Abstract
The diagnosis of poorly differentiated synovial sarcoma (PD-SS) may be challenging due to overlapping morphologic features with other undifferentiated round cell sarcomas (URCS). Particularly relevant is the histologic overlap and shared BCOR overexpression between a subset of SS and URCS with various BCOR genetic abnormalities. Here we report a case of PD-SS lacking the canonical SS18-SSX gene fusion, but showing strong BCOR immunoreactivity and BCOR gene abnormalities by FISH which were misinterpreted as a URCS with BCOR gene rearrangements. The tumor had an unusual clinical presentation arising as an intraneural tumor in the ankle of a 29-year-old female. The tumor displayed a mixture of fascicular spindle cells and undifferentiated round cell components. FISH studies showed no SS18 gene abnormality; however, RNA sequencing identified a fusion transcript involving SS18L1 (a paralog gene of SS18 at 20q13.33) and SSX1. Further FISH testing validated rearrangements in SSX1 and SS18L1 genes, in addition to complex structural abnormalities of the Xp11.22-4 region. This is the second reported SS case harboring an SS18L1-SSX1 alternative fusion variant, similarly occurring in association with a large nerve. The lack of SS18 gene rearrangements by FISH corroborated with the BCOR overexpression at both mRNA and protein level may result in diagnostic pitfalls with URCS with BCOR gene abnormalities. Our results further suggest that BCOR upregulation is emerging as a common downstream pathway for SS with either typical SS18-SSX transcript or with rare fusion variants, such as SS18L1-SSX.
Keywords: synovial sarcoma, intraneural, SS18L1, SSX1, BCOR
INTRODUCTION
The genetic hallmark of synovial sarcomas (SS) is a recurrent t(X;18) translocation resulting in the fusion of SS18 (18q11.2) to one of the SSX genes, including SSX1 (Xp11.23), SSX2 (Xp11.22), or rarely SSX4 (Xp11.23). As the SS18-SSX canonical fusion is present in the overwhelming majority of cases (>95%) (Ladanyi et al., 2002), detection of the fusion transcript by RT-PCR or demonstration of the SS18 gene rearrangement by FISH have become the gold-standard in the diagnosis of SS. Particularly relevant is the molecular diagnosis in the setting of poorly differentiated synovial sarcoma (PD-SS), where the histologic overlap with undifferentiated round cell sarcomas (URCS) in the spectrum of Ewing sarcoma-like tumors is significant. Our group has recently shown that half of SS display BCOR overexpression at mRNA and protein levels, an overlapping finding with URCS harboring a variety of BCOR-genetic abnormalities (Kao et al., 2016b). Here we report a challenging diagnostic case of a PD-SS lacking the typical SS18 gene abnormalities and showing strong and diffuse BCOR immunoreactivity suggesting an alternative diagnosis of URCS.
MATERIALS AND METHODS
A 29 year-old female was diagnosed with a left ankle mass and underwent an incisional biopsy, followed by a wide en bloc resection. Intraoperatively, the mass appeared to be circumscribed and originating from the posterior tibial nerve (Fig. 1). The tumor measured grossly 3 cm in largest dimension and had a white-tan homogenous cut surface. Histologically, the tumor was composed of undifferentiated monomorphic spindle cells arranged in intersecting fascicles (Fig. 2A) alternating with sheets of primitive round cells (Fig. 2B). The mitotic activity ranged from 5 MF/10 HPFs in the spindle cell component to 10 MF/10HPFs in the primitive round cell areas. Small foci of necrosis were noted. The tumor cells had scant cytoplasm and uniform nuclei with fine chromatin and indistinct nucleoli. No benign neurogenic component was identified. The study was approved by the Institutional Review Board.
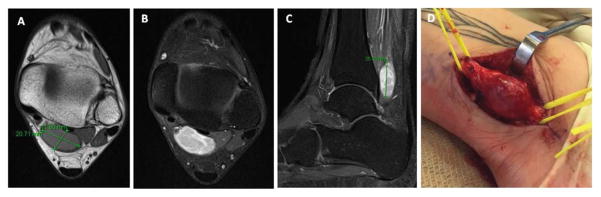
Figure 1.
