Abstract
With advances in screening, the incidence of detection of premalignant breast lesions has increased in recent decades; however, treatment options remain limited to surveillance or surgical removal by lumpectomy or mastectomy. We hypothesized that disease progression could be blocked by RNA interference (RNAi) therapy and set out to develop a targeted therapeutic delivery strategy. Using computational gene network modeling, we identified HoxA1 as a putative driver of early mammary cancer progression in transgenic C3(1)-SV40TAg mice. Silencing this gene in cultured mouse or human mammary tumor spheroids resulted in increased acinar lumen formation, reduced tumor cell proliferation, and restoration of normal epithelial polarization. When the HoxA1 gene was silenced in vivo via intraductal delivery of nanoparticle-formulated small interfering RNA (siRNA) through the nipple of transgenic mice with early-stage disease, mammary epithelial cell proliferate rates were suppressed, loss of estrogen and progesterone receptor expression was prevented, and tumor incidence was reduced by 75%. This approach that leverages new advances in systems biology and nanotechnology offers a novel non-invasive strategy to block breast cancer progression through targeted silencing of critical genes directly within the mammary epithelium.
INTRODUCTION
The most common non-invasive lesion of the breast is ductal carcinoma in situ (DCIS), in which abnormal ductal epithelial cells proliferate inside the mammary duct but do not penetrate through the basement membrane to invade adjacent tissue. In the United States, approximately 25% of newly diagnosed breast lesions are classified as DCIS, and more than one million women will be living with DCIS in the United States alone by 2020 (1, 2). Atypical ductal hyperplasia represents approximately 10% of mammographically detected breast lesions. These are marked by the presence of abnormal epithelial cells and elevated cell numbers compared with normal mammary ducts (3). All classes of early lesions are highly heterogeneous and, although only a fraction will progress to invasive breast tumors (estimates range from 14 to 53% for DCIS) (4–6), there are currently no biomarkers to aid in identifying which tumors will become invasive. Surveillance (so called, “watch-and-wait”) may be recommended for earlier stages of hyperplasia with or without atypia.
Treatment of premalignant disease is typically aggressive for DCIS lesions and options include mastectomy, lumpectomy, and radiation. All of these have serious systemic side effects and impact quality of life. Some patients with hormone receptor–positive DCIS will also receive five years of endocrine therapy, which has been shown to reduce recurrence, although no survival benefit has been demonstrated. The side effects of endocrine therapy may be life threatening and include stroke, blood clots, bone loss and elevated risks of uterine and endometrial cancers (7). For these reasons, a recent survey of breast cancer professionals identified the need for minimally invasive therapies that can be selectively targeted to the ductal epithelium to prevent progression of premalignant breast lesions without producing systemic toxicity as one of the highest priorities of translational breast cancer research (8).
Small interfering RNA (siRNA) has been used as a therapeutic agent in the treatment of a variety of tumor types in rodent models, including human mammary tumor xenografts in mice(9–11), and it is well-tolerated with few adverse side effects. Therapeutic RNA interference (RNAi) is also currently being tested in human clinical trials in patients with liver cancer, metastatic liver disease, pancreatic ductal adenocarcinoma, advanced colon cancer and familial adenomatous polyposis (FAP) which leads to colon tumorigenesis (12). RNAi based approaches are interesting because they offer the possibility for a more personalized therapy in that the same technology and approach can be adapted to a variety of tumor-specific gene targets. In addition, RNAi is useful for silencing undruggable targets that are not amenable to traditional small molecule– or antibody-based inhibition (13–15). Nevertheless, development of RNAi-based cancer therapeutics has been hampered by two major obstacles: 1) identification of appropriate target genes that are causally involved in cancer development, and 2) selection of an effective delivery method with minimal systemic side effects.
Here, we addressed the first challenge through gene network inference—a computational biology approach that evaluates gene expression changes in context of the entire gene regulatory network (GRN). Through this analysis, we identified the HoxA1 gene as a critical mediator of mammary tumor progression. With increasing availability of genome-wide characterization of clinical samples, we envision the broad application of this approach to multiple types of premalignant lesions, in the breast and other organs. To validate the importance of this gene for breast cancer progression and to address the second challenge, we delivered HoxA1 siRNA into mammary epithelial cells in vivo using lipidoid nanoparticles, which have been shown to facilitate interaction with the cell membrane and endosomal escape in other tissues, including an ovarian tumor model in mice (16). In contrast to earlier studies in which lipidoid-formulated siRNA was injected intratumorally, we delivered the nanoparticles to premalignant lesions within the lining epithelium of intact mammary glands of virgin transgenic mice by direct injection through the nipple. Local delivery is non-invasive and avoids the uptake and accumulation of nanoparticles in the liver—an issue that has proven challenging in systemic administration (17).
HoxA1 silencing was tested in human mammary spheroids as well as in a transgenic mouse model of mammary tumorigenesis. The FVB C3(1) SV40TAg transgenic mouse was chosen as a relevant model of human breast cancer because: 1) tumors form spontaneously in the correct tissue microenvironment, 2) tumor onset and formation are slow, occurring over a period of 4–5 months, 3) lesions robustly progress through all of the typical stages of breast tumorigenesis seen in humans, including hyperplasia ductal carcinoma in situ (DCIS), and locally invasive carcinomas that can metastasizd to the lung and liver, 4) the immune system remains intact. and 5) tumor onset does not require exogenous hormone intervention or pregnancy is required in other transgenic breast cancer models (18, 19).
RESULTS
Putative target identification by computational gene network inference
To identify candidate gene targets with a high likelihood of driving tumor progression in the transgenic mouse FVB C3(1)-SV40TAg model, we constructed models of GRN connectivity with the Mode-of-action by Network Identification (MNI) tool (20, 21). A detailed description of the MNI algorithm may be found in Supplementary Methods. In summary, the MNI tool was first trained on 3,000 gene microarray datasets from various mouse tissues and organs under diverse conditions to develop a basal GRN connectivity model—a directed graph that relates the concentration of each gene transcript to that of every other transcript across the genome (Fig. 1; fig. S1). Genes are deemed to be connected in this network if the activity of one gene influences the transcriptional state of the other, regardless of whether the effect occurs at transcriptional or post-transcriptional levels and independently of the type or directionality of the interaction. Hierarchical clustering (by Euclidean distance with complete linkage performed with the Z-score–normalized expression value) revealed that samples clustered based on disease progression (fig. S1A). Expression profiles of transgenic glands from early stages of cancer progression (<16 weeks) were most similar, whereas the profile of the 20-week glands that contained fulminant cancers was clearly distinct (Pearson correlation coefficient > 0.99 at 20 weeks versus ~0.92 at earlier times; fig. S1B and fig. S1C).
Figure 1. Gene network inference pipeline.
