector (KO+EV) showed no detectable STAT3 expression. Moreover, IFN treatment resulted in STAT3 tyrosine phosphorylation in KO+STAT3 MEFs, but not in KO+F705−STAT3 transfectants.
Figure 1. The role of STAT3 in IFN-induced antiproliferative and antiviral activities.
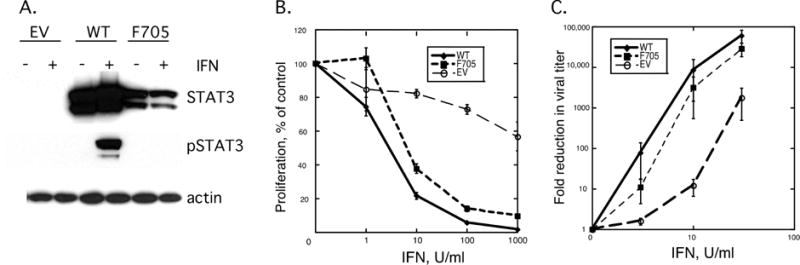
(Panel A) Lysates prepared from the indicated STAT3-KO MEFs before and after IFN addition (1,000 IU/mL, 30 min) were resolved by SDS-PAGE, blotted onto PVDF membranes and probed with anti-STAT3, anti-Y705-STAT3 or β-actin. (Panel B) The indicated STAT3-KO MEFs were treated with varying IFN concentrations (1–1000 IU/mL) and after 2 days the cell counts were determined in a Coulter Counter. (Panel C) The indicated STAT3-KO MEFs were treated overnight with varying IFN concentrations (0–30 IU/mL), infected with VSV (0.1 pfu/cell), and at 24 h post-infection the viral titer was assayed on Vero cells. The results of at least three independent experiments, each carried out in duplicate, were averaged.
To determine the role of STAT3 in the antiproliferative activity of IFN, KO+EV, KO+STAT3, and KO+F705−STAT3 MEFs were cultured for two days in the presence of varying concentrations of IFN. As shown in Figure 1B, KO+EV MEFs were relatively resistant to the antiproliferative action of IFN, with ~40% reduction in cell proliferation observed at the highest concentration of IFN examined (1,000 IU/mL). In contrast, KO+F705−STAT3 MEFs were highly sensitive to the antiproliferative action of IFN with greater than 50% reduction observed at 10 IU/mL IFN, and >90% reduction in cell proliferation observed at 1000 IU/mL. In addition, KO+F705−STAT3 MEFs were as sensitive to IFN as WT-MEFs, suggesting that unphosphorylated STAT3 restored IFN sensitivity. We then assessed the sensitivity of the various MEFs to the antiviral action of IFN by determining the ability of IFN to inhibit VSV replication. As shown in Figure 1C, STAT3-KO MEFs expressing F705-STAT3 were significantly more sensitive to IFN’s ability to inhibit VSV replication than STAT3-KO MEFs transfected with empty vector. The apparent leftward shift in the dose-response curve suggests that STAT3-expressing MEFs were ~ one log more sensitive to IFN’s antiviral action, as compared STAT3-KO MEFs. Moreover, WT-MEFs were equally sensitive to IFN’s antiviral action as cells expressing the F705-STAT3 mutant, suggesting that unphosphorylated STAT3 play a crucial role in IFN action.
The role of STAT3 in the induction of gene expression by IFN
To define the role of STAT3 in ISG expression, STAT3-KO MEFs were transfected with constructs corresponding to EV, wild-type STAT3 or unphosphorylated STAT3, RNA was isolated from MEFs cultured in the absence or presence of IFNβ, and subjected to microarray analysis. As shown in Figure 2, 168 ISGs were identified (fold change >2, p<0.05). While the expression of ~40% of the ISGs was STAT3-independent (cluster 1), many ISGs were STAT3-regulated (cluster 2 and 3). Among the STAT3-regulated ISGs (31/101), ~30% were IFN-induced in KO+STAT3 and KO+F705−STAT3 MEFs, indicating that their expression was Y705 phosphorylation independent (cluster 3). To further characterize the role of STAT3 in ISG induction, RNA was prepared from KO+EV, KO+STAT3 and KO+F705−STAT3 MEFs treated with IFNβ (5 h, 1000 IU/mL), and gene expression determined by qPCR. Consistent with the microarray data, ISGs such as Cxcl11, Gadd45γ, Myd88 and T2bp were induced to higher levels by IFN in KO+F705−STAT3 MEFs than in KO+EV MEFs. These STAT3-regulated ISGs do not require Y705 phosphorylation for their IFN-induction and were denoted unphosphorylated STAT3-regulated ISGs. In addition, ISGs such as Adar, Cxcl10, Irf1 and Stat2 were induced by IFN to similar extents in both KO+EV and KO+F705−STAT3 MEFs (Fig. 4), indicating that the induction of these genes was independent of STAT3 expression.