Radiographic images and intra-operative photograph of the lesion. (A) Axial T1 shows a well defined lesion measuring 2 x 1.8 cm, located posterior to the distal tibia; (B, C) Axial and sagittal T1 fat-saturated with gadolinium images showing a bright circumscribed lesion, with heterogenous septations; (D) Gross photograph intra-operatively of a circumscribed fusiform tumor, encasing and displacing the fascicles of posterior tibial nerve, mimicking a neural tumor; the tumor measured about 3 cm in largest dimension.
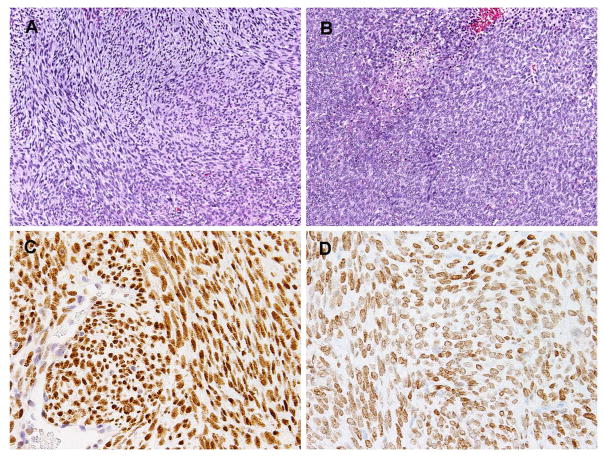
Figure 2.
(A–B) Microscopically the tumor showed a mixture of monomorphic spindle cells arranged in intersecting fascicles (A) and areas of primitive round cells in sheets, with focal areas of necrosis (B). (C) BCOR immunostain showed diffuse and strong nuclear staining in contrast to the endothelial cells. (D) SATB2 stain also showed diffuse reactivity with moderate intensity.
Fluorescence in situ hybridization (FISH)
Interphase FISH for SS18 and BCOR gene abnormalities were performed on 4 μm-thick formalin-fixed paraffin-embedded (FFPE) tissue sections. Custom bacterial artificial chromosomes (BAC) clones flanking the target genes were designed according to UCSC genome browser (http://genome.ucsc.edu) and obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (Oakland, CA; http://bacpac.chori.org) (Supplementary Table 1). DNA from each BAC was isolated according to the manufacturer’s instructions. The BAC clones were labeled with different fluorochromes (fluorescent-labeled dUTPs, Enzo Life Sciences, NY, USA) by nick translation and validated on normal metaphase chromosomes. For the BCOR assay, we employed a combined BCOR-CCNB3 inversion-fusion design that can detect both BCOR gene abnormalities, including break-apart and inversion/fusion with CCNB3. The BACs flanking centromeric and telomeric areas of BCOR were labeled with orange and green fluorescence, respectively, while BACs flanking both sides of CCNB3 were labeled with red. The slides were deparaffinized, pretreated, and hybridized with denatured probes. After overnight incubation, the slides were washed, stained with DAPI, mounted with an antifade solution, and then examined on a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany) controlled by Isis 5 software (Metasystems). As the initial results suggested a BCOR break-apart signal, additional FISH testing included known BCOR fusion partners, i.e. ZC3H7B and MAML3 (Specht et al., 2016). Subsequent to the RNA sequencing findings, further investigation of the 20q13.33 and Xp11.2 loci was performed with individual BACs interrogating SS18L1, SSX1, and SSX2 genes being designed. The BACs used for SSX1 gene flanked both SSX1 and SSX4 genes, as well as other SSX genes not previously reported in the pathogenesis of SS.
Immunohistochemistry
Immunohistochemical stains were performed on 4 μm-thick FFPE tissue sections for TLE1 (Santa Cruz Biotech, clone Poly; sc-9121; 1:100 dilution), BCL2 (Ventana: Ready to Use, clone 124Mo/Mo), H3K27me3 stain (clone C36B11; Cell Signaling Technology, Danvers, MA; 1:200 dilution), BCOR (clone C10; sc-514576; Santa Cruz, Dallas, TX; 1:150 dilution) and SATB2 (clone EP281; CellMarque, California, USA; 1:200 dilution). The antigen retrieval and staining protocols were as reported previously (Kao et al., 2016b; Prieto-Granada et al., 2016).