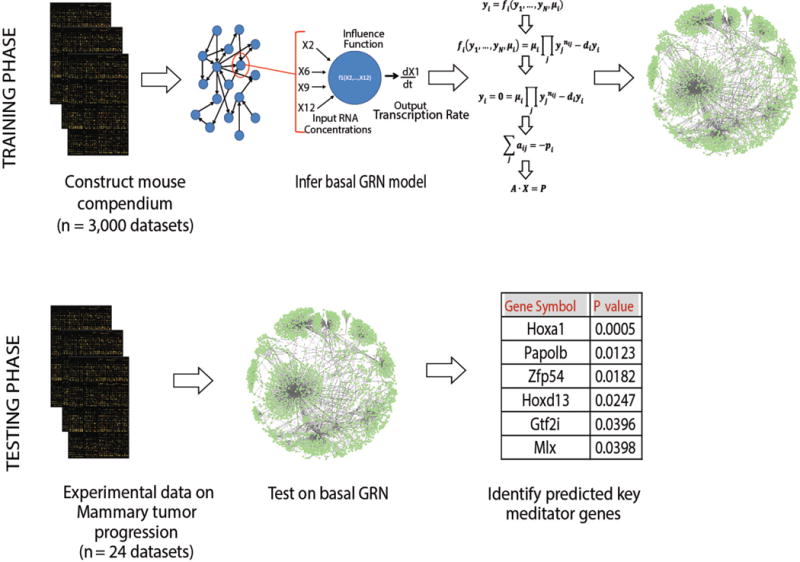
In the first phase, the MNI algorithm was trained on a compendium of 3000 mouse microarray data sets to construct a basal network connectivity model. Genome-wide expression data from wild type and 8 week-old transgenic mammary glands were then interrogated using the GRN model to pinpoint alterations in gene behavior that were unique to particular stages of tumorigenesis. The significance of the predicted key mediator genes of the disease stage of interest is quantified with a z-score. Two-tailed p-values are calculated based on the z-score value. Genes are ranked according to the p-value, and top-ranked genes are selected as probable key mediators of the disease state.
The trained GRN model was then tested with transgenic and wild type mammary gland transcriptome data to identify the earliest changes that rewire the GRN during tumorigenesis. In female transgenic C3(1)-SV40TAg mice, breast tumor formation progresses in a highly reproducible manner, with hyperplasia first appearing at ~ 12 weeks of age, DCIS-like lesions at ~ 16 weeks, and invasive carcinomas at 20 weeks (Fig. 2A) (19). To focus on early events in tumorigenesis, whole genome transcriptome profiles were obtained for mammary glands isolated from 8 week-old transgenic mice when the transgene is highly expressed, but the glands remain histologically normal. Candidate transcription factors were ranked by two-tailed p-value and the HoxA1 gene emerged as the most significant potential key mediator of early cancer progression (p = 0.0005) (Fig. 1). We focused on HoxA1 because past work in human tissues also suggested that it might play an important role in breast cancer development (22–24).
Figure 2. In vitro silencing of HoxA1 normalizes mouse mammary tumor cell spheroid cultures, restoring polarity and reducing proliferative index.
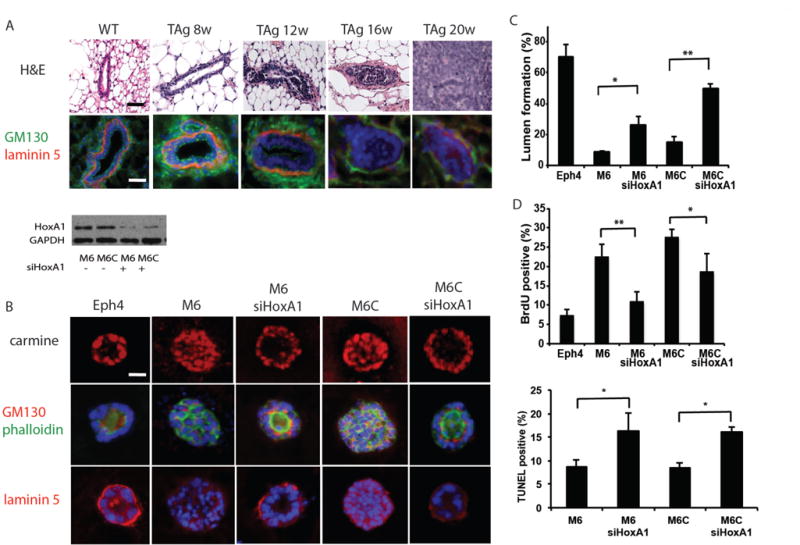
(A) Disease progression in the C3(1)-SV40TAg glands is marked by atypia and hyperplasia, ductal filling and tumor invasion (H&E) with a progressive loss of GM130 (green) and laminin (red) polarity by 12 weeks and basement membrane thinning by 16 weeks. Scale bars, 100 μm. (B) Confocal images through the center of carmine-labeled normal mouse mammary (Eph4) and mouse mammary tumor (M6 and M6C) spheroid cultures reveal filled lumens in tumor cell cultures and a hollow organized lumen morphology when HoxA1 is silenced. Cell polarity was visualized by immunocytochemistry of sectioned spheroids with staining of an apical Golgi marker GM130 and the basal basement membrane protein, laminin. Nuclei are stained with DAPI (blue) and F-actin is labeled with phalloidin (green). Scale bars, 20 μm. Images representative of n = 3 studies, 150–300 spheroids imaged per sample) Downregulation of HoxA1 protein expression was confirmed by immunoblotting in lysates prepared from M6 and M6C cells (n=3). (C and D) The effects of silencing HoxA1 on hollow lumen formation (C) and cell proliferation (D) in collagen-Matrigel spheroid cultures of mouse M6 and M6C tumor cells. Eph4 mouse mammary cells served as normal controls. Data are means +/− SD (n = 3, 150–300 spheroids imaged per sample; *p < 0.05, **p<0.01, Fisher’s exact t-test). (E) Quantitation of apoptosis by TUNEL staining after treatment of M6 and M6C cells with HoxA1 siRNA (n=3, 150–300 spheroids imaged per sample) *p<0.05, Fisher’s exact t-test.
Validation of targets in three-dimensional mouse mammary epithelial cell spheroids
To assess the functional relevance of HoxA1 to breast cancer biology, we then explored whether down-regulating its expression would alter the growth or differentiation of cultured mammary epithelial tumor cells that were originally isolated from primary (M6) or metastatic (M6C) tumors in the same transgenic C3(1)-SV40-Tag mice (25). HoxA1 was silenced with siRNA in M6 and M6C cells, or in normal mouse Eph4 mammary epithelial cells, which were cultured within extracellular matrix (ECM) gels composed of Matrigel and type I collagen. More than 75% of the normal Eph4 mammary cells formed well-differentiated, hollow acinar spheroids lined by a polarized epithelium with a central lumen (Fig. 2B).
