Abstract
Stress induced by accumulation of misfolded proteins in the endoplasmic reticulum (ER) is observed in many physiological and pathological conditions. To cope with ER stress, cells activate the unfolded protein response (UPR), a dynamic signaling network that orchestrates the recovery of homeostasis or triggers apoptosis, depending on the level of damage. Here we provide an overview of recent insights into the mechanisms cells employ to maintain proteostasis and how the UPR determines cell fate under ER stress.
The endoplasmic reticulum (ER) orchestrates the synthesis, folding, and structural maturation of at least a third of all cellular proteins. Most proteins that are ultimately secreted or become resistant in the ER, Golgi apparatus, lysosomes, and plasma membrane are translated on ER membrane-associated ribosomes and transported into the ER lumen. As such, the ER is a key contributor to proteostasis -- a network of interconnected quality-control processes in the cell that maintains the functional proteome1. Chaperones, oxidoreductases, and glycosylating enzymes ensure that secretory proteins are properly folded, modified, and assembled into multi-protein complexes in the ER before they traffic farther downstream in the secretory pathway. Despite the existence of these protein-folding machines, there is evidence to suggest that at least a third of all polypeptides translocated into the ER fail to satisfy the quality control mechanisms that ensure proper folding, and for some proteins the success rate is much lower2. Given that client proteins of the secretory pathway often carry out crucial signaling functions (e.g., polypeptide hormones, cell surface receptors), improperly folded forms are recognized and disposed of by stringent quality control systems such as ER-associated degradation (ERAD), which removes unfolded proteins to the cytosol for subsequent ubiquitinylation and degradation by the 26S proteasome. Moreover, inherited mutations in an individual protein can compromise its folding efficiency and lead to disease from its deficiency. For example, mutations in the transmembrane conductance regulator (CFTR), an ion channel transporting chloride across epithelial tissues, disrupt its folding within the ER, leading to Cystic Fibrosis.
The protein-folding capacity within the ER greatly differs among cells types. Specialized secretory cells contain a large, well-developed ER. For example, each βcell of the endocrine pancreas is capable of synthesizing and secreting up to one million molecules of insulin per minute; in insulin-resistant states, this enormous protein synthetic load becomes even greater3. Regardless of the size of their ER, cells seem to perform near the functional limits of their secretory pathway capacity and often face conditions during which the load imposed on the ER protein-folding machinery overwhelms capability, a condition referred to as “ER stress.” A wide range of cellular perturbations can induce ER stress, including hypoxia, nutrient deprivation, point mutations in secreted proteins that promote misfolding, redox changes, and loss of calcium homeostasis with harmful effects on ER resident calcium-dependent chaperones. Therefore, cells have evolved a robust surveillance system to respond to fluctuations of ER homeostasis before they become a threat to survival.
ER stress engages an adaptive signal transduction pathway called the unfolded protein response (UPR). However, irremediable ER stress turns signaling toward a “terminal UPR” that drives cells into apoptosis. Cell injury due to chronic ER stress is being increasingly recognized as a common contributor to a wide range of prevalent human diseases, including neurodegeneration, diabetes, cancer, stroke, pulmonary fibrosis, viral infection, inflammatory and metabolic disorders, and heart disease4. The shared thread among these seemingly dissimilar diseases is the presence of intracellular and/or extracellular conditions that compromise protein folding and cause ER stress. Here we discuss recent advances in our understanding of how the UPR network maintains proteostasis in healthy cells, but actively promotes cell loss when ER homeostasis cannot be restored.
Key players in the Unfolded Protein Response
The discovery of an ER stress-induced adaptive response in the yeast S. cerevisiae in the late 80’s5–7, led to much speculation about a potential role for a UPR pathway in mammals. The field exploded with the discovery of the three main mammalian UPR sensors IRE1α8, PERK9 and ATF6α10 in 1993, 1998 and 1999, respectively. The signaling pathways activated downstream of these sensors constitute an adaptive response to allow cells to cope with protein misfolding by temporarily reducing de novo protein synthesis, and improving the folding and clearance capacity of the ER11. However, if these adaptive measures are inadequate to resolve ER stress, the mammalian UPR shifts signals toward the engagement of apoptosis12. In this section we discuss the main signaling mechanisms involved in the UPR and the downstream consequences in terms of the cellular processes affected.
Detection of misfolded protein species by the three UPR sensors mentioned above has been shown to be partly dependent on BiP/GRP78, a key ER chaperone. Under basal conditions, BiP constitutively binds to the luminal domains of the three sensors thus preventing their activation. However, when misfolded proteins accumulate in the ER, BiP strongly binds to their exposed hydrophobic domains, and dissociates from the UPR sensors, thereby priming IRE1α and PERK for oligomerization and auto-transphosphorylation13 and revealing an ER export motif in ATF6α14. BiP binding to UPR sensors was recently shown to be independent of its chaperon activity, suggesting an allosteric regulation15. The analysis of the ER luminal structure of IRE1p in S. cerevisiae revealed an MHC-like structure16 that appears to operate as a direct sensing motif that binds to hydrophobic domains of unfolded proteins 17. A similar model has been proposed for IRE1β18, but may not operate for IRE1α based on structural and in vitro analysis15, 19–21.
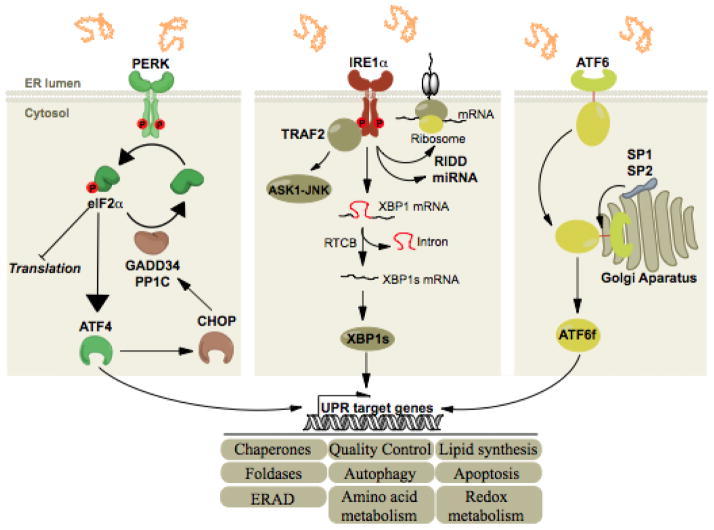
Stress signaling leads to selective activation of downstream cascades (Figure 1), in which active PERK phosphorylates the translation initiation factor eIF2α, leading to attenuation of global protein synthesis22, 23. Inhibition of eIF2α allows the selective expression of ATF4, a transcription factor controlling genes involved in protein folding, antioxidant responses, autophagy, amino acid metabolism and apoptosis24, 25. Moreover, active PERK phosphorylates and activates NRF2, a transcription factor involved in the control of the antioxidant pathway26–28. Finally, ATF6α belongs to a family of transmembrane transcription factors of about 10 members that function in different ways as stress response elements29. Following ER stress, ATF6α is exported from the ER10 and activated in the Golgi complex through cleavage by the proteases S1P and S2P11. The ATF6α cytosolic domain (ATF6f) then translocates to the nucleus where it activates specific transcriptional programs that promote adaptation, including upregulation of ERAD components30.
