Abstract
What changes need to occur in a primary tumor to make it metastatic? Denny et al. address this question for small cell lung cancer (SCLC), finding that changes in genomic accessibility mediated by a single transcription factor, NFIB, comprise at least one mechanism influencing metastasis.
Small cell lung cancer (SCLC), representing 15% of all lung cancers, is a devastating neuroendocrine cancer caused by cigarette smoking that usually presents with widespread metastases to lymph nodes, liver, bone marrow, brain, adrenal gland, and other sites. It has an average survival of about 1 year and a 5 year survival of 5% or less (Bunn et al., 2016; NCI-Translational-Research-Advisory-Committee, 2014). Despite many decades of cell and molecular biologic study, we still do not understand why SCLC exhibits such aggressive metastatic behavior nor why the initial dramatic response to chemotherapy is inevitably followed by resistance. A surprising insight into the molecular changes that drive metastasis is now reported by Denny et al. (Denny et al., 2016), who have identified the transcription factor NFIB as an agent that can cause metastatic behavior by reconfiguring the regions of open chromatin in SCLC cells (Figure 1).
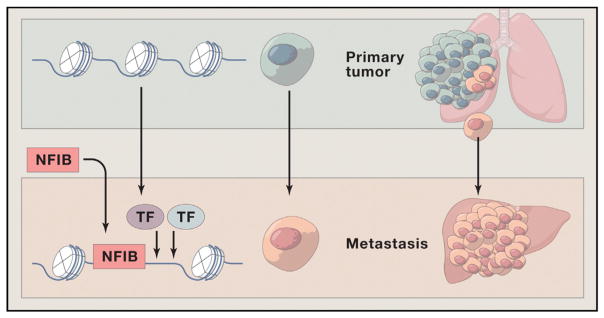
Figure 1. NFIB Promotes Metastasis through Increasing Chromatin Accessibility.
Small cell lung carcinoma (SCLC) is a neuroendocrine tumor that is highly metastatic to many sites including the liver. Denny et al. use a genetically engineered mouse model of SCLC to show that some cells of the primary tumor in the lung acquire elevated levels of NFIB, a transcription factor (in some cases by gene amplification). These cells selectively disseminate and form metastases in the liver. Chromatin in the metastatic tumor has widespread increases in accessibility in gene distal regions that resemble those seen in neural tissue. They show that NFIB is responsible for opening these chromatin regions, is found bound to these sites, is required for maintenance of the open sites, may influence the binding of other transcription factors to alter gene expression, and leads to a program generating metastases. Overall they show that NFIB alone is both necessary and sufficient to cause liver metastases in SCLC through this change in chromatin accessibility and thus represents both a previously unrecognized mechanism underlying metastasis and an important new therapeutic target.
Denny et al. deployed a genetically engineered mouse model (featuring induced loss of Tp53, Rb1, and Rbl2 and GFP labeling triggered by Cre inhalation into the lung) that develops disease histologically and molecularly resembling human SCLC and behaving like it as well, including the development of widespread neuroendocrine metastatic disease (Schaffer et al., 2010). This allowed them to isolate primary tumors and liver metastases and identify changes in chromatin accessibility upon transition of a primary tumor to metastases using ATAC-seq. In brief, they found a dramatic change in chromatin accessibility in nearly all of the liver metastases compared to the matched primary tumor, with 24% of the accessible regions in the metastases showing increased openness. These differentially accessible regions were “gene distal” and in “gene deserts,” but were evolutionary conserved and enriched in NFI transcription factor binding motifs. Of great interest, the hyper-accessible samples from liver metastases had increased levels of NFIB, often in association with Nfib gene amplification. While cells were heterogeneous for high levels of NFIB in the primary tumors, metastatic tumors were more homogeneous for high NFIB. The authors show that Nfib overexpression was necessary and sufficient to generate metastatic disease in vivo, was required for clonal growth and invasive ability, and that NFIB was associated with the newly open chromatin and maintained the hyper-accessible chromatin state. The distal regions that became accessible upon NFIB upregulation were similar to open regions in neural tissue. Thus, SCLC gains metastatic ability through dramatic remodeling of their chromatin state through the auspices of one transcription factor, NFIB.