Targeted RNA sequencing and analysis
RNA was extracted from FFPE tissue using Amsbio’s ExpressArt FFPE Clear RNA Ready kit (Amsbio LLC, Cambridge, MA). Fragment length was assessed with an RNA 6000 chip on an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA-seq libraries were prepared using 20–100 ng total RNA with the Trusight RNA Fusion Panel (Illumina, San Diego, CA), which targets a list of N genes of interest. Each sample was subjected to targeted RNA sequencing on an Illumina MiSeq at 8 samples per flow cell (approximately 3 million reads per sample). All reads were independently aligned with STAR (ver 2.3) and BowTie2 against the human reference genome (hg19) for Manta-Fusion and TopHat-Fusion analysis, respectively, for fusion discovery. The mRNA expression levels of certain genes of interest (BCOR, SS18L1, SSX1, etc.) were evaluated and compared to those of other samples analyzed in the same batch of targeted RNA sequencing platform, including 3 fusion-negative URCS, 2 epithelioid hemangioendotheliomas, one SS with SS18-SSX2 fusion, and one inflammatory myofibroblastic tumor.
RESULTS
Diagnostic dilemma in a PD-SS lacking SS18 gene abnormalities by FISH
Based on the morphologic appearance of an undifferentiated sarcoma with spindle and round cell phenotype, showing involvement/origin from the posterior tibial nerve, the main differential diagnosis included PD-SS, high grade malignant peripheral nerve sheath tumor, and less likely an URCS with a spindle cell component. Immunohistochemically, the tumor cells were positive for TLE1 and Bcl2 stains, while H3K27me3 expression was retained. Although this immunoprofile was in keeping with a diagnosis of PD-SS, FISH studies for SS18 break-apart were negative. As URCS with BCOR gene abnormalities (i.e. BCOR-CCNB3, BCOR-MAML3)(Puls et al., 2014; Peters et al., 2015; Specht et al., 2016) have been described as having spindle cell areas in addition to a primitive round cell phenotype, further immunostains and FISH analysis were performed to exclude a BCOR-fusion positive URCS. BCOR immunostaining showed diffuse and strong nuclear reactivity (Fig. 2C), while SATB2 was positive with moderate intensity (Fig. 2D). FISH showed complex gene rearrangements involving both BCOR and CCNB3 genes (Fig. 3A), however, no fusion signal between BCOR and CCNB3 was noted. No FISH break-apart was identified in the two known BCOR fusion gene partners: ZC3H7B and MAML3 genes.
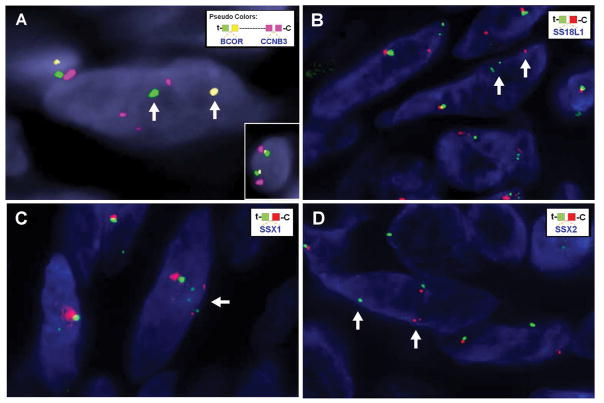
Figure 3.
Fragmentation of the Xp11.22–4 region detected by FISH. (A) BCOR rearrangement is detected with split yellow (centromeric) and green (telomeric) signals (arrows), while CCNB3 shows multiple smaller red signals, in keeping with complex rearrangements and fragmentation. Inset for comparison shows a normal cell with BCOR (green and yellow) and CCNB3 (red) signals are close to each other, being separated only by a 9 Mb gap. (B–D) Complex rearrangements of SS18L1, SSX1, and SSX2 genes as demonstrated by break apart signals as well as additional multiple signals, smaller in size compared to normal allele (arrows).