Figure 1. The Three Arms of the UPR.
All three ER stress sensors (PERK, IRE1α, ATF6) initially activate signaling events that increase protein-folding capacity and reduce protein load on the ER. These transcriptional and translational outputs tend to re-establish protein-folding homeostasis in the ER and promote cell survival. PERK phosphorylates eIF2alpha, which in turn shut down global translation and in the mean time increases the expression of the transcription factor ATF4. The latter induces the transcription of select genes whose functions are to restore proteostasis (grey boxes) and of CHOP, itself inducing the transcription of GADD34, a regulatory subunit of PP1C. This creates a feedback mechanism leading to the dephosphorylation of eIF2alpha and translation is reinitiated. IRE1α signals through i) the recruitment of TRAF2 leading the activation of the ASK1-JNK cascade and ii) through its RNase via the splicing of XBP1 mRNA or the degradation of RNAs, thereby regulating gene expression at transcriptional and post-transcriptional levels. Finally, upon ER stress, ATF6 is exported from the ER to the Golgi complex where it is cleaved by the proteases S1P and S2P, releasing its cytosolic domain which is a potent transcription factor.
The most striking advances have been recently made in understanding IRE1α signaling. IRE1α is an ER transmembrane protein with kinase and endoribonuclease (RNase) activities associated with its cytoplasmic tail. In response to luminal activation, IRE1α dimerizes and trans-autophosphorylates, inducing a conformational change that activates the RNase domain. IRE1α’s RNase catalyzes the excision of a 26-nucleotide intron within the XBP1 mRNA, shifting the reading frame to translate a stable and active transcription factor known as XBP1s31–34. XBP1s controls genes involved in protein folding, secretion, ERAD and lipid synthesis35–37. In addition, XBP1s may heterodimerize with ATF6f to control distinct gene expression patterns38. The unspliced XBP1u protein has been suggested to have a regulatory activity in the efficient delivery of its mRNA to the ER for processing39, 40, in addition to controlling the degradation of XBP1s41, 42. IRE1α activity is involved in the degradation of RNAs (known as Regulated IRE1-dependent decay or RIDD43), including ER-localized mRNAs44–46, ribosomal RNA47 and microRNAs48, 49. New insights into the mechanisms regulating IRE1α’s switch from XBP1 splicing to RIDD activities have been recently reported. IRE1α’s RNase substrate specificity can be controlled by its oligomeric state50. When IRE1α exists in lower order oligomers (e.g., dimer/tetramer), its RNase activity is largely restricted to XBP1 splicing. However, under high or chronic ER stress, IRE1α surpasses an oligomerization threshold that expands its RNase substrate repertoire to many ER-localized mRNAs, leading to RIDD. In contrast to this view, a recent study suggested that XBP1 splicing requires obligate IRE1α oligomers, but that IRE1α dimers suffice for RIDD51. Of note, this later report was largely restricted to in vitro analyses of IRE1α activity under saturating protein concentrations, which do not mimic the continuum of IRE1α oligomerization states that likely exist in vivo. However, the latter model was also supported in cell-based systems in which peptide-induced IRE1 high order oligomers correlated with enhanced IRE1-mediated XBP1 mRNA splicing and reduced RIDD52. Furthermore, IRE1α RNase activity was also linked to its phosphorylation status with key residues involved (i.e., Ser724); other phosphorylation sites mediate its inactivation in yeast53, 54, whereas the remaining identified phosphorylation sites have not yet been functionally tested55. Very recently, the missing link in IRE1α-mediated XBP1 mRNA splicing was uncovered. While the S cerevisiae ligase responsible for HAC1 mRNA (the yeast equivalent of XBP1) ligation was identified in 200856, the mammalian counterpart had remained unknown. Four studies have now uncovered the nature of the ligase as the tRNA ligase RtcB57–60 and demonstrated its physiological role in plasma cell differentiation57. Altogether, these discoveries shed light on the activation and signaling mechanisms of the three UPR sensors and on the biological networks that could be involved in their regulation.
The main biological consequences of UPR activation are the recovery of ER proteostasis (adaptive response) and the induction of a terminal UPR (apoptotic phase). One of the first responses to ER homeostasis imbalance involves global translational attenuation to reduce entry of newly synthesized proteins into the ER. This is in part achieved through PERK-mediated phosphorylation of eIF2α, but also through RIDD of mRNAs encoding secretory proteins or ribosomal RNAs. Recently, a general mechanism leading to translation shutdown was uncovered that results in massive detachment of translating ribosomes from the ER membrane upon ER stress61. Another safeguard set in place by the ER to alleviate stress involves an increased clearance of misfolded proteins. Although this has been shown to be controlled by ERAD and autophagy 62, 63, a recent study has demonstrated that misfolded GPI-anchored proteins can escape ER retention and translocate to the plasma membrane where they are then degraded by the lysosomal pathway64. Finally, in order to re-establish homeostasis within the ER lumen, ER stress also leads to the activation of amino-acid metabolism and improvement of antioxidant responses, in addition to the reinforcement of folding and quality control mechanisms65. Altogether, these events reduce the misfolding burden on the ER thereby allowing the organelle to recover homeostasis.
Cell death control under ER stress
When the buffering capacity of the UPR proves inadequate to restore ER proteostasis, the pathway alternates its signaling towards a terminal UPR66. Whereas the molecular details are still being solved and may differ depending on cell type, accumulating evidence suggests that a distinct program of pro-apoptotic signals activate the cell death machinery if ER stress cannot be remedied (Figure 2)12, 67. For example, while a temporary halt in protein translation due to eIF2α phosphorylation can be advantageous for cells under ER stress, a prolonged block in translation from sustained PERK activation is incompatible with survival. Under chronic ER stress, sustained PERK activation upregulates the transcription factor CHOP/GADD153, which inhibits expression of the anti-apoptotic BCL-2 to promote cell death68, 69. Moreover, it has been shown that CHOP and ATF4 cooperate to transcriptionally activate targets that enhance protein synthesis, and contribute to cell death through ROS production and ATP depletion70. A recent report also suggests that PERK activation increases expression of death receptor 5 (DR5) to trigger caspase-8 induced cell death71.