Figure 2. Microarray analysis of ISG expression.
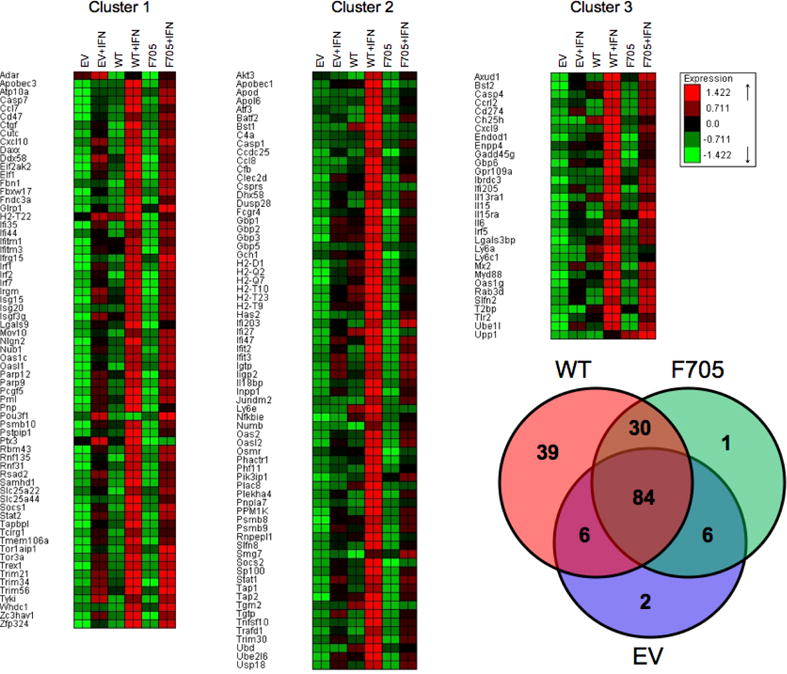
The Venn diagram shows the distribution of the 168 ISGs (Fold induction by IFN >2, P<0.05) between KO+EV, KO+WT and KO+F705−STAT3 MEF lines. Based on expression patterns, the ISGs were partitioned into three clusters: cluster 1 represents STAT3-independent ISGs; cluster 2 represents tyrosine phosphorylated STAT3-regulated ISGs; and cluster 3 represents unphosphorylated STAT3-regulated ISGs.
Figure 4. The effects of IFN on STAT binding to the promoters of STAT3-regulated ISGs.
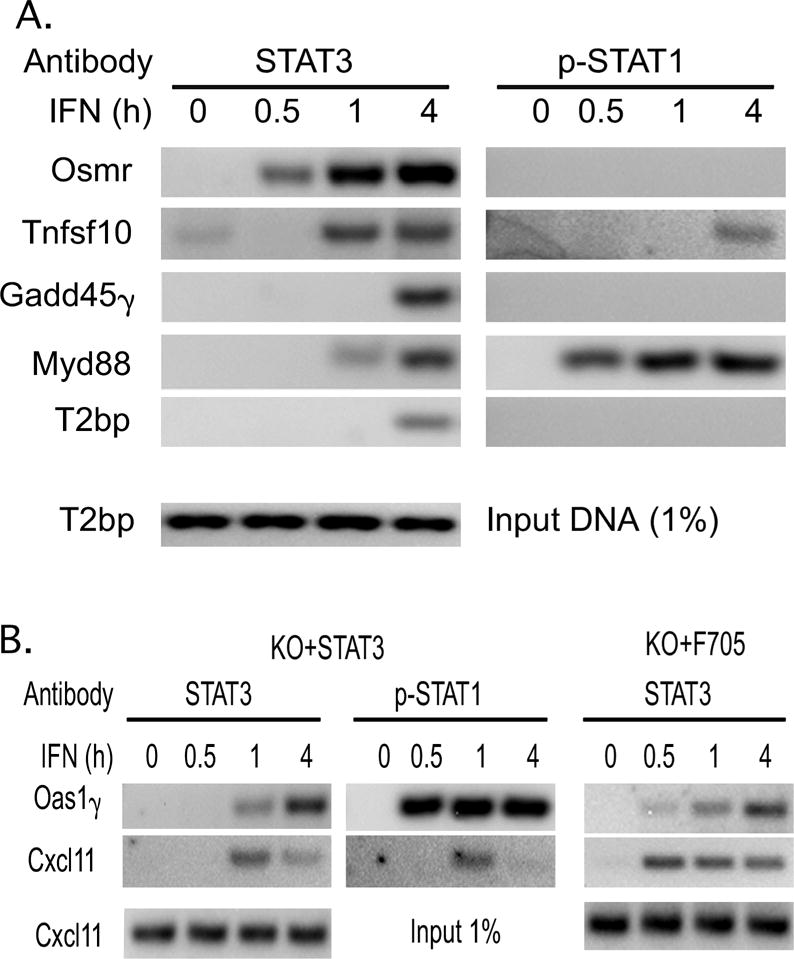
ChIP assays were carried out on extracts from control and IFN-treated STAT3-KO MEFs transfected with WT-STAT3 (Panel A), and WT-STAT3 or F705-STAT3 (Panel B) using the indicated antibodies for precipitation and primers that targeted the promoters of these STAT3-regulated ISGs. Similar results were obtained in at least three independent experiments.
To gain a greater understanding into the role of unphosphorylated STAT3 in the induction of STAT3-regulated ISGs, the promoters of these genes were subjected to bioinformatic analysis. The promoter sequences that are 1-kb upstream of the transcription start site of STAT3-regulated ISGs were retrieved using the UCSC Genome Browser (http://genome.ucsc.edu) and potential ISRE (STAT1/STAT2) and SIE (STAT1/STAT3) binding sites were identified by the JASPAR CORE database (http://jaspar.genereg.net/). Potential STAT1/STAT3 binding sites were enriched in the promoters of 83.9% unphophorylated STAT3-regulated ISGs as compared to only being present in 43.3% of the promoters of STAT3-independent ISGs (p<0.001, Fisher’s Exact test). The enrichment of SIE sites in the promoters of unphosphorylated STAT3-regulated ISGs led us to investigate whether STAT3 directly bound to the promoters of these ISGs.