These normal mammary cells were also highly polarized as indicated by the accumulation of a circumferential basement membrane containing a linear distribution of laminin-5 along their basal membranes, and a preferentially apical distribution of GM130 after 2 weeks of culture, which mimics normal polarized mammary epithelial architecture in vivo (Fig. 2, A and B). In contrast, M6 and M6C tumor cells failed to form hollow lumens and instead grew as disorganized spheroids filled with cells in vitro (Fig. 2, B and C), much as they do when they form solid tumors during later stages of cancer progression in vivo (Fig. 2A). As expected from their in vivo behavior, M6 and M6C cells also did not form a polarized epithelium or accumulate a linear basement membrane along their periphery (Fig. 2B).
Gene silencing using HoxA1 siRNA (siHoxA1) (fig. S2A) induced both M6 and M6C cancer cells to reorganize into hollow acinar structures lined by a polarized epithelium, with restoration of both apical GM130 staining and a continuous, laminin-5–containing basement membrane at their basal cell surface (Fig. 2, B and C). Thus, silencing of a single gene identified by the MNI algorithm — HoxA1 — resulted in breast cancer differentiation including reconstitution of the normal tissue architecture of wild type mammary gland that is gradually lost in transgenic glands during disease progression (Fig. 2B). Spheroid differentiation induced by gene silencing was also accompanied by a significant reduction in the percentage of proliferating tumor cells in vitro (Fig. 2D), while the overall cell number per spheroid remained the same. This is consistent with the finding that a concomitant increase in tumor cell apoptosis was observed within the newly formed acinar lumens (Fig. 2E). Treatment of M6 and M6C cells with siHoxA1 reduced DNA synthesis by more than 50% and 32%, respectively, and apoptosis increased by approximately 2-fold (~16.5% TUNEL-positive cells in siHoxA1-treated cells vs. 8.5% in control M6 and M6C cells).
Silencing HoxA1 in human breast cancer
To examine the relevance of HoxA1 for human breast cancer, we performed differential expression analysis using the Oncomine compendium of cancer transcriptome profiles obtained from patient tumor samples. Human patient data from ten previously published gene expression analyses, each consisting of 22 to 150 samples representing healthy breast tissue and tumors (26–31) revealed that HoxA1 was over-expressed by greater than 2-fold in human breast lesions (Fig. 3A), which supports the relevance of the present results for human disease. To more directly explore this possibility, we then tested whether silencing of its expression using siRNA would alter the proliferation or differentiation of human MDA-MB-468 and HCC1937 breast tumor cells when grown within the same ECM gels used for the mouse mammary tumor 3D cultures. Upon silencing HoxA1, we observed a major increase in the ability of both human breast tumor cell lines to organize into spheroids containing hollow central lumens under these 3D gel culture conditions (Fig. 3B), with siHoxA1-treated MDA-MB-468 cells and HCC1937 cells exhibiting approximately 6-fold and > 2-fold increases relative to spheroids treated with non-targeting control siRNA, respectively (Fig. 3C). Between 30 and 38% of spheroids formed hollow lumens in the siHoxA1-treated human breast cancer cell cultures, compared with ~50% of spheroids formed by normal human mammary epithelial MCF10A cells (Fig. 3, B and C). Analysis of DNA synthesis under these culture conditions revealed that the growth of the MDA-MB-468 cells and HCC1937 cells were reduced by 60 and 50%, respectively, upon silencing HoxA1 (Fig.3D). Similar results were also observed in human breast cancer cells cultured in 2D on conventional cell culture plastic (fig. S2,A and B). Thus, silencing HoxA1 suppressed growth and restored differentiation, as measured by normal acinar morphology and lumen formation, to two human breast tumor cell lines.
Figure 3. Silencing HoxA1 in human breast cancer cells reduces cell growth and restores lumen organization in tumor cell spheroid cultures.
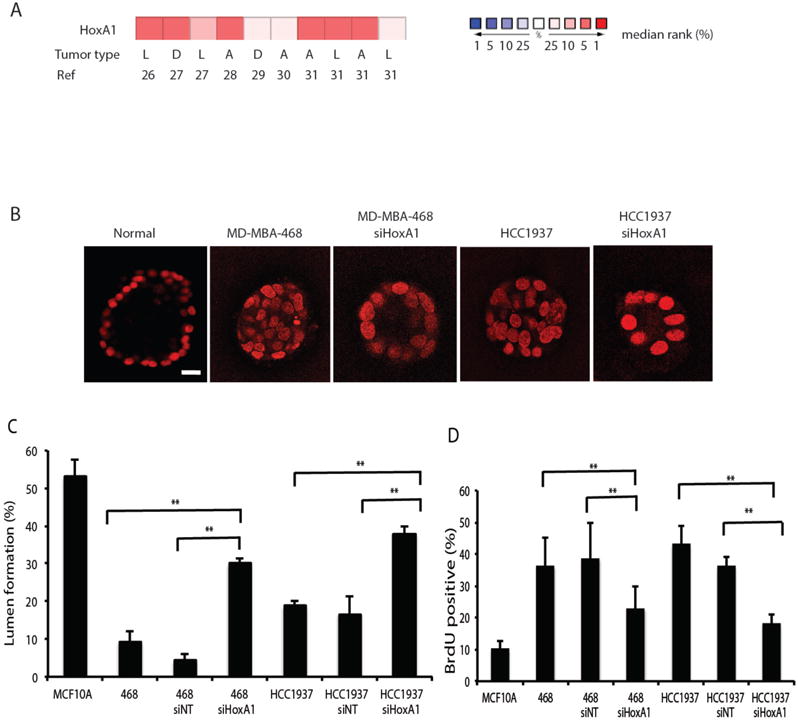
(A) The Oncomine compendium of cancer transcriptome profiles was used for analysis and visualization of HoxA1 expression data in human breast tumors and normal breast tissue. Differential expression analysis identified 10 patient datasets in which HoxA1 was over-expressed in human breast lesions by greater than 2-fold (see references 26–31). The type of tumor represented in the datasets is categorized as D, ductal breast carcinomas; L, lobular breast carcinomas; A; all breast carcinomas. The rank order of HoxA1 in each analysis is displayed (red, increased rank %; blue, decreased rank %). (B) Confocal images through the center of carmine-labeled spheroid cultures of normal human breast epithelial cells (MCF10A) and human breast cancer MD-MBA-468 and HCC1937 cell lines reveal. Breast cancer cells were also treated withsiHoxA1. Scale bar, 20 μm. (C) Hollow lumen formation and (D) proliferation of cancer cells in collagen-Matrigel spheroid cultures. Quantitation of lumen formation in non-tumorigenic human MCF10A breast epithelial cells and cells treated with scrambled siRNA controls are shown for comparison. Data are means +/− SD (n = 3, 150–300 cells per sample) **p<0.01, Fisher’s exact t-test.