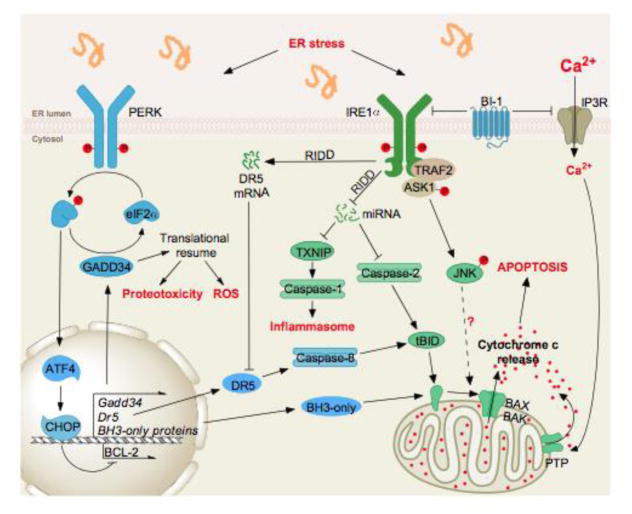
Figure 2. Under irremediable ER stress, the UPR actively promotes proteotoxicity, sterile inflammation and apoptosis.
When exposed to chronically high levels of ER stress, PERK and IRE1α both have multiple signaling outputs that lead to cell dysfunction, activation of the inflammasome, and apoptosis. Among the ATF4 targets downstream of PERK (see Figure 1) are also found genes whose products are involved in the control of cell death (DR5 or BH3-only) through the activation of signaling pathways from the plasma membrane or the mitochondria. Similarly, IRE1α signals through the JNK and mRNA/miRNA degradation pathways to control cell survival outputs. Interestingly, specific cross-talks between IRE1α and PERK signaling, notably at the level of DR5, that tightly control cell fate.
As mentioned above, chronic ER stress causes IRE1α to transition from a homodimeric state into higher order oligomeric structures, which appear to be the critical step in switching to its apoptotic program50. Sustained RIDD activity might deplete protein-folding components to further worsen ER stress46. IRE1α also induces the activation of a number of pro-inflammatory and pro-death proteins. For example, the RNase activity of IRE1α decreases the levels of select microRNAs that normally suppress pro-apoptotic targets such as pro-oxidant protein TXNIP (thioredoxin-interacting protein) and caspase-2, leading to their upregulation48, 49. Increased TXNIP then activates the inflammasome and caspase-148. Finally, under sustained engagement, IRE1α assembles into an activation platform for apoptosis signal-regulating kinase 1 (ASK1) and its target c-Jun NH2-terminal kinase (JNK)72, 73.
Many of the pro-death signals emerging from UPR sensors ultimately regulate the canonical mitochondrial apoptotic pathway. This pathway is initiated when pro-apoptotic mitochondrial proteins, such as cytochrome c, are actively released into the cytoplasm to triggerthe proteolytic activation of effector caspases such as caspase-3. Pro- and anti- death proteins of the large BCL-2 family govern the intrinsic apoptotic pathway by regulating the permeability of the outer mitochondrial membrane74. This pathway is set in motion when cell injury is sensed and leads to the transcriptional upregulation and/or post-translational activation of one or more BH3-only proteins, a group of pro-death proteins all containing an alpha helix known as the BCL-2 homology 3 (BH3) domain that is necessary for their killing activity74, 75. Once activated, BH3-only proteins bind to and neutralize mitochondrial-protecting proteins (e.g., BCL-2, BCL-XL, MCL-1) and in some cases directly engage pro-apoptotic BAX and BAK proteins, causing their homo-oligomerization and resultant permeabilization of the outer mitochondrial membrane.
The terminal UPR activates at least four distinct BH3-only proteins (BID, BIM, NOXA, PUMA) that then signal mitochondrial apoptosis 66. In fact, BIM/PUMA/BID triple knockout cells are much more resistant to ER stress than cells deficient in any one BH3-only protein76, and resemble the phenotype of BAX/BAK double deficiency77. Each of of the BH3-only proteins seems to be activated by ER stress in a unique way66. However, it remains unclear if these BH3-only proteins are simultaneously engaged by all forms of severe ER stress or if only a subset can be activated under specific insults. Another group of ancient cell death regulators, known as the TMBIM or Bax-inhibitor 1 (BI-1) family78 also has relevant activities against ER stress (i.e. BI-1/TMBIM6 and GRINA/TMBIM3), possibly through the modulation of ER calcium release by the IP3 receptors79, 80. Although multiple mechanisms mediating ER stress-induced apoptosis are available, their individual impact on cell viability is partial, suggesting that combinatorial mechanisms or so called cell death networks control apoptosis when the ER is irreversibly damaged67.
Cell fate decisions under ER stress: fine tuning of the UPR
The UPR operates as a highly dynamic signaling network that integrates information about the intensity and duration of the stress stimuli and possibly the type of perturbation to the secretory pathway. Depending on the experimental system, distinct UPR signaling branches operate independently in terms of their activation, signaling kinetics and downstream cellular outputs. In this section, we discuss recent advances in defining how the UPR is fine-tuned, and its impact on determining the transition from adaptive to pro-apoptotic programs.
Analysis of the signaling kinetics of the three main UPR sensors has revealed that the temporal activation of each branch can vary depending on the nature of the pharmacological stressor used to perturb ER function81, 82. Moreover, ATF6α can be selectively activated by the overload of the ER lumen with proteins83, and its activity is also specifically modulated by its N-glycosylation state and the redox status of the ER84, 85. In addition, depending on the intensity of the stress, the signaling outputs of the UPR differ. Indeed low concentrations of ER stress agents engage all UPR signaling branches to initiate adaptive outputs, whereas high levels of ER stress results in apoptotic signals86. This is consistent with the observation that certain specialized secretory cells, such as pancreatic β-cells and B lymphocytes, are normally able to function under sustained levels of physiological ER stress without apoptosis induction87.
The differential modulation of UPR signaling may underlay the mechanism behind the integration of global responses toward proteostasis recovery and the threshold to induce apoptosis. Despite the fact that the ER-sensing domains of IRE1α and PERK have similar primary structure, and they are functionally interchangeable88, the temporal pattern of their signaling drastically differs depending on the experimental setting. Under prolonged ER stress, XBP1 mRNA splicing is diminished, whereas PERK signaling is maintained leading to the expression of downstream proapoptotic components. This may contribute to apoptosis also by attenuating the survival effects of XBP1s, sensitizing cells to ER stress. Besides, PERK-induced CHOP expression could also upregulate GADD34, a component of the eIF2α phosphatase complex that reverts translational inhibition, triggering oxidative stress and proteotoxicity69, 89. In other experimental settings, the sustained activation of IRE1α occurs under high ER stress to trigger cell death43, 46, 50. Using single cell imaging, a recent report also suggested that the relative kinetics of PERK and IRE1α signaling determines the induction of apoptosis rather than a switch between both branches90. These observations emphasize the highly dynamic and complex nature of the UPR regulatory network.