The binding of phosphorylated and unphosphorylated STAT3 to the promoters of STAT3-regulated ISGs
To investigate the binding of STAT1 and STAT3 to the promoter of STAT3-regulated genes, ChIP assays were carried out on formaldehyde-fixed chromatin prepared from KO+STAT3 MEFs. IFN induced the binding of STAT3 to the promoters of Osmr, Tnsfs10, Gadd45γ, Myd88 and T2bp within 4 hr, with gene-specific differences in binding kinetics (Fig 4A). IFN also induced the binding of STAT1 to the Tnsfs10 and Myd88 promoters. Most importantly, Gadd45γ, Myd88 and T2bp were shown to be STAT3-regulated ISGs that do not require STAT3 phosphorylation (Fig. 3), suggesting that nonphosphorylated STAT3 bound to the promoters of STAT-dependent genes. To further investigate the binding of unphosphorylated STAT3 to STAT3-regulated ISGs promoters, ChIP assays were carried out on chromatin prepared from KO+STAT3 and KO+F705−STAT3 MEFs. Cxcl11 and Oas1γ were selected for analysis as they are well described ISGs that we found to be dependent on unphosphorylated STAT3. As shown in Figure 4B, IFN induced the binding of STAT3 to the Cxcl11 and Oas1γ promoters in MEFs expressing WT-STAT3. Moreover, STAT1 was also recruited to the promoters of these genes. We then carried out ChIP assays with extracts prepared from KO+F705−STAT3 MEFs and found low but detectable IFN-induced binding of STAT3 to the Cxcl11 and Oas1γ promoters. To increase the sensitivity of the assay we carried out a double crosslinking of the chromatin prepared from KO+F705−STAT3 cells. As shown in Figure 4B, IFN induced the binding of unphosphorylated STAT3 to the Cxcl11 and Oas1γ promoters with similar kinetics as observed with MEFs reconstituted with WT-STAT3, demonstrating that unphosphorylated STAT3 is recruited to the promoters of STAT3-regulated ISGs.