Synovial sarcoma with SS18L1-SSX1 fusion by targeted RNA sequencing
To better characterize the complex genetic abnormalities identified by FISH, targeted RNA sequencing was performed which identified an in-frame SS18L1-SSX1 fusion candidate by both TopHat-Fusion and Manta-Fusion algorithms (Fig. 4). The fusion reads showed the SS18L1 (nBAF chromatin remodeling complex subunit, 20q13.33) exon 10 fused to SSX1 (SSX family member 1, Xp11.23) exon 6. The projected chimeric transcript retained most of SS18L1 coding sequence, except for the last exon (exon 11), similar to the canonical SS18 breakpoint seen in SS18-SSX fusions. In addition, multiple intra- and inter- chromosomal rearrangements were identified involving chromosomes 20 and X, involving TMX4, ZFP64, GPR64, MSN, TENM1 genes and other unannotated regions (Fig. 4). No specific BCOR gene abnormalities were identified by RNAseq, even after manual inspection of the reads.
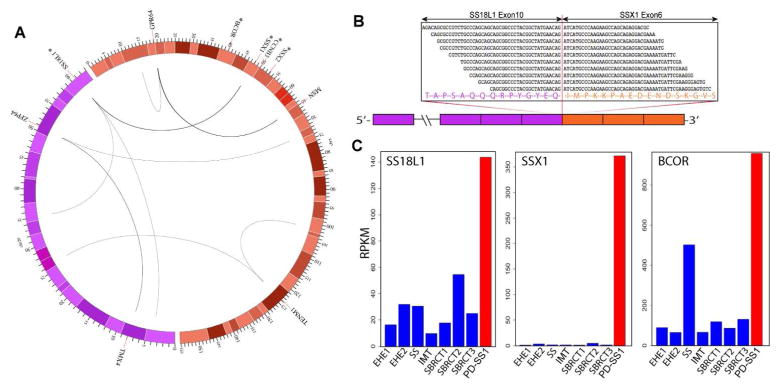
Figure 4.
(A) Circos plot showing complex genetic abnormalities in chromosomes 20 (purple) and X (orange) by RNA sequencing. Curved lines indicate inter- and intra-chromosomal rearrangements between genes and/or unannotated regions. The line thickness is proportional to the numbers of supporting reads. The chromosome scales correspond to megabases (Mb) away from the telomeric end of p-arm. The color shades represent chromosome bandings. Asterisks indicate genes tested by FISH. (B) Fusion junction reads supporting the in-frame fusion of SS18L1 exon 10 to SSX1 exon 6. (C) Index case (PD-SS1, red) showing significant mRNA up-regulation of SS18L1, SSX1 and BCOR genes, compared to 7 control samples (blue). One of the control samples, an SS18-SSX2 fusion-positive SS showed moderately elevated BCOR expression.
Subsequent confirmatory FISH assays demonstrated rearrangements of SS18L1 and SSX1 genes (Fig. 3B–C). Further complex gene abnormalities, including multiple smaller/fragmented signals, were observed by FISH at 20q13.33 and Xp11.22–23 loci, including the SSX2 locus (Fig. 3D).
Expression analysis of the RNA sequencing data showed up-regulated expression of BCOR, SS18L1 and SSX1 compared to a control group of 7 soft tissue tumors (Fig. 4). A SS with SS18-SSX2 fusion in the control group also showed moderately elevated expression of BCOR, but not for SS18L1 and SSX1. Other genes with rearrangements identified by RNA sequencing, including TMX4, ZFP64, GPR64, MSN, TENM1, did not show significant mRNA overexpression.
Corroborating the clinical presentation, pathologic and molecular findings, the results are in keeping with an intraneural PD-SS harboring a rare SS18L1-SSX1 fusion along with complex secondary genomic changes and activation of BCOR mRNA and protein.