This finding is important for understanding metastasis in SCLC, particularly as the ability to molecularly analyze primary and metastatic tumor cells from patients is extremely challenging. Interestingly, a related study using another genetically engineered mouse model for SCLC also provides evidence for a role of NFIB in driving metastasis in SCLC and reports that NFIB expression correlates with advanced stage neuroendocrine carcinomas in humans (Semenova et al., 2016). The chromatin-related findings of Denny et al. also reveal a mechanism that may be broadly applicable to understanding metastatic potential of other tumor types, through elevated levels of expression of NFI factors, or other chromatin altering transcription factors. Thus, a major priority going forward is to analyze the chromatin state of metastases in tumors other than SCLC to assess the generality for NFIB function in metastases, or to identify other factors playing an analogous role in altering chromatin accessibility. Likewise, as shown by the isolated examples of metastases without NFIB overexpression, there must be other metastatic driver mechanisms we need to discover. We know at diagnosis that SCLCs have already, or will metastasize. It is possible the discovery of tumor cells in primary tumors with open chromatin are those that will metastasize, providing a rationale and mechanistic approach to prediction of metastatic behavior.
Importantly, NFIB, which appeared to displace nucleosomes in the Denny et al. study, is not itself a pioneering factor (a factor that can bind closed chromatin), but rather seems to need its sites to be at least loosely available for binding. Other factors with known pioneering factor function present in SCLC, such as FOXA2 or even ASCL1, may be involved together with NFIB to influence the sites with increased chromatin accessibility (Zaret and Mango, 2016). The combination of lineage-specific pioneering factors plus elevated levels of factors such as NFIB may enable the opening of sites that allow transcriptional activation of genes influencing metastatic cell behaviors. These studies lead to many questions that when addressed will provide additional targets for therapeutic options. For example, what mechanisms allow elevated NFIB in the first place to drive the metastases and which genes associated with the NFIB-dependent chromatin changes are required for metastatic behavior of the cells? In addition, in the SCLC metastatic model presented by Denny et al., they find genes associated with the chromatin changes are enriched in those in neuronal tissue. Will this neural program be a general feature of metastases in other tumor types or does this reflect the neural related lineage of the neuroendocrine cell of origin in SCLC with a prefigured chromatin landscape? The application of the findings reported by Denny et al., plus answers to these additional questions, are of great interest in clinical translation of these findings for early detection, prevention, and treatment of SCLC.
Acknowledgments
The authors are supported by NCI SPORE in Lung Cancer P50CA70907.
References
- Bunn PA, Jr, Minna JD, Augustyn A, Gazdar AF, Ouadah Y, Krasnow MA, Berns A, Brambilla E, Rekhtman N, Massion PP, et al. J Thorac Oncol. 2016;11:453–474. doi: 10.1016/j.jtho.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny SK, Yang D, Chuang C-H, Brady JJ, Lim JS, Grüner BM, Chiou S-H, Schep AN, Baral J, Hamard C, et al. Cell. 2016;166(this issue):328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCI-Translational-Research-Advisory-Committee. Small Cell Lung Cancer: Seizing on Opportunities to Translate Recent Research into the Clinic for New Diagnostics and Interventions. 2014 http://deainfo.nci.nih.gov/advisory/ctac/0614/SCLCworkshopReport.pdf.
- Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, Sweet-Cordero EA, Sage J. Cancer Res. 2010;70:3877–3883. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova EA, Kwon MC, Monkhorst K, Song JY, Bhaskaran R, Krijgsman O, Kuilman T, Peters D, Buikhuizen WA, Smit EF, et al. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.06.020. Published online on June 30, 2016. http://dx.doi.org/10.1016/j.celrep.2016.06.021. [DOI] [PMC free article] [PubMed]
- Zaret KS, Mango SE. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]