Figure 3. The role of STAT3 in ISG expression.
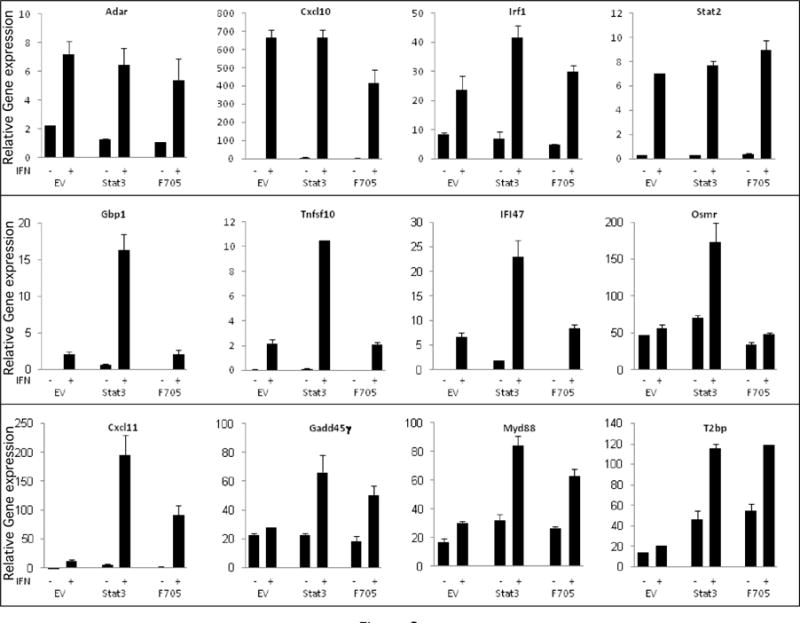
qPCR for several representative STAT3-independent ISGs (Upper Panel), tyrosine phosphorylated STAT3-regulated ISGs (Middle Panel), and unphosphorylated STAT3-regulated ISGs (Lower Panel) was carried out on cDNAs prepared from the three MEF lines (EV-transfected cells, WT-STAT3 transfected cells, and F705-STAT3-transfected cells) treated in the absence or presence of IFN (1,000 IU/mL for 5 hr). Gene expression was normalized to actin expression and presented as mean values +/− SEM (n=3).
DISCUSSION
The present study explored the involvement of phosphorylated and unphosphorylated STAT3 in the biological response to IFN in MEFs with a germ line deletion of STAT3. We show that STAT3 expression enhances the sensitivity of MEFs to the antiviral and antiproliferative actions of IFN, irrespective of the phosphorylation state of STAT3. These findings are consistent with our previous studies on a STAT3-defective human Daudi lymphoblastoid DRST3 cell line, which was highly resistant to the antiproliferative and antiproliferative actions of IFN [15]. Expression of WT-STAT3 rescued sensitivity to IFN’s antiproliferative and antiviral actions. In the present study, we found that the tyrosine phosphorylation of Y705 of STAT3 is not required for the increased sensitivity to IFN action of STAT3-KO MEFs reconstituted with STAT3. Taken together, these results demonstrate that unphosphorylated STAT3 plays an important role in the induction of IFN’s antiproliferative and antiviral effects in very different cell types, i.e. fibroblasts and B-lymphoblastoid cells.
To further characterize STAT3′s role we carried out gene expression profiling on STAT3-KO MEFs reconstituted either with WT-STAT3, F705-STAT3, or empty vector. This approach showed that STAT3 regulates the induction of ISGs, and that a subset of the STAT3-regulated ISGs is independent of Y705 phosphorylation. STAT3 forms homodimers as well as heterodimers with STAT1 that bind to promoter elements in target genes to regulate their transcription. To define how STAT3 regulates ISG expression, we analyzed the putative transcription factor binding sites in the proximal promoter regions (1-kb upstream of the transcription start site) of the three groups of ISGs that we identified by gene expression profiling (STAT3-independent ISGs, phosphorylation-dependent STAT3-regulated ISGs, and unphosphorylated STAT3-regulated ISGs). This analysis revealed that ISRE binding sites are present at a similar frequency in the promoters of STAT3-regulated ISGs and STAT3-independent ISGs. However, GAS binding sites are significantly enriched in the promoters of STAT3-regulated ISGs, suggesting that STAT3 regulates ISG expression primarily through GAS sites. To determine whether STAT3 was bound to the promoters of` STAT3-regulated ISGs that are dependent on Y705 phosphorylation, we carried out ChIP assays on chromatin prepared from IFN treated STAT3-KO cells reconstituted with WT-STAT3, and found that STAT3 was recruited to GAS sites in phosphorylation-dependent STAT3-regulated genes such as Tnsf10 and Osmr. Moreover, IFN also induced the recruitment of STAT3 to unphosphorylated STAT3-regulated genes such as Gadd45γ, T2bp, and Myd88. STAT1, as well as STAT3, was also bound to the promoters of several STAT3-regulated ISGs, such as Tnsf10 and Myd88, which differ in the requirement for Y705 phosphorylation.