DISCUSSION
SS is characterized histologically by fascicles of monomorphic spindle cells with or without an epithelial/glandular component. In contrast, PD-SS display variable areas of round cell morphology, mimicking Ewing sarcomas or other round cell sarcomas (de Silva et al., 2003). Ancillary techniques, such as nuclear immunoreactivity for TLE1, detected in the majority of SS (82%), are useful to support the diagnosis of SS (Foo et al., 2011). Furthermore, the presence of the SS18-SSX fusion in the overwhelming majority of SS have provided pathologists with a reliable molecular assay in confirming the diagnosis, either by RT-PCR amplification of the fusion transcript or more often by demonstration of SS18 break-apart by FISH. A negative molecular result in the setting of an undifferentiated sarcoma with spindle and round cell morphology is often used against a diagnosis of SS.
One entity recently emerging as having morphologic overlap with SS is the URCS with BCOR genetic abnormalities, including both fusions and internal tandem duplications (ITD) (Pierron et al., 2012; Kao et al., 2016a; Specht et al., 2016). URCS harboring BCOR-ITDs appear to be limited to the infantile age group, while tumors with BCOR-related fusions occur in both children or young adults (Pierron et al., 2012; Kao et al., 2016a; Specht et al., 2016). In particular, URCS with BCOR-CCNB3 intra-chromosomal inversions have a mixed round and spindle cell phenotype, reminiscent of SS (Peters et al., 2015). Although BCOR immunohistochemistry has been recently shown as a highly sensitive marker for URCS with various BCOR genetic alterations, showing strong and diffuse reactivity, it is also positive in about half of SS cases tested (Kao et al., 2016b). Further immuno overlap between these two entities has been reported, including TLE1 positivity stain in a subset (3 out of 4 cases) of BCOR-CCNB3 fused tumors (Li et al., 2016) and SATB2 immunoreactivity in most (71 %) BCOR-CCNB3 fused tumors and a small subset of SS (12%) (Kao et al., 2016b).
Located telomeric to the SSX gene cluster on the short arm of X chromosome, BCOR (Xp11.4) is a sarcoma-associated gene, whose rearrangements are associated with round cell sarcomas (BCOR-CCNB3, BCOR-MAML3, ZC3H7B-BCOR), ossifying fibromyxoid tumors (ZC3H7B-BCOR) or endometrial stromal sarcoma (ZC3H7B-BCOR)(Pierron et al., 2012; Panagopoulos et al., 2013; Antonescu et al., 2014; Specht et al., 2016).
There is one prior reported case of a biphasic SS showing an identical SS18L1-SSX1 fusion. Interestingly, similar to our case, this lesion was also described as being associated with a nerve (i.e. peroneal nerve) and occurred in the lower leg of a 36-year-old man, which required opening the nerve for a marginal excision (Storlazzi et al., 2003). Although most intraneural SS have been described to harbor typical SS18-SSX1/2 fusions (Scheithauer et al., 2011), it is quite remarkable that both cases so far reported with this unusual SS18L1-SSX fusion variant have occurred within the nerves, further simulating a malignant peripheral nerve sheath tumor in the setting of negative SS18 gene abnormalities.
SS18L1 encodes a calcium-responsive transactivator, an essential subunit of a neuron-specific chromatin-remodeling complex. SS18L1 is a paralog of SS18, sharing 54% amino acid homology (Aizawa et al., 2004), both being subunits of the chromatin remodeling Brg/Brm-associated factor (BAF) complexes. During neuronal development, mitotic exit of neurons is associated with the switch of npBAF (neural progenitor BAF) to nBAF (neuron-specific BAF), which involves replacement of SS18 in the npBAF complex by CREST, the protein encoded by SS18L1 (Staahl et al., 2013; Kadoch and Crabtree, 2015). SS18L1 plays an important role in neuronal development, especially in dendrite growth and branching (Aizawa et al., 2004). Mutations of SS18L1 has been identified as one of the genetic risk factors related to amyotrophic lateral sclerosis, a motor neuron disease (Chesi et al., 2013; Teyssou et al., 2014). The chimeric transcripts of SS18L1-SSX1 in our case and the previously reported case were identical, with exon 10 of SS18L1 fused to exon 6 of SSX1, which is also the most common fusion junction of typical SS18-SSX1/2 (Storlazzi et al., 2003; Przybyl et al., 2012). In the chimeric proteins, the same C-terminal 8 amino acids from exon 11 of SS18 or SS18L1 were replaced with the C-terminus 78 amino acids of SSX1.