In vivo HoxA1 silencing within mammary gland by intraductal delivery of siRNA-lipidoid nanoparticles
To explore whether HoxA1 represents a feasible target for treating the early stages of breast cancer progression, HoxA1 and control, non-targeting siRNAs (siNTs) were formulated with lipidoid nanoparticles (16, 32). These particles were delivered intraductally via injection through the nipple to reach the transformed mammary epithelial cells that give rise to early localized lesions within the lining ductal epithelium. Injection of a fluorescently tagged control siRNA formulated with lipidoid confirmed that a volume of 20 μl of siRNA-nanoparticle solution filled the intact mammary tree and that the nipple and mammary ducts remained intact throughout the treatment (Fig. 4A; fig. S3A). In high-magnification confocal images of injected mammary ducts, fluorescent nanoparticles were observed within mammary epithelial cells at 48 h and 1 week post-injection, and they were not observed in the stromal cells that surround the epithelium (Fig. 4B). No fluorescent particles were observed in the liver, spleen, kidney, heart, lung, or peripheral blood at 48 h, 72 h, or 1 week post-injection.
Figure 4. Silencing HoxA1 in the mammary epithelium reduces tumor incidence, reduces proliferative index, and prevents the loss of hormone receptor expression.
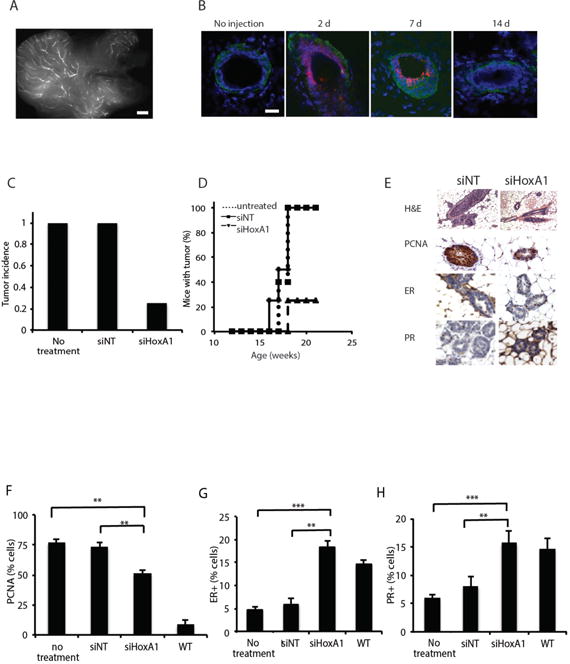
(A) Virgin C3(1)-SV40TAg mammary glands injected in parallel with 20 μl of a fluorescent control siRNA-nanoparticle to visualize the efficiency of its delivery throughout the ductal tree. The entire injected gland was removed and whole mounted for fluorescent imaging. Scale bar, 1 mm. (B) Confocal images of mammary tissue sections show distribution of fluorescent control siRNA (red) in the duct at 2, 7, and 14 days post-injection. Tissues were counterstained with laminin (green) and DAPI (blue). Bar= 100 μm. (C) Tumor incidence measured at 21 weeks in control C3(1)-SV40TAg mice versus mice injected with siHoxA1 bi-weekly for 9 weeks (n=10 siNT, n=8 siHoxA1, n=5 untreated). (D) Timing of onset of tumor formation in untreated, siNT control, and siHoxA1 treated mice. (E) H&E and F) immunohistochemistry for PR, ER, and PCNA in mammary glands. Scale bar, 20 μm. (G, H, I) Percentage of PCNA+, ER+, PR+ cells after 9 weeks of treatment are shown in Data are mean +/− SEM, n=5 animals, 400–550 cells scored per animal, **p<0.01, ***p< 0.001, Pearson’s chi-square test. (For each sample, data).
To determine the potential therapeutic value of this approach in vivo, virgin adult female transgenic mice were injected with siHoxA1 or non-targeting control siRNA biweekly for 9 weeks beginning at 12 weeks of age. Gene silencing was confirmed by immunoblotting of protein isolated from mammary glands after the 9th week of treatment (fig. S3B). While 100% of control transgenic mice (n = 5) or mice treated with non-targeting siRNA (n = 10) developed macroscopic tumors by 21 weeks of age, only 25% of mice treated with intraductal injection of siHoxA1 (n = 8) exhibited tumor formation (p < 0.0001, log rank test) (Fig. 4C). Tumor onset in those mice was delayed by approximately 3 weeks, as detected by weekly ultrasound monitoring (Fig. 4D). Immunohistological characterization of the siHoxA1-treated mammary glands revealed the presence of hyperplasia and ductal filling at 21 weeks (Fig. 4E); however, these lesions did not progress to invasive macroscopic tumors in glands treated with siHoxA1 (Fig. 4, C to E). Cell proliferation also was lower in glands injected with siHoxA1 in vivo compared with controls, as measured by PCNA (Fig. 3F), but there was no significant change in the levels of apoptosis (P > 0.05, Fisher’s t test) (fig. S3C), suggesting that HoxA1 inhibition acted by increasing (or preventing loss of) cell differentiation, rather than by inducing mammary tumor apoptosis.
Mammary tumorigenesis in C3(1)-SV40TAg transgenic glands is also marked by progressive loss of expression of hormone receptors, most notably the estrogen receptor (ER) and progesterone receptor (PR). This is a common characteristic of the most highly aggressive forms of breast cancer in humans as well. Interestingly, while only 4.7% and 5.9% of cells were ER+ in untreated and siNT-treated glands, respectively, at 21 weeks of age, siHoxA1-treated glands continued to express ER at high levels, with 18.4% of cells staining positive for ER (p < 10−7 by Pearson’s chi-square test) (Fig. 4, E and G). A similar response was observed for PR, with 15.7% of PR+ cells in siHoxA1-treated glands versus only 5.9 % and 7.9% of in untreated and siNT-treated glands (p < 10−7 by Pearson’s chi-square test) (Fig. 4, E and H).
FVB C3(1)-SV40TAg mice receiving intraductal injections of siRNA-lipidoid nanoparticles were also monitored closely for signs of local tissue damage, toxicity or systemic side effects. No weight loss was observed in the siHoxA1- or siNT-treated populations (fig. S4A). We also failed to detect an inflammatory response, either when systemic responses were analyzed by measuring spleen size or by quantitating changes in interleukin-6 and interferon-γ cytokine levels in response to administration of siHoxA1- or siNT-containing nanoparticles (P > 0.05, chi-square test) (fig. S4, B–D). Hence, mammary cancer progression was suppressed in this robust transgenic model without producing detectable generalized toxicities by localized delivery of siHoxA1.
HoxA1 silencing downregulates MAPK signaling
Activation of the MAP kinase pathway regulating cell proliferation and survival is a frequent event in tumorigenesis, and modulation of the p44/42 MAP kinase signaling pathway has previously been shown to contribute to the mechanism by which HoxA1 mediates oncogenic transformation and anchorage-independent growth in cultured human mammary tumor cells (33). We therefore investigated the response of p44/42 signaling to local HoxA1 silencing in mouse mammary glands in vivo. Gene expression analysis of transgenic mammary glands revealed alterations in expression of 13 core genes involved in the p44/42 MAPK signaling pathway as the disease progressed from early to late stages (Fig. 5A). Intraductal delivery of siHoxA1 reduced expression of Kras, Erk1/2, Src, Grb2, and Ier3 (Fig. 5B) in the transgenic mammary gland.