To define the role of unphosphorylated STAT3 in ISG expression at the level of promoter interaction ChIP assays were carried out on STAT3-KO cells reconstituted with F705-STAT3. Although IFN-induced binding of phosphorylated STAT3 to ISG promoters was detected by standard ChIP assays using formaldehyde cross-linked chromatin, binding of unphosphorylated STAT3 could not be detected using standard formaldehyde crosslinking. We resorted to a two-step cross-linking procedure with both EGS and formaldehyde, which was used previously to reproducibly detect the inducible binding of STAT3 to the promoter of angiotensinogen [21]. We detected the IFN-induced binding of unphosphorylated STAT3 to the promoter of a number of STAT3-regulated genes (Cxcl11 and Oas1γ) that did not require Y705 phosphorylation. These observations indicate that unphosphorylated STAT3 may directly bind to select ISG promoters, but that the binding is of lower affinity when compared to phosphorylated STAT3. Alternatively, the binding of unphosphorylated STAT3 may involve an indirect interaction mediated by the binding of other transcription factors. Since STAT1 was also recruited to the promoter regions of some STAT3-regulated genes, it is possible that unphosphorylated STAT3 is recruited through its interaction with STAT1. However, STAT1 was not bound to all STAT3-regulated genes, suggesting that the recruitment of unphosphorylated STAT3 to STAT3-regulated ISG promoters may also involve protein binding to sites that are adjacent to GAS elements, such as ISRE, IRF and NF-κB binding sites. We previously found that NF-κB binding sites are frequently found near ISRE or GAS elements in ISG promoters [22, 23]. Moreover, STAT3 and NF-κB proteins were co-recruited upon IFN treatment to the promoter of human Cxcl11 [16]. A number of studies indicate that there is molecular crosstalk between STAT3 and NF-κB pathways [15, 24–26]. It is interesting that many previously identified NF-κB-regulated ISGs [22, 23] are also STAT3-regulated ISGs. Future detailed studies on the regulatory elements adjacent to GAS elements are necessary to determine how interaction with other transcription factors such as NF-κB affects the binding of STAT3 to ISG promoters.
Highlights.
Expression of STAT3, and of a STAT3 mutant in which canonical Y705 tyrosine phosphorylation site was mutated restored sensitivity to the antiviral and antiproliferative of interferon-α/β
The expression of ~60% of the interferon-stimulated genes was STAT3-dependent and ~30% of these STAT3-regulated genes were not dependent on STAT3 tyrosine phosphorylation
STAT3 that is not tyrosine phosphorylated (unphosphorylated STAT3) is recruited to the promoters of STAT3-regulated interferon-stimulated genes
These results suggest that unphosphorylated STAT3 plays an important role in the interferon response pathway
Acknowledgments
We thank Dr. Darren Baker (Biogen) for rat IFNß, and Yi Zheng (UTHSC) for the STAT3-KO MEFs. This work was supported in part by NIH R01CA133322 and the Muirhead Chair Endowment at UTHSC (LP).