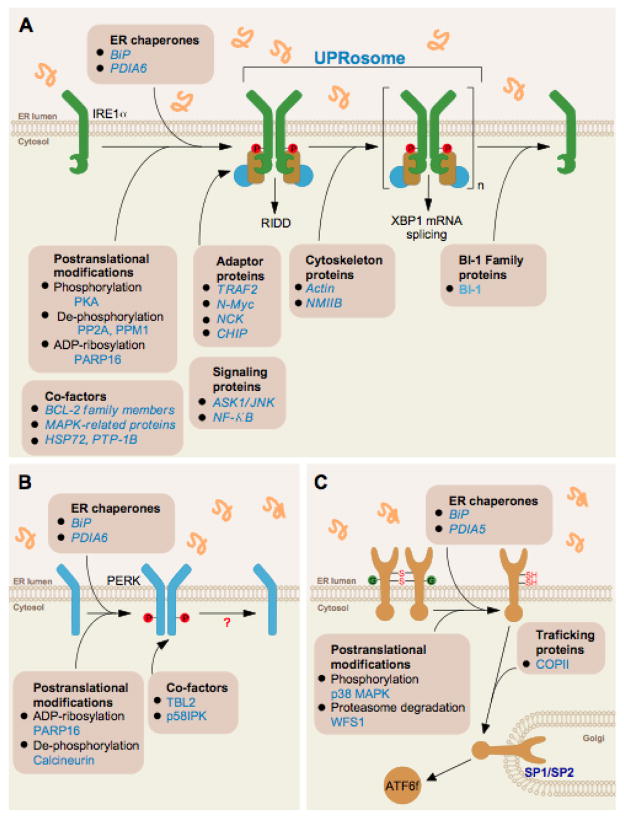
Given that the ER sensing domains of IRE1α and PERK are similar, the temporal behavior of UPR signaling may be in part mediated by intrinsic conformational changes of the sensors, post-translational modifications and/or the binding to specific positive and negative regulators (Figure 3). Most of the studies addressing the molecular basis of fine-tuning UPR activity have been performed with IRE1α. The concept of the UPRosome envisions IRE1α as a scaffold where many components assemble to selectively regulate its activity (amplitude and kinetics) and the control of specific downstream signaling outputs (Figure 3A)91. IRE1α activation and inactivation are instigated by a direct binding of several factors to the UPRosome that modulate the amplitude of UPR responses without affecting PERK92. Interestingly, many regulators of IRE1α have relevant roles in apoptosis93, including members of the BCL-2 family such as BAX, BAK94, some BH3-only proteins95, components of the MAP kinase pathway96–98, and its negative regulator BI-199, 100. Although the list of IRE1-binding partners is increasing and a few screens have been recently reported, systematic interactome studies are still missing. Here we highlight several recent discoveries of interacting proteins that modulate the sustained signaling of IRE1α under prolonged ER stress. N-MYC Interactor (NMI) protein is an IRE1α-binding partner identified through an interactome screen, which selectively enhanced JNK activation and apoptosis, but does not affect XBP1 mRNA splicing101. Another proteomic screen also found non-muscle myosin heavy chain IIB (NMIIB), and possibly actin cytoskeleton, as a specific regulator of IRE1α clustering, without affecting its dimerization and phosphorylation process102; a mechanism validated also in yeast103. Furthermore, specific protein complexes may be assembled at the level of the IRE1α ER luminal domain. BiP has been known for more than a decade to operate as a negative regulator of IRE1α13. The disulfide isomerase PDIA6 was recently shown to control activation and/or inactivation of IRE1α signaling through a direct binding104, 105. Hence, IRE1α signaling is fine-tuned by the assembly of distinct signaling complexes at the level of its cytosolic and ER-luminal regions.
Figure 3. Fine-tuning the UPR through protein-protein interactions and/or post-translational modifications.
(A) Fine-tuning IRE1 signaling through the dissociation of ER chaperones in the activation phase, recruitment of the UPRosome that controls signaling outputs and inactivation phase. (B) Fine-tuning PERK activation through the dissociation of BiP in the activation phase and the control of phosphorylation and ADP-ribosylation. (C) ATF6α activation depends on its N-glycosylation and redox status in addition to the dissociation from BiP to allow egress from the ER and proteolytic cleavage in the Golgi apparatus by the S1P/S2P proteases. In addition, ATF6α protein stability plays a key role in its signaling outputs.
Recent discoveries also indicate that PERK and ATF6 are regulated through the binding of specific factors (Figure 3A and B). For example, the kinase activity of PERK is selectively reduced by the binding of p58IPK106, a regulation antagonized by a cytosolic variant of BiP known as GRP78va107. An interactome analysis recently identified transducin (beta)-like 2 (TBL2) as a protein that associates with phosphorylated PERK under stress conditions, determining optimal signaling to drive ATF4 expression and stress mitigation108. The small GTPase Rheb was also recently shown to bind and activate PERK, repressing protein translation109. Finally, PDIA6 also binds to PERK and negatively modulates the decay of its signaling104. In the case of ATF6α, a few direct regulators have been reported such as Wolfram syndrome 1 (WFS1), which mediates the degradation of ATF6 by the proteasome110. Since the reduction of intra- and inter-luminal disulfide bonds in ATF6α modulates its translocation to the Golgi, a cell-based RNAi screen was performed to target most PDIs and ER oxidoreductases111. This study identified PDIA5 as a selective and critical regulator of ATF6α activation. Overall, these novel studies highlight the concept that all UPR branches are modulated by protein-protein interactions through the assembly of dynamic complexes or UPRosomes that control the amplitude and temporal behavior of their signaling.