Figure 5. siHoxA1 reduces mammary tumor cell proliferation in vivo through modulation of p44/42 MAPK signaling.
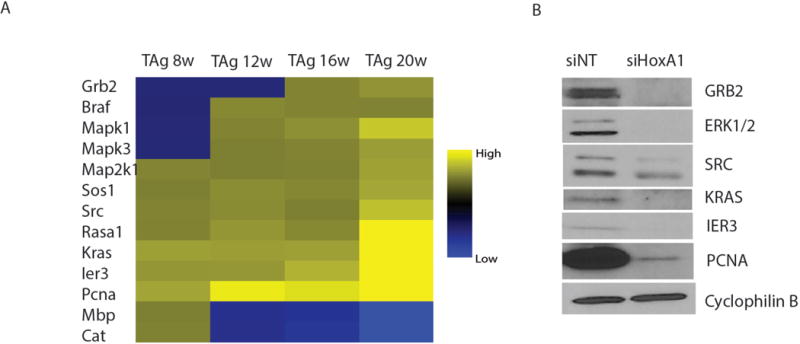
(A) Microarray analysis of MAPK pathway signature genes in untreated transgenic C3(1)-SV40TAg mammary glands (n = 3) at progressive disease stages. (B) The effect of bi-weekly siHoxA1 injections on protein expression of MAPK components, ERK1/2, SRC, IER3, KRAS, PCNA and GRB2, in mammary glands of 21-week old transgenic animals (n = 3) compared with injection of non-targeting control siRNA (siNT) (n = 3).
DISCUSSION
These data show that a GRN computational approach designed to define GRN connectivity can be used to successfully identify genes that are physiologically relevant to human breast tumor progression, and that can serve as potential therapeutic targets with RNAi. Intraductal delivery of siRNA encapsulated in lipidoid nanoparticles enabled localization to the relevant cells lining the mammary ducts that give rise to the majority of invasive breast tumors. To our knowledge, this is the first example of minimally invasive local delivery of RNAi-based therapeutic agents through the nipple.
This approach can be generalized to target any candidate gene, allowing the treatment of breast lesions in a tumor-specific and patient-specific manner. For example, it may be possible to personalize siRNA treatments for patients with known gene mutations. In this study, we determined that HoxA1 was a critical transcriptional regulator during early stages of mammary tumorigenesis; thus, we tailored RNAi therapy to this target. Hox genes act as master regulators of development in the embryo and they are expressed in a tightly regulated spatiotemporal fashion. In humans, HoxA1 plays a critical role in early embryonic development and is critical in hindbrain segmentation, skull morphogenesis, and inner ear organogenesis (34). Alterations in Hox genes also have been associated with a variety of human cancers, including breast (22, 35), skin (36), lung (37), prostate (38), and hematopoietic tumors (39). HoxA1 is not expressed in adult human mammary gland, but several previous studies have revealed upregulation in mammary carcinomas (22, 23, 40, 41). Although additional studies will be required to clarify the mechanism by which HoxA1 contributes to breast tumor formation in humans, previous work has shown that over-expression of HoxA1 in normal human mammary epithelial cells is sufficient to initiate oncogenic transformation (24). Furthermore, we demonstrate here that silencing HoxA1 suppresses growth and induces differentiation of human breast cancer spheroids and that suppression of this gene in the early stages of the disease is associated with reduced MAPK signaling in mice, suggesting that it may also be a relevant target in human tumors that respond to MEK inhibitors.
The promise of less-invasive, intraductal alternatives for human breast cancer patients has been widely discussed in recent years, and small pilot studies have demonstrated the feasibility of cannulating diseased ducts in humans (42) for the delivery of chemotherapeutic agents to specific ducts (43). Conventional chemotherapeutic agents, such as 5-FU, carboplatin, and pegylated liposomal doxorubicin, have also been shown to be effective when injected intraductally in rodent models of chemically induced carcinogenesis (44, 45). However, long-term follow up (> 30 weeks) revealed that intraductal delivery of this form of doxorubicin also induced formation of malignant mammary tumors in normal FVB/N mice (46). Our results show that RNAi-based therapies can be similarly delivered intraductally in a transgenic mouse mammary cancer model, which both provides a more relevant animal model for breast cancer progression and opens up the possibility for streamlined development of this class of non-cytotoxic siRNA-based agents as a new minimally-invasive, localized treatment option.
All untreated SV40 TAg animals developed tumors by 21 weeks of age and hence, they needed to be sacrificed at this time according to humane animal care procedures. Therefore, the 21-week endpoint was chosen to permit comparison of the tumor burden in untreated and siRNA HoxA1 treated groups. So while this study demonstrated safety and efficacy throughout the 21-week study period, we can not comment on the long-term effects of RNAi therapy. In any case, additional studies will be needed to characterize the pharmacokinetics, long-term safety, and long-term dosing and efficacy of intraductally administered RNAi therapeutics before this therapeutic strategy is explored for clinical use. Unlike mice, human breasts contain multiple ductal systems with separate openings on the nipple. Thus, a large animal model with similar mammary gland anatomy, such as rabbit, may provide additional insight. It is important to note, however, that the diameter and volume of the human breast ducts are larger than mouse, but still lined by a single layer of epithelial cells; this may allow administration of higher doses or use of less frequent dosing. In the end, appropriate dosing in humans would have be determined and optimized in Phase I clinical trials.
If this therapeutic strategy is found to be useful in humans in the future, the therapeutic RNAi could be administered in a prophylactic capacity to high-risk patients, prior to the detection of DCIS or breast lesions. Alternatively, it could be administered with chemotherapy to women who have tumors removed and who would normally receive chemotherapy (or radiation therapy) to prevent progression of remaining small undetected lesions. The feasibility, absence of side effects over several months, and the overall prevention of tumor progression observed in this study support the further investigation of intraductal delivery of patient-specific siRNA therapy for breast cancer prevention and treatment. However, safety studies in humans will be critical in determining the potential extension of these findings to patients.
METHODS
Study design
At 21 weeks of age, FVB C3(1) SV40 Tag animals typically have mammary tumors greater than 10% of body weight and must be sacrificed according to animal care protocols. We therefore chose 21 weeks as an endpoint to allow comparison of the tumor burden in treated animals to the control group (without siHoxA1 treatment). Animals were assigned randomly to experimental groups by the veterinary technician. Quantitation and scoring of immunohistochemistry of PR, ER, PCNA were performed by blinded individuals, and tumor scoring was confirmed by a blinded pathologist.