Abbreviations
- STAT
signal transducers and activators of transcription
- IFN
interferon
- WT
wild-type
- ISRE
IFN-stimulus response element
- ISG
IFN-stimulated gene
- KO
knockout
- MEFs
mouse embryo fibroblasts
- EV
empty vector
- DCS
defined calf serum
- SIE
c-sis inducible element
- VSV
vesicular stomatitis virus
- qPCR
quantitative real time-PCR
- ChIP
chromatin Immunoprecipitation
- EGS
Ethylene glycolbis(succinimidylsuccinate)
- DAVID
Database of Annotation, Visualization and Integrated Discovery
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiological specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 2.Darnell JEJ, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular Cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhong Z, Wen Z, Darnell JEJ. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 5.Yang CH, Shi W, Basu L, Murti A, Constantinescu SN, Blatt L, Croze E, Mullersman JE, Pfeffer LM. Direct association of STAT3 with the IFNAR1 signal transducing chain of the type I IFN receptor. J Biol Chem. 1996;271:8057–8061. doi: 10.1074/jbc.271.14.8057. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 7.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, Larner AC. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 11.Bromberg JF, Darnell JE., Jr Potential roles of Stat1 and Stat3 in cellular transformation. Cold Spring Harb Symp Quant Biol. 1999;64:425–428. doi: 10.1101/sqb.1999.64.425. [DOI] [PubMed] [Google Scholar]
- 12.Grandis JR, Drenning SD, Zeng Q, Watkins SC, Melhem MF, Endo S, Johnson DE, Huang L, He Y, Kim JD. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci U S A. 2000;97:4227–4232. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q, Briggs J, Park S, Niu G, Kortylewski M, Zhang S, Gritsko T, Turkson J, Kay H, Semenza GL, Cheng JQ, Jove R, Yu H. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 14.Pfeffer LM, Mullersman JE, Pfeffer SR, Murti A, Shi W, Yang CH. STAT3 as an adapter to couple phosphatidylinositol-3 kinase to the IFNAR-1 chain of the type I IFN receptor. Science. 1997;276:1418–1420. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- 15.Yang CH, Murti A, Pfeffer LM. STAT3 complements defects in an interferon-resistant cell line: Evidence for an essential role for STAT3 in interferon signaling and biological activities. Proc Natl Acad Sci USA. 1998;95:5568–5572. doi: 10.1073/pnas.95.10.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang CH, Wei L, Pfeffer SR, Du Z, Murti A, Valentine WJ, Zheng Y, Pfeffer LM. Identification of CXCL11 as a STAT3-dependent gene induced by IFN. J Immunol. 2007;178:986–992. doi: 10.4049/jimmunol.178.2.986. [DOI] [PubMed] [Google Scholar]
- 17.Arduini RM, Li Z, Rapoza A, Gronke R, Hess DM, Wen D, Miatkowski K, Coots C, Kaffashan A, Viseux N, Delaney J, Domon B, Young CN, Boynton R, Chen LL, Chen L, Betzenhauser M, Miller S, Gill A, Pepinsky RB, Hochman PS, Baker DP. Expression, purification, and characterization of rat interferon-beta, and preparation of an N-terminally PEGylated form with improved pharmacokinetic parameters. Protein Expr Purif. 2004;34:229–242. doi: 10.1016/j.pep.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Debidda M, Wang L, Zang H, Poli V, Zheng Y. A role of STAT3 in Rho GTPase-regulated cell migration and proliferation. J Biol Chem. 2005;280:17275–17285. doi: 10.1074/jbc.M413187200. [DOI] [PubMed] [Google Scholar]
- 19.Pfeffer LM, Kim JG, Pfeffer SR, Carrigan DJ, Baker DP, Wei L, Homayouni R. The role of NF-κB in the antiviral action of interferon and interferon-regulated gene expression. J Biol Chem. 2004;279:31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- 20.Eisenkraft BL, Nanus DM, Albino AP, Pfeffer LM. a-Interferon down-regulates epidermal growth factor receptors on renal carcinoma cells: Relation to cellular responsiveness to the antiproliferative action of a-interferon. Cancer Res. 1991;51:5881–5887. [PubMed] [Google Scholar]
- 21.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 22.Wei L, Fan M, Xu L, Heinrich K, Berry MW, Homayouni R, Pfeffer LM. Bioinformatic analysis reveals cRel as a regulator of a subset of interferon-stimulated genes. J Interferon Cytokine Res. 2008;28:541–551. doi: 10.1089/jir.2007.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L, Sandbulte MR, Thomas PG, Webby RJ, Homayouni R, Pfeffer LM. NFκB negatively regulates interferon-induced gene expression and anti-influenza activity. J Biol Chem. 2006;281:11678–11684. doi: 10.1074/jbc.M513286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squarize CH, Castilho RM, Sriuranpong V, Pinto DS, Jr, Gutkind JS. Molecular cross-talk between the NFkappaB and STAT3 signaling pathways in head and neck squamous cell carcinoma. Neoplasia. 2006;8:733–746. doi: 10.1593/neo.06274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]