Although less explored, the UPR is also fine-tuned by post-translational modifications at the level of ER sensors and transcription factors. In the context of glucose metabolism, IRE1α function is directly instigated through the phosphorylation of Ser724 by PKA112. IRE1α phosphorylation is attenuated by different phosphatases, including PP2A113 and PPM1114, 115, which were identified through proteomic screens. Protein Tyrosine Phosphatase-1B (PTP-1B) also has a selective effect on bursting IRE1α signaling116. Moreover, ADP-ribosylation of PERK and IRE1α enhances their activities117, and IRE1α ubiquitination is mediated by the E3 ligase CHIP increases JNK signaling without affecting XBP1 mRNA splicing118. p38MAPK also phosphorylates ATF6α119 and XBP1s120, having a positive effect on gene expression. XBP1s activity is also modulated by acetylation and sumoylation121, 122. Moreover, the association of ATF6f, ATF4 and XBP1s with other transcription factors determines the establishment of stimuli- and tissue-specific transcriptional patterns92, 123. In agreement with this concept, genome-wide transcriptional profiling under ER stress indicated that the genetic background highly influences the pattern of gene regulation124, 125. Finally, Nonsense Mediated RNA Decay also determines the threshold of stress necessary to activate the UPR126,127, in addition to adjusting the amplitude of downstream responses and the termination phase by controlling the mRNA stability of UPR components128, highlighting the dynamic crosstalk between mRNA metabolism and the proteostasis network. These examples illustrate that the strength and temporal activation/inactivation of each UPR signaling arm is independently regulated at different stages depending on the stress condition and the tissue/cell type affected.
Although our understanding of how the UPR network is regulated is constantly growing, the mechanisms underlying the integration of the stress intensity and its duration are not well described. It is becoming clear that the assembly of UPRosome regulatory complexes determines the threshold of activation of UPR sensors and modulates the final outcome of the pathway. One of the major limitations in the field is the fact that most studies use acute stresses that irreversibly damage the ER, leading to apoptosis. These “non-physiological” conditions normally trigger the upregulation of pro-survival and pro-apoptotic components with virtually similar kinetics. Importantly, a new approach was recently developed to model “resolvable” ER stress, leading to full activation of the adaptive UPR in the absence of apoptosis features129. Using this approach, a novel signaling crosstalk was identified that mediates adaptation to ER stress involving estrogen signaling129. We believe these technological advances will enable the examination of cell fate mechanisms under ER stress in a tractable system.
Novel functions of the UPR
Secreted factors can control extracellular proteostasis. There is a continuum between the ER lumen and the extracellular space; and stressed cells have developed ways to control their microenvironment by secreting key enzymes. The most recent example of this mechanism is illustrated by the secretion of ERdj3, which extracellularly binds misfolded proteins, inhibits their aggregation, and attenuates proteotoxicity of disease-associated prion protein130. Similarly, enforced expression of XBP1s or ATF6α in a stress-independent manner was recently shown to reduce extracellular aggregation of amyloidogenic immunoglobulin variants131. Other chaperones of the GRP and PDI family are also secreted through a regulation dependent on ER stress132. This phenomenon is also observed with redox proteins such as QSOX1, which controls the structure of the extracellular matrix133 most likely in conjunction with secreted PDI family members such as ERp57134.
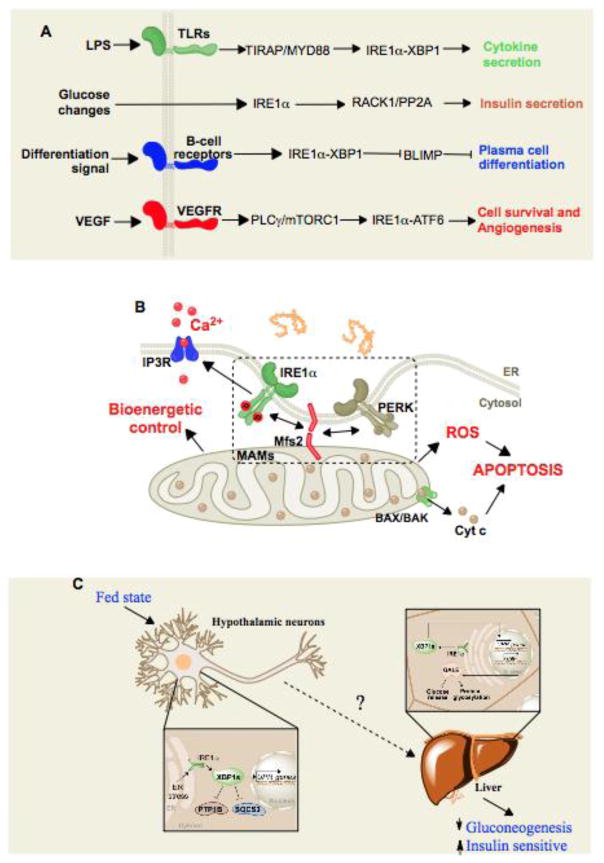
Stress-independent functions of the UPR have been identified. The crosstalk between plasma membrane signaling and the ER is illustrated by cell surface receptors and UPR components (Figure 4A). In macrophages, optimal secretion of pro-inflammatory cytokines by Toll-like receptor (TLR) is mediated by XBP1, where TLR stimulation engages IRE1α in the absence of global ER stress markers, possibly mediated by NADPH oxidase 2 signaling135. Moreover, TLR signaling may even signal to the UPR under ER stress to repress ATF4/CHOP expression136, 137. Similarly, the well-known function of XBP1s in plasma cell differentiation138 has been suggested to occur in a stress-independent manner initiated by the signaling of B cell receptor modulating cell differentiation programs139. In agreement with this concept, XBP1 modulates the activity of MIST1, a master regulator of cell differentiation in different cell types140. IRE1α has been also proposed to monitor fluctuation in glucose levels in the absence of ER stress, mediated by its phosphorylation independent of the release of BiP from the luminal domain113, 141. Similarly, CD40 signaling increases XBP1 mRNA splicing and thus protects the secretory pathway of hepatocytes from ER stress induced by either tunicamycin or oleic acid142. In endothelial cells, VEGF was recently shown to activate all UPR mediators via signaling through a PLCγ-mediated crosstalk with mTORC1 in an ER stress-independent manner, contributing to endothelial cell survival and angiogenesis143. Thus, UPR signaling modules may have important cellular functions unrelated to ER stress.
Figure 4. ER stress-independent functions of the UPR.
(A) Signaling (LPS, glucose, differentiation, VEGF)-dependent and protein misfolding-independent activation of the canonical UPR sensors. (B) Control of mitochondrial function by UPR signaling at the ER-mitochondrion interface. (C) Cell-nonautonomous UPR activation. ER stress is triggered in neurons that subsequently prompt the activation of select UPR signals in the non-stressed liver through yet uncharacterized mediators.
Functional and physical connections between the ER and mitochondria have long been described in the literature, especially in the field of apoptosis and in mitochondrial dynamics. PERK is located at mitochondrial associated membranes (MAMs), and is required to maintain the ER-mitochondria juxtapositions and contributes to ROS production to engage mitochondrial-dependent apoptosis144, 145 (Figure 4B). These observations were associated with a physical interaction between PERK and the ER tethering protein mitofusin 2145. Interestingly, mitofusin 2 deficiency leads to chronic ER stress146. IRE1α has been also detected in MAMs147, and a recent study suggested that IRE1α might contribute to apoptosis by modulating calcium transfer and ROS production by the mitochondria148. In the liver, obesity leads to a marked reorganization of MAMs, resulting in mitochondrial calcium overload and compromised mitochondrial oxidative capacity149. Whether the localization of UPR stress sensors at MAMs affects mitochondrial bioenergetics and its dynamics remains an open question.