Animals
For transcriptome analysis, whole mammary glands were harvested from wild type FVB mice at 8 weeks of age and transgenic FVB C3(1)-SV40Tag mice (The Jackson Laboratory, Bar Harbor, ME) at 8, 12, 16, and 20 weeks of age (n=5 per time point). All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Children’s Hospital Boston.
Gene Expression Analysis
Tumor gene expression profiling was carried out using Mouse Genome MG 430 2.0 Affymetrix GeneChip arrays. In brief, total RNA was extracted from tissue with the RNeasy kit including DNase digestion (Qiagen). Biotinlabeled cDNA was obtained from 1 μg total RNA with the GeneChip One-Cycle labeling kit (Affymetrix). According to the manufacturer’s instructions, 1.5 μg of cDNA were fragmented and hybridized to Affymetrix MG 430 2.0 GeneChip arrays. Arrays were processed by the Harvard Genotyping and Microarray Center (Boston, MA). DNA chips were washed, stained and scanned using an Affymetrix Fluidics device and a GCS3000 scanner, and the images obtained were analyzed using GCOS software. The experiment was performed in 5 replicates for wild type mice at 8 weeks of age and transgenic mice at 8, 12, 16, and 20 weeks of age. Data normalization was calculated with the robust multichip average (RMA) algorithm (33) implemented in BioConductor (www.bioconductor.org). MG 430 2.0 mRNA data from each individual sample have been deposited with the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE50813.
Compendium Generation for MNI
Approximately 11,000 mouse microarrays data were collected from the NIH Gene Expression Omnibus (GEO). All the arrays were processed based on the Affymetrix 430 2.0 chip, which contains more than 45,000 probe sets representing more than 34,000 validated mouse genes. Sequences used in the design of the array were selected from GenBank, dbEST, and RefSeq. Eleven pairs of oligonucleotide probes measure the level of transcription of each sequence represented on the GeneChip Mouse Genome 430 2.0 Array. Data normalization on the total compendium was calculated with the robust multichip average (RMA) algorithm implemented in BioConductor (www.bioconductor.org) K-mean clustering method was applied on the total mouse compendium data and approximately 3000 microarrays were selected as final training input data set for MNI.
MNI Network Inference
A detailed description of the MNI algorithm and underlying assumptions has been published previously (34). Here we provide a brief summary with additional details available in Supplementary Methods. The algorithm consists of two phases. In the first (the training phase), a model of regulatory influences in the cell is learned from an NxM data matrix, X, consisting of measurements of steady-state expression ratios of N genes in M experiments. To estimate the network model A, the MNI algorithm uses a recursive strategy. The algorithm begins by using the data matrix, X, and a naive model of the regulatory structure (i.e. no genes regulate any other) to estimate P, an NxM matrix of external influences on the genes. The estimate of P is then used, with A, to determine A by principal components regression analysis. Estimates of A and P are then used to estimate one another recursively until the estimates converge. In the second phase of the algorithm, the A matrix, representing a model of regulatory influences in the cell, is used to estimate the “key mediator genes” of the disease stage of interest. The disease stage then becomes an Nx1 vector, p, of gene-specific influences that result in the log-transformed expression ratios, x, measured for that disease state. The p vector is then calculated directly from the log-linear regulatory model as: P=Ax. The significance of each element of the p vector is then calculated as a z-score. Genes are ranked according to the z-score of their corresponding element in the p vector and top-ranked genes and pathways are selected as probable key mediators of the disease state.
Cell Culture
M6 and M6C cell lines were a kind gift from C. Jorczyk (Boise State University) and Eph4 cells were a gift from Kevin Freeman (Boston Children’s Hospital). M6, M6C, and Eph4 cells were maintained in high-glucose (4.5 g/mL) DMEM growth media supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin. Human breast and breast cancer cell lines (MCF10A, HCC1937 and MDA-MB-468) were purchased from ATCC. MCF10A cells were maintained in DMEM/F12 growth media containing 5% equine serum, 20ng/ml EFG, 0.5 μg/ml hydrocortisone, 0.1 μg/ml cholera toxin, 10 μg/ml insulin and 1% penicillin/streptomycin. HCC1937 were maintained in RPMI-1640 medium with 10%FBS and 1% penicillin/streptomycin whereas MDA-MB-468 cells were grown in Leibovitz’s L15 medium with 10% FBS and 1% penicillin/streptomycin. Whereas all other cells were maintained at 37°C and 5% CO2, MDA-MB-468 cells were grown at 37°C and atmospheric air (0% CO2).
In vitro silencing
siRNA targeting HoxA1 was purchased from Dharmacon and transfected into M6, M6C, MD-MBA-468 or HCC1937 cells using 25 nM using DharmaFECT1 reagent according to the manufacturer’s protocols. Target sequences were as follows: siRNA HoxA1_1: 5′ CAACAAGUACCUUACACGA 3′; siRNA HoxA1_ 2: 5′ CCAUAGGAUUACAACUUUCT 3′. A non-targeting sequence (ON-TARGETplus D-001810-01) was transfected as a negative control. To confirm the effectiveness of the silencing reagent in 3D cell culture, ON-TARGET GAPDH positive control siRNA was also transfected. Cells were maintained in 3D culture for 14 days and then collected for analysis by dissolution of the gel with collagenase. Immunoblotting confirmed effective silencing in cells using rabbit anti-HoxA1 (Abcam).
Spheroid Culture in 3D Gels
Mammary epithelial and tumor cells described above were trypsinized, centrifuged and resuspended in a 1:1 solution of type I collagen (3 mg/mL) and Matrigel and added to the top section of a 12-mm transwell plate with 0.4-μm polycarbonate membrane, such that the final cell number was 100,000 cells per well. Gels were allowed to harden at 37°C for 30 min before growth media was added to the top and bottom sections of the transwell plate. Gel cultures were maintained at 37°C, 5% CO2 for 8–14 days. Gels were then fixed in 10% formalin overnight and the transwell membrane containing the gel was removed from the culture chamber prior to carmine staining or sectioning.
In vivo RNAi Delivery
siRNA was formulated with lipid-like amino alcohols (lipidoids) as described previously (47). Particle sizes were measured by dynamic light scattering using a Malvern Zetasizer NanoZS (Malvern). siRNA (20 μg) was delivered intraductally to the thoracic gland of the mouse through the nipple using a 33 G needle while visualizing the nipple site under a stereoscope (48). Fluorescently tagged siRNA (Dharmafect siGLO) and Evans blue vital dye were used to determine the volume needed to fill the mammary ductal tree completely without perforating the wall of the duct). Following injection, glands were imaged weekly with a VEVO 2100 VisualSonic ultrasound machine in order to determine the onset, number, size, and location of tumors.