In addition to the SS18L1-SSX1 fusion detected by RNA sequencing, the tumor further displayed complex genetic abnormalities in both chromosomes 20 and X, including numerous gene rearrangements. This finding was further supported by FISH studies using probes flanking SS18L1 (20q13.33), SSX1 (Xp11.23), BCOR (Xp11.4), CCNB3 (Xp11.22), SSX2 (Xp11.22), and TFE3 (Xp11.23), which identified signal fragmentation of SS18L1, SSX1, CCNB3, SSX2 and TFE3. Similar to our case, cytogenetic and FISH analyses performed on the prior SS with SS18L1-SSX1 fusion showed two marker chromosomes consisting of genetic materials from both chromosomes 20 and X (Storlazzi et al., 2003). The complex rearrangements occurring in these two cases may be explained by the opposite directions of transcription of the two gene partners, with a simple, reciprocal translocation event not being able to form a functional fusion transcript.
Our case demonstrates that BCOR up-regulation and overexpression is not only present in SS with SS18-SSX1/2 as previously shown (Kao et al., 2016b), but also with SS18L1-SSX1 fusions. The mechanism of BCOR expression in SS remains unclear and requires further investigation.
In summary, we report an unusual case of intraneural PD-SS harboring a SS18L1-SSX1 fusion, resulting in BCOR upregulation at mRNA and protein levels. This case further illustrates the emerging overlap between SS and URCS with BCOR genetic alterations, at histologic, immunohistochemical, and transcriptional levels, implicating BCOR oncogenic activation as a potential downstream common core between SS with various gene fusions and BCOR-related URCS. This case also illustrates the fact that using a single molecular method in isolation might be misleading, and corroborating additional genetic/genomic methods can help in the correct classification.
Supplementary Material
Acknowledgments
Supported in part by: P50 CA140146-01 (CRA); P30-CA008748 (CRA); Kristen Ann Carr Foundation (CRA); Cycle for Survival (CRA)
Grant Support: P30 CA008748
Footnotes
Conflicts of interest: None
References
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Sung YS, Chen CL, Zhang L, Chen HW, Singer S, Agaram NP, Sboner A, Fletcher CD. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors--molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer. 2014;53:183–193. doi: 10.1002/gcc.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi A, Staahl BT, Jovicic A, Couthouis J, Fasolino M, Raphael AR, Yamazaki T, Elias L, Polak M, Kelly C, Williams KL, Fifita JA, Maragakis NJ, Nicholson GA, King OD, Reed R, Crabtree GR, Blair IP, Glass JD, Gitler AD. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva MV, McMahon AD, Paterson L, Reid R. Identification of poorly differentiated synovial sarcoma: a comparison of clinicopathological and cytogenetic features with those of typical synovial sarcoma. Histopathology. 2003;43:220–230. doi: 10.1046/j.1365-2559.2003.01668.x. [DOI] [PubMed] [Google Scholar]
- Foo WC, Cruise MW, Wick MR, Hornick JL. Immunohistochemical staining for TLE1 distinguishes synovial sarcoma from histologic mimics. Am J Clin Pathol. 2011;135:839–844. doi: 10.1309/AJCP45SSNAOPXYXU. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YC, Sung YS, Zhang L, Huang SC, Argani P, Chung CT, Graf NS, Wright DC, Kellie SJ, Agaram NP, Ludwig K, Zin A, Alaggio R, Antonescu CR. Recurrent BCOR internal tandem duplication and YWHAE-NUTM2B fusions in soft tissue undifferentiated round cell sarcoma of infancy: overlapping genetic features with clear cell sarcoma of kidney. Am J Surg Pathol. 2016a;40:1009–1020. doi: 10.1097/PAS.0000000000000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YC, Sung YS, Zhang L, Jungbluth AA, Huang SC, Argani P, Agaram NP, Zin A, Alaggio R, Antonescu CR. BCOR overexpression is a highly sensitive marker in round cell sarcomas with BCOR genetic abnormalities. Am J Surg Pathol. 2016b;40:1670–1678. doi: 10.1097/PAS.0000000000000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladanyi M, Antonescu CR, Leung DH, Woodruff JM, Kawai A, Healey JH, Brennan MF, Bridge JA, Neff JR, Barr FG, Goldsmith JD, Brooks JS, Goldblum JR, Ali SZ, Shipley J, Cooper CS, Fisher C, Skytting B, Larsson O. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res. 2002;62:135–140. [PubMed] [Google Scholar]