Classically, the mechanisms controlling responses to altered proteostasis are considered “cell intrinsic” (also termed cell-autonomous). Recent studies in the field have provided evidence for regulation of stress responses at a distance, largely mediated by the nervous system, on a cell non-autonomous manner150. One of the first examples for a cell non-autonomous control of the UPR came from aging studies in C. elegans. Enforced expression of XBP1s in neurons increases life span of this model organism151. Remarkably, neuronal XBP1s is able to promote IRE1α activation and XBP1 mRNA splicing in peripheral tissues, and this stress signaling propagation was crucial to protect against aging151. An analogous cell non-autonomous regulatory circuit was only recently uncovered in mammals where hypothalamic XBP1s expression modulates global metabolism through a cell non-autonomous propagation of UPR signals to the liver152 (Figure 4C). In all these studies, whether the activation of the UPR in the distant cell is ER stress-independent remains to be clarified. The activation of a cell non-autonomous UPR is emerging as an evolutionary conserved mechanism to prevent uncontrolled protein misfolding on an organismal level. Thus, the UPR may be also fine-tuned at the level of complex tissues and even the whole organism.
Concluding remarks
The UPR is an evolutionarilyconserved signal transduction pathway activated when cells fail to keep up with the protein folding demands on the ER. In response to mild ER stress, the UPR promotes adaptive outputs that reduce unfolded protein load and improve the ability of the secretory pathway to restore proteostasis. However, under irreversible ER stress, the UPR assembles into an alternate platform that engages a complex network of signals to hasten cell demise. Cell injury due to ongoing ER stress has emerged as a central contibutor of the pathophysiology of a wide range of common human diseases and aging. Recent advances in our knowledge of how the UPR alternates between life and death signaling, and the development of small molecule inhibitors of the UPR153, are the stepping stones for new strategies to combat these ER stress-associated diseases. Despite the immense progress made over the past decade into the biological relevance of the UPR, many questions remain open. The next few years promise to shed much needed light on the coordination and specific contribution of the three arms of the UPR in health and disease, what role each plays in the absence ER stress, and how they are regulated at the level of the whole organism. Many novel stress-independent functions of UPR signaling modules are also emerging as relevant contributors to cell physiology and disease, which may involve allosteric and posttranslational modifications to stress sensors through the assembly of distinct UPRosome complexes. It is becoming clear that this field is gaining complexity each year and the UPR can no longer be viewed as a linear pathway, as multiple modulatory steps and dynamic cross talk operate to integrate the UPR with the global proteostasis network and other relevant signaling pathways.
Acknowledgments
Funded by ECOS-CONICYT C13S02 (CH, EC), Ring Initiative-ACT1109, FONDEF-D11I1007, Millennium Institute-P09-015-F, the Frick Foundation, FONDECYT no. 1140549, Michael J.Fox Foundation for Parkinson Research, COPEC-UC Foundation, CONICYT grant USA2013-0003 (CH), the Institut National du Cancer (PLBio and ICGC ; INCa) and La Ligue Nationale Contre le Cancer (LARGE) (EC); NIH: RO1CA136577 and R01-DK095306, American Cancer Society Research Scholar Award, Harrington Discovery Institute Scholar-Innovator Award, Michael J. Fox Foundation for Parkinson's Research, Caring for Carcinoid Foundation and American Association for Cancer Research (S.A.O.).
References
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 3.Seino S, Shibasaki T, Minami K. Dynamics of insulin secretion and the clinical implications for obesity and diabetes. J Clin Invest. 2011;121:2118–2125. doi: 10.1172/JCI45680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 6.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 7.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 8.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 10.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 12.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 14.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 15.Carrara M, Prischi F, Nowak PR, Kopp MC, Ali MM. Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. Elife. 2015;4 doi: 10.7554/eLife.03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner BM, Walter P. Unfolded Proteins Are Ire1-Activating Ligands that Directly Induce the Unfolded Protein Response. Science. 2011 doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oikawa D, Kitamura A, Kinjo M, Iwawaki T. Direct association of unfolded proteins with mammalian ER stress sensor, IRE1beta. PLoS One. 2012;7:e51290. doi: 10.1371/journal.pone.0051290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr Opin Cell Biol. 2011;23:135–142. doi: 10.1016/j.ceb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Oikawa D, Kimata Y, Kohno K, Iwawaki T. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res. 2009;315:2496–2504. doi: 10.1016/j.yexcr.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheuner D, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 23.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 24.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 25.Ye J, Koumenis C. ATF4, an ER stress and hypoxia-inducible transcription factor and its potential role in hypoxia tolerance and tumorigenesis. Curr Mol Med. 2009;9:411–416. doi: 10.2174/156652409788167096. [DOI] [PubMed] [Google Scholar]
- 26.Del Vecchio CA, et al. De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. PLoS Biol. 2014;12:e1001945. doi: 10.1371/journal.pbio.1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 28.Cullinan SB, et al. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asada R, Kanemoto S, Kondo S, Saito A, Imaizumi K. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J Biochem. 2011;149:507–518. doi: 10.1093/jb/mvr041. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Shen X, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 33.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 35.Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 36.Acosta-Alvear D, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 37.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoulders MD, et al. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanagitani K, et al. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 40.Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CY, et al. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J. 2014;33:2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurel M, Chevet E, Tavernier J, Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 45.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han D, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwawaki T, et al. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nature cell biology. 2001;3:158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 48.Lerner AG, et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Upton JP, et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh R, et al. Allosteric Inhibition of the IRE1alpha RNase Preserves Cell Viability and Function during Endoplasmic Reticulum Stress. Cell. 2014 doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tam AB, Koong AC, Niwa M. Ire1 Has Distinct Catalytic Mechanisms for XBP1/HAC1 Splicing and RIDD. Cell Reports. 2014;9:1–9. doi: 10.1016/j.celrep.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouchecareilh M, Higa A, Fribourg S, Moenner M, Chevet E. Peptides derived from the bifunctional kinase/RNase enzyme IRE1{alpha} modulate IRE1{alpha} activity and protect cells from endoplasmic reticulum stress. FASEB J. 2011;25:3115–3129. doi: 10.1096/fj.11-182931. [DOI] [PubMed] [Google Scholar]