Protein isolation and immunoblotting
Tissues were washed once in 500 μl of ice-cold Bio-Rad BioPlex Cell Wash Buffer, cut into 3×3 mm pieces, and homogenized in 500 μl of ice-cold Bio-Rad BioPlex Cell Lysis Buffer using an IKA T10 Basic Ultra-Turrax homogenizer (dispersing element S10N-5G) for 30 s at minimum speed and 15 s at maximum speed. Homogenates were frozen at −80°C to complete the lysis, thawed, and centrifuged at 4°C, 8000 rcf, for 20 min, followed by isolation of the soluble layer. Protein concentrations were measured using Bio-Rad DC Protein Assay on NanoDrop 2000. The following primary antibodies were used for immunoblotting: 1:10,000 rabbit anti-PCNA (Novus Biologicals), 1:2000 rabbit anti-Src (Cell Signaling Technology), 1:500 goat anti-Grb2 (R&D Systems), 1:1000 rabbit anti-ERK1/2 (R&D Systems), 1:2000 rabbit anti-IER3 (Novus Biologicals), 1:5000 rabbit anti-KRAS (Novus Biologicals), 1:1000 rabbit-anti-HoxA1 (Abcam).
Immunohistochemistry
Parallel gel samples were processed for frozen and paraffin embedded sections. Following formalin fixation to obtain frozen sections, gels were washed with PBS and placed in OCT (TissueTek) with 30% sucrose at 4°C overnight. Gels were then flash frozen on liquid N2 and 8–10 μm sections were prepared using a cryostat (Leica). Staining of gels was carried out with the following antibodies and concentrations: 1:100 mouse anti-GM130 (BD Biosciences), 1:100 rabbit anti-laminin-5 (Abcam), 1:200 rabbit anti-ZO1 (Invitrogen), and TUNEL stain (Roche kit). Paraffin-embedded gel sections were stained with 1:200 rat anti-BrdU (Abcam). Paraffin tissue sections were prepared by the Harvard Medical School histology core facility and stained with 1:100 anti-ER (Abcam), 1:400 anti-PR (Dako), 1:100 anti-PCNA (Dako) and TUNEL (Roche). Stained sections were imaged at 40X using a Zeiss Imager and cells from at least 5 representative fields were quantified per animal.
Microscopic Analysis and Quantitation
Confocal images of 3D spheroids in collagen/Matrigel were obtained on a Leica white laser Leica SP5 X multispectral-multiphoton confocal system. Images were acquired throughout the intact gel with 2-μm resolution to verify formation of a hollow central lumen. Sections and 2D cultures were imaged on a Zeiss Axio Observer Z1 with a CoolSNAP camera. Brightfield imaging was carried out with a Zeiss Axio Observer with Nuance color camera and multispectral imaging system. Inform software package (CRI) was used to perform automated quantitation of proliferative and apoptotic indices. MetaMorph software was used to perform morphometric analysis of lumen formation. Spheroids from three independent experiments were quantified for each sample and 180–200 cells were scored per condition for each assay.
Statistical analysis
All in vitro assays were performed in triplicate. Data are reported as means +/− SD for n=3 assays. In vivo quantitation was performed on samples from n=5 animals to account for potential heterogeneity. Data are reported as means +/− SEM for assays in which n>5 samples were quantified. For non-continuous variables, p-values were determined by Pearson’s chi-squared test for large sample sizes and by Fisher’s exact t-test for smaller samples sizes.
Supplementary Material
Fig. S1. Conventional bioinformatics analysis of mammary gland gene expression changes in C3(1)-SV40Tag–driven tumorigenesis.
Fig. S2. Silencing of HoxA1 in human cell lines in vitro.
Fig. S3. In vivo delivery of siRNA to the mammary ductal tree.
Fig. S4. Monitoring systemic response to in vivo RNAi.
One-sentence summary.
Gene network reverse engineering revealed that HoxA1 is an important mediator of breast cancer progression, and silencing of HoxA1 through intraductal delivery of nanoparticle-formulated siRNA in a transgenic mouse breast cancer model prevented loss of estrogen and progesterone receptor expression, suppressed cell proliferation, and reduced mammary tumor incidence by 75%.
Acknowledgments
The authors thank C. Jorcyk (Boise State University) for M6 and M6C cells, K. Freeman for Eph4 cells, and H. Tobin and H. Lurvey for excellent technical assistance.
Funding: This study was funded by Department of Defense Breast Cancer Innovator Award W81XWH-08-1-0659 (to DEI) and the Wyss Institute for Biologically Inspired Engineering at Harvard University. JJC and HL were supported by the Howard Hughes Medical Institute, SysCODE (Systems-based Consortium for Organ Design & Engineering), and NIH grant RL1DE019021. The postdoctoral training of SK was supported by the Susan G Komen Foundation (KG101329). H.L is supported by Mayo Clinic Center for Individualized Medicine. Microarray analysis was performed at MRDDRC Molecular Genetics Core Facility at Children’s Hospital Boston (NIH-P30-HD18655) and has been deposited with the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE50813.
Footnotes
Supplementary materials
Materials and Methods
AUTHOR CONTRIBUTIONS: AB designed and performed experiments, and drafted the manuscript with input from the other authors; SK performed animal studies; HL performed the computational analyses; MSG prepared siRNA lipidoid nanoparticles; MK performed IHC staining; DEI and JJC conceived the study and led the manuscript preparation effort.
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
Data and materials availability: No data or materials require purchase; MTA required for transfer of materials.