- Li WS, Liao IC, Wen MC, Lan HH, Yu SC, Huang HY. BCOR-CCNB3-positive soft tissue sarcoma with round-cell and spindle-cell histology: a series of four cases highlighting the pitfall of mimicking poorly differentiated synovial sarcoma. Histopathology. 2016;69:792–801. doi: 10.1111/his.13001. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Thorsen J, Gorunova L, Haugom L, Bjerkehagen B, Davidson B, Heim S, Micci F. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation. Genes Chromosomes Cancer. 2013;52:610–618. doi: 10.1002/gcc.22057. [DOI] [PubMed] [Google Scholar]
- Peters TL, Kumar V, Polikepahad S, Lin FY, Sarabia SF, Liang Y, Wang WL, Lazar AJ, Doddapaneni H, Chao H, Muzny DM, Wheeler DA, Okcu MF, Plon SE, Hicks MJ, Lopez-Terrada D, Parsons DW, Roy A. BCOR-CCNB3 fusions are frequent in undifferentiated sarcomas of male children. Mod Pathol. 2015;28:575–586. doi: 10.1038/modpathol.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre JM, Delattre O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Prieto-Granada CN, Wiesner T, Messina JL, Jungbluth AA, Chi P, Antonescu CR. Loss of H3K27me3 expression is a highly sensitive marker for sporadic and radiation-induced MPNST. Am J Surg Pathol. 2016;40:479–489. doi: 10.1097/PAS.0000000000000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyl J, Sciot R, Rutkowski P, Siedlecki JA, Vanspauwen V, Samson I, Debiec-Rychter M. Recurrent and novel SS18-SSX fusion transcripts in synovial sarcoma: description of three new cases. Tumour Biol. 2012;33:2245–2253. doi: 10.1007/s13277-012-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls F, Niblett A, Marland G, Gaston CL, Douis H, Mangham DC, Sumathi VP, Kindblom LG. BCOR-CCNB3 (Ewing-like) sarcoma: a clinicopathologic analysis of 10 cases, in comparison with conventional Ewing sarcoma. Am J Surg Pathol. 2014;38:1307–1318. doi: 10.1097/PAS.0000000000000223. [DOI] [PubMed] [Google Scholar]
- Scheithauer BW, Amrami KK, Folpe AL, Silva AI, Edgar MA, Woodruff JM, Levi AD, Spinner RJ. Synovial sarcoma of nerve. Hum Pathol. 2011;42:568–577. doi: 10.1016/j.humpath.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Specht K, Zhang L, Sung YS, Nucci M, Dry S, Vaiyapuri S, Richter GH, Fletcher CD, Antonescu CR. Novel BCOR-MAML3 and ZC3H7B-BCOR gene fusions in undifferentiated small blue round cell sarcomas. Am J Surg Pathol. 2016;40:433–442. doi: 10.1097/PAS.0000000000000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl BT, Tang J, Wu W, Sun A, Gitler AD, Yoo AS, Crabtree GR. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J Neurosci. 2013;33:10348–10361. doi: 10.1523/JNEUROSCI.1258-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi CT, Mertens F, Mandahl N, Gisselsson D, Isaksson M, Gustafson P, Domanski HA, Panagopoulos I. A novel fusion gene, SS18L1/SSX1, in synovial sarcoma. Genes Chromosomes Cancer. 2003;37:195–200. doi: 10.1002/gcc.10210. [DOI] [PubMed] [Google Scholar]
- Teyssou E, Vandenberghe N, Moigneu C, Boillee S, Couratier P, Meininger V, Pradat PF, Salachas F, Leguern E, Millecamps S. Genetic analysis of SS18L1 in French amyotrophic lateral sclerosis. Neurobiol Aging. 2014;35:1213.e1219–1213 e1212. doi: 10.1016/j.neurobiolaging.2013.11.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.