- 53.Rubio C, et al. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J Cell Biol. 2011;193:171–184. doi: 10.1083/jcb.201007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chawla A, Chakrabarti S, Ghosh G, Niwa M. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J Cell Biol. 2011;193:41–50. doi: 10.1083/jcb.201008071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prischi F, Nowak PR, Carrara M, Ali MM. Phosphoregulation of Ire1 RNase splicing activity. Nat Commun. 2014;5:3554. doi: 10.1038/ncomms4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harding HP, et al. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA. 2008;14:225–232. doi: 10.1261/rna.859908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jurkin J, et al. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 2014 doi: 10.15252/embj.201490332. [DOI] [PMC free article] [PubMed]
- 58.Kosmaczewski SG, et al. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 2014 doi: 10.15252/embr.201439531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu Y, Liang FX, Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell. 2014;55:758–770. doi: 10.1016/j.molcel.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ray A, Zhang S, Rentas C, Caldwell KA, Caldwell GA. RTCB-1 Mediates Neuroprotection via XBP-1 mRNA Splicing in the Unfolded Protein Response Pathway. J Neurosci. 2014;34:16076–16085. doi: 10.1523/JNEUROSCI.1945-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell. 2014;158:1362–1374. doi: 10.1016/j.cell.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vidal R, Caballero B, Couve A, Hetz C. Converging pathways in the occurrence of endoplasmic reticulum (ER) stress in Huntington's disease. Curr Mol Med. 2011;11:1–12. doi: 10.2174/156652411794474419. [DOI] [PubMed] [Google Scholar]
- 64.Satpute-Krishnan P, et al. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell. 2014;158:522–533. doi: 10.1016/j.cell.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 66.Shore GC, Papa FR, Oakes SA. Signaling cell death from the endoplasmic reticulum stress response. Curr Opin Cell Biol. 2011;23:143–149. doi: 10.1016/j.ceb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urra H, Dufey E, Lisbona F, Rojas-Rivera D, Hetz C. When ER stress reaches a dead end. Biochim Biophys Acta. 2013;1833:3507–3517. doi: 10.1016/j.bbamcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 68.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marciniak SJ, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu M, et al. Cell death. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 73.Nishitoh H, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 75.Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 76.Ren D, et al. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–1393. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei MC, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rojas-Rivera D, Hetz C. TMBIM protein family: ancestral regulators of cell death. Oncogene. 2015;34:269–280. doi: 10.1038/onc.2014.6. [DOI] [PubMed] [Google Scholar]
- 79.Rojas-Rivera D, et al. TMBIM3/GRINA is a novel unfolded protein response (UPR) target gene that controls apoptosis through the modulation of ER calcium homeostasis. Cell Death Differ. 2014;19:1013–1026. doi: 10.1038/cdd.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chae HJ, et al. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 81.Yoshida H, et al. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 82.DuRose JB, Tam AB, Niwa M. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol Biol Cell. 2006;17:3095–3107. doi: 10.1091/mbc.E06-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maiuolo J, Bulotta S, Verderio C, Benfante R, Borgese N. Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc Natl Acad Sci U S A. 2011;108:7832–7837. doi: 10.1073/pnas.1101379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hong M, et al. Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J Biol Chem. 2004;279:11354–11363. doi: 10.1074/jbc.M309804200. [DOI] [PubMed] [Google Scholar]
- 85.Nadanaka S, Okada T, Yoshida H, Mori K. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol. 2007;27:1027–1043. doi: 10.1128/MCB.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rutkowski DT, et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cornejo VH, Pihan P, Vidal RL, Hetz C. Role of the unfolded protein response in organ physiology: lessons from mouse models. IUBMB Life. 2013;65:962–975. doi: 10.1002/iub.1224. [DOI] [PubMed] [Google Scholar]
- 88.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 89.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walter F, Schmid J, Dussmann H, Concannon CG, Prehn JH. Imaging of single cell responses to ER stress indicates that the relative dynamics of IRE1/XBP1 and PERK/ATF4 signalling rather than a switch between signalling branches determine cell survival. Cell Death Differ. 2015 doi: 10.1038/cdd.2014.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 93.Woehlbier U, Hetz C. Modulating stress responses by the UPRosome: a matter of life and death. Trends Biochem Sci. 2011;36:329–337. doi: 10.1016/j.tibs.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 94.Hetz C, et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 95.Rodriguez Da, et al. BH3-only proteins are part of a regulatory network that control the sustained signalling of the unfolded protein response sensor IRE1α. The EMBO journal. 2012;31:2322–2335. doi: 10.1038/emboj.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luo D, et al. AIP1 is critical in transducing IRE1-mediated endoplasmic reticulum stress response. J Biol Chem. 2008;283:11905–11912. doi: 10.1074/jbc.M710557200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yoneda T, et al. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem. 2001;276:13935–13940. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- 98.Oono K, et al. JAB1 participates in unfolded protein responses by association and dissociation with IRE1. Neurochem Int. 2004;45:765–772. doi: 10.1016/j.neuint.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 99.Lisbona F, et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Castillo K, et al. BAX inhibitor-1 regulates autophagy by controlling the IRE1alpha branch of the unfolded protein response. EMBO J. 2011;30:4465–4478. doi: 10.1038/emboj.2011.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brozzi F, et al. A combined “omics” approach identifies N-Myc interactor as a novel cytokine-induced regulator of IRE1 protein and c-Jun N-terminal kinase in pancreatic beta cells. J Biol Chem. 2014;289:20677–20693. doi: 10.1074/jbc.M114.568808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.He Y, et al. Nonmuscle myosin IIB links cytoskeleton to IRE1alpha signaling during ER stress. Dev Cell. 2012;23:1141–1152. doi: 10.1016/j.devcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ishiwata-Kimata Y, Yamamoto YH, Takizawa K, Kohno K, Kimata Y. F-actin and a type-II myosin are required for efficient clustering of the ER stress sensor Ire1. Cell Struct Funct. 2013;38:135–143. doi: 10.1247/csf.12033. [DOI] [PubMed] [Google Scholar]