REFERENCES AND NOTES
- 1.Simpson JF. Update on atypical epithelial hyperplasia and ductal carcinoma in situ. Pathology. 2009 Jan;41:36. doi: 10.1080/00313020802568097. [DOI] [PubMed] [Google Scholar]
- 2.Allegra CJ, et al. National Institutes of Health State-of-the-Science Conference statement: Diagnosis and Management of Ductal Carcinoma In Situ September 22–24, 2009. J Natl Cancer Inst. 2010 Feb 3;102:161. doi: 10.1093/jnci/djp485. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK, et al. Atypical ductal hyperplasia: interobserver and intraobserver variability. Mod Pathol. 2011 Jul;24:917. doi: 10.1038/modpathol.2011.66. [DOI] [PubMed] [Google Scholar]
- 4.Betsill WL, Jr, Rosen PP, Lieberman PH, Robbins GF. Intraductal carcinoma. Long-term follow-up after treatment by biopsy alone. Jama. 1978 May 5;239:1863. doi: 10.1001/jama.239.18.1863. [DOI] [PubMed] [Google Scholar]
- 5.Eusebi V, et al. Long-term follow-up of in situ carcinoma of the breast. Semin Diagn Pathol. 1994 Aug;11:223. [PubMed] [Google Scholar]
- 6.Sanders ME, Schuyler PA, Dupont WD, Page DL. The natural history of low-grade ductal carcinoma in situ of the breast in women treated by biopsy only revealed over 30 years of long-term follow-up. Cancer. 2005 Jun 15;103:2481. doi: 10.1002/cncr.21069. [DOI] [PubMed] [Google Scholar]
- 7.Fisher B, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998 Sep 16;90:1371. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 8.Dowsett M, et al. International Web-based consultation on priorities for translational breast cancer research. Breast Cancer Res. 2007;9:R81. doi: 10.1186/bcr1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tekedereli I, et al. Therapeutic Silencing of Bcl-2 by Systemically Administered siRNA Nanotherapeutics Inhibits Tumor Growth by Autophagy and Apoptosis and Enhances the Efficacy of Chemotherapy in Orthotopic Xenograft Models of ER (−) and ER (+) Breast Cancer. Molecular therapy Nucleic acids. 2013;2:e121. doi: 10.1038/mtna.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno S, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013 Jan;21:185. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aliabadi HM, et al. Effective response of doxorubicin-sensitive and -resistant breast cancer cells to combinational siRNA therapy. Journal of controlled release : official journal of the Controlled Release Society. 2013 Aug 30; doi: 10.1016/j.jconrel.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnology journal. 2011 Sep;6:1130. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001 May 24;411:494. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 14.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998 Feb 19;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 15.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009 Feb;8:129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg MS, et al. Nanoparticle-mediated delivery of siRNA targeting Parp1 extends survival of mice bearing tumors derived from Brca1-deficient ovarian cancer cells. Proc Natl Acad Sci U S A. 2011 Jan 11;108:745. doi: 10.1073/pnas.1016538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, et al. Elimination pathways of systemically delivered siRNA. Mol Ther. 2011 Feb;19:381. doi: 10.1038/mt.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green JE, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000 Feb 21;19:1020. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 19.Shibata MA, et al. The C3(1)/SV40 T antigen transgenic mouse model of prostate and mammary cancer. Toxicol Pathol. 1998 Jan-Feb;26:177. doi: 10.1177/019262339802600121. [DOI] [PubMed] [Google Scholar]
- 20.di Bernardo D, et al. Chemogenomic profiling on a genome-wide scale using reverse-engineered gene networks. Nat Biotechnol. 2005 Mar;23:377. doi: 10.1038/nbt1075. [DOI] [PubMed] [Google Scholar]
- 21.Xing H, Gardner TS. The mode-of-action by network identification (MNI) algorithm: a network biology approach for molecular target identification. Nature protocols. 2006;1:2551. doi: 10.1038/nprot.2006.300. [DOI] [PubMed] [Google Scholar]
- 22.Cantile M, et al. In vivo expression of the whole HOX gene network in human breast cancer. Eur J Cancer. 2003 Jan;39:257. doi: 10.1016/s0959-8049(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 23.Chariot A, Castronovo V. Detection of HOXA1 expression in human breast cancer. Biochem Biophys Res Commun. 1996 May 15;222:292. doi: 10.1006/bbrc.1996.0737. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Human growth hormone-regulated HOXA1 is a human mammary epithelial oncogene. J Biol Chem. 2003 Feb 28;278:7580. doi: 10.1074/jbc.M212050200. [DOI] [PubMed] [Google Scholar]
- 25.Holzer RG, et al. Development and characterization of a progressive series of mammary adenocarcinoma cell lines derived from the C3(1)/SV40 Large T-antigen transgenic mouse model. Breast Cancer Res Treat. 2003 Jan;77:65. doi: 10.1023/a:1021175931177. [DOI] [PubMed] [Google Scholar]
- 26.Ma XJ, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer cell. 2004 Jun;5:607. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000 Aug 17;406:747. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 28.Pollack JR, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002 Oct 1;99:12963. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radvanyi L, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2005 Aug 2;102:11005. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nature genetics. 2003 Jan;33:49. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 31.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001 Sep 11;98:10869. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang YH, et al. Claudin-3 gene silencing with siRNA suppresses ovarian tumor growth and metastasis. Proc Natl Acad Sci U S A. 2009 Mar 3;106:3426. doi: 10.1073/pnas.0813348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohankumar KM, et al. HOXA1-stimulated oncogenicity is mediated by selective upregulation of components of the p44/42 MAP kinase pathway in human mammary carcinoma cells. Oncogene. 2007 Jun 7;26:3998. doi: 10.1038/sj.onc.1210180. [DOI] [PubMed] [Google Scholar]
- 34.Barrow JR, Stadler HS, Capecchi MR. Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development. 2000 Mar;127:933. doi: 10.1242/dev.127.5.933. [DOI] [PubMed] [Google Scholar]
- 35.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010 May;10:361. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 36.Svingen T, Tonissen KF. Altered HOX gene expression in human skin and breast cancer cells. Cancer Biol Ther. 2003 Sep-Oct;2 doi: 10.4161/cbt.2.5.441. [DOI] [PubMed] [Google Scholar]
- 37.Abe M, et al. Disordered expression of HOX genes in human non-small cell lung cancer. Oncol Rep. 2006 Apr;15:797. [PubMed] [Google Scholar]
- 38.Chen JL, et al. Deregulation of a hox protein regulatory network spanning prostate cancer initiation and progression. Clin Cancer Res. 2012 Aug 15;18:4291. doi: 10.1158/1078-0432.CCR-12-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice KL, Licht JD. HOX deregulation in acute myeloid leukemia. J Clin Invest. 2007 Apr;117:865. doi: 10.1172/JCI31861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedmann Y, Daniel CA, Strickland P, Daniel CW. Hox genes in normal and neoplastic mouse mammary gland. Cancer Res. 1994 Nov 15;54:5981. [PubMed] [Google Scholar]
- 41.Lewis MT. Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2000;2:158. doi: 10.1186/bcr49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flanagan M, Love S, Hwang ES. Status of Intraductal Therapy for Ductal Carcinoma in Situ. Curr Breast Cancer Rep. 2010 Jun;2:75. doi: 10.1007/s12609-010-0015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love SM, et al. A feasibility study of the intraductal administration of chemotherapy. Cancer prevention research. 2013 Jan;6:51. doi: 10.1158/1940-6207.CAPR-12-0228. [DOI] [PubMed] [Google Scholar]
- 44.Stearns V, et al. Preclinical and clinical evaluation of intraductally administered agents in early breast cancer. Sci Transl Med. 2011 Oct 26;3:106ra108. doi: 10.1126/scitranslmed.3002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murata S, et al. Ductal access for prevention and therapy of mammary tumors. Cancer Res. 2006 Jan 15;66:638. doi: 10.1158/0008-5472.CAN-05-4329. [DOI] [PubMed] [Google Scholar]
- 46.Chun YS, et al. Intraductally administered pegylated liposomal doxorubicin reduces mammary stem cell function in the mammary gland but in the long term, induces malignant tumors. Breast Cancer Res Treat. 2012 Aug;135:201. doi: 10.1007/s10549-012-2138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008 May;26:561. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krause S, Brock A, Ingber D. Intraductal Injection for Localized Drug Delivery to the Mouse Mammary Gland. J Vis Exp. 2013:e50692. doi: 10.3791/50692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Conventional bioinformatics analysis of mammary gland gene expression changes in C3(1)-SV40Tag–driven tumorigenesis.
Fig. S2. Silencing of HoxA1 in human cell lines in vitro.