- 104.Eletto D, Dersh D, Gidalevitz T, Argon Y. Protein disulfide isomerase A6 controls the decay of IRE1alpha signaling via disulfide-dependent association. Mol Cell. 2014;53:562–576. doi: 10.1016/j.molcel.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Groenendyk J, et al. Interplay between the oxidoreductase PDIA6 and microRNA-322 controls the response to disrupted endoplasmic reticulum calcium homeostasis. Sci Signal. 2014;7:ra54. doi: 10.1126/scisignal.2004983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yan W, et al. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ni M, Zhou H, Wey S, Baumeister P, Lee AS. Regulation of PERK signaling and leukemic cell survival by a novel cytosolic isoform of the UPR regulator GRP78/BiP. PLoS One. 2009;4:e6868. doi: 10.1371/journal.pone.0006868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsukumo Y, et al. TBL2 Is a Novel PERK-Binding Protein that Modulates Stress-Signaling and Cell Survival during Endoplasmic Reticulum Stress. PLoS One. 2014;9:e112761. doi: 10.1371/journal.pone.0112761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tyagi R, et al. Rheb Inhibits Protein Synthesis by Activating the PERK-eIF2alpha Signaling Cascade. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fonseca SG, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest. 2010;120:744–755. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Higa A, et al. Endoplasmic reticulum stress-activated transcription factor ATF6alpha requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol. 2014;34:1839–1849. doi: 10.1128/MCB.01484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mao T, et al. PKA phosphorylation couples hepatic inositol-requiring enzyme 1alpha to glucagon signaling in glucose metabolism. Proc Natl Acad Sci U S A. 2011;108:15852–15857. doi: 10.1073/pnas.1107394108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qiu Y, et al. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci Signal. 2010;3:ra7. doi: 10.1126/scisignal.2000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu G, et al. PPM1l encodes an inositol requiring-protein 1 (IRE1) specific phosphatase that regulates the functional outcome of the ER stress response. Mol Metab. 2014;2:405–416. doi: 10.1016/j.molmet.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ren S, et al. IRE1 Phosphatase PP2Ce Regulates Adaptive ER Stress Response in the Postpartum Mammary Gland. PLoS One. 2014;9:e111606. doi: 10.1371/journal.pone.0111606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gu F, et al. Protein-tyrosine phosphatase 1B potentiates IRE1 signaling during endoplasmic reticulum stress. J Biol Chem. 2004;279:49689–49693. doi: 10.1074/jbc.C400261200. [DOI] [PubMed] [Google Scholar]
- 117.Jwa M, Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1alpha-mediated unfolded protein response. Nat Cell Biol. 2012;14:1223–1230. doi: 10.1038/ncb2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhu X, et al. Ubiquitination of Inositol-requiring Enzyme 1 (IRE1) by the E3 Ligase CHIP Mediates the IRE1/TRAF2/JNK Pathway. J Biol Chem. 2014;289:30567–30577. doi: 10.1074/jbc.M114.562868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gade P, et al. Regulation of the death-associated protein kinase 1 expression and autophagy via ATF6 requires apoptosis signal-regulating kinase 1. Mol Cell Biol. 2014;34:4033–4048. doi: 10.1128/MCB.00397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee J, et al. p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nat Med. 2011 doi: 10.1038/nm.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang FM, Ouyang HJ. Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem J. 2010 doi: 10.1042/BJ20101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen H, Qi L. SUMO modification regulates the transcriptional activity of XBP1. Biochem J. 2010;429:95–102. doi: 10.1042/BJ20100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 124.Chow CY, Wang X, Riccardi D, Wolfner MF, Clark AG. The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse. PLoS Genet. 2015;11:e1004924. doi: 10.1371/journal.pgen.1004924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dombroski BA, et al. Gene expression and genetic variation in response to endoplasmic reticulum stress in human cells. Am J Hum Genet. 2010;86:719–729. doi: 10.1016/j.ajhg.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sakaki K, et al. RNA surveillance is required for endoplasmic reticulum homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8079–8084. doi: 10.1073/pnas.1110589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Carreras-Sureda A, Hetz C. RNA metabolism: Putting the brake on the UPR. EMBO Rep. 2015 doi: 10.15252/embr.201540227. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Karam R, et al. The unfolded protein response is shaped by the NMD pathway. EMBO rep. 2015 doi: 10.15252/embr.201439696. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Raina K, et al. Targeted protein destabilization reveals an estrogen-mediated ER stress response. Nat Chem Biol. 2014;10:957–962. doi: 10.1038/nchembio.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Genereux JC, et al. Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J. 2014 doi: 10.15252/embj.201488896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cooley CB, et al. Unfolded protein response activation reduces secretion and extracellular aggregation of amyloidogenic immunoglobulin light chain. Proc Natl Acad Sci U S A. 2014;111:13046–13051. doi: 10.1073/pnas.1406050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–10634. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 133.Ilani T, et al. A secreted disulfide catalyst controls extracellular matrix composition and function. Science. 2014;341:74–76. doi: 10.1126/science.1238279. [DOI] [PubMed] [Google Scholar]
- 134.Dihazi H, et al. Secretion of ERP57 is important for extracellular matrix accumulation and progression of renal fibrosis, and is an early sign of disease onset. J Cell Sci. 2014;126:3649–3663. doi: 10.1242/jcs.125088. [DOI] [PubMed] [Google Scholar]
- 135.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Woo CW, et al. Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat Cell Biol. 2009;11:1473–1480. doi: 10.1038/ncb1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Woo CW, Kutzler L, Kimball SR, Tabas I. Toll-like receptor activation suppresses ER stress factor CHOP and translation inhibition through activation of eIF2B. Nat Cell Biol. 2012;14:192–200. doi: 10.1038/ncb2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 139.Hu CC, Dougan SK, McGehee AM, Love JC, Ploegh HL. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28:1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Huh WJ, et al. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology. 2010;139:2038–2049. doi: 10.1053/j.gastro.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lipson KL, et al. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4:245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 142.Villeneuve J, et al. A protective role for CD154 in hepatic steatosis in mice. Hepatology. 2010;52:1968–1979. doi: 10.1002/hep.23935. [DOI] [PubMed] [Google Scholar]
- 143.Karali E, et al. VEGF Signals through ATF6 and PERK to promote endothelial cell survival and angiogenesis in the absence of ER stress. Mol Cell. 2014;54:559–572. doi: 10.1016/j.molcel.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 144.Verfaillie T, et al. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012;19:1880–1891. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Munoz JP, et al. Mfn2 modulates the UPR and mitochondrial function via repression of PERK. EMBO J. 2013;32:2348–2361. doi: 10.1038/emboj.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ngoh GA, Papanicolaou KN, Walsh K. Loss of mitofusin 2 promotes endoplasmic reticulum stress. J Biol Chem. 2012;287:20321–20332. doi: 10.1074/jbc.M112.359174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mori T, Hayashi T, Hayashi E, Su TP. Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS One. 2013;8:e76941. doi: 10.1371/journal.pone.0076941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Son SM, Byun J, Roh SE, Kim SJ, Mook-Jung I. Reduced IRE1alpha mediates apoptotic cell death by disrupting calcium homeostasis via the InsP3 receptor. Cell Death Dis. 2014;5:e1188. doi: 10.1038/cddis.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Arruda AP, et al. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat Med. 2014 doi: 10.1038/nm.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mardones P, Martinez G, Hetz C. Control of systemic proteostasis by the nervous system. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 151.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Williams KW, et al. Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metab. 2014;20:471–482. doi: 10.1016/j.cmet.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov. 2013;12:703–719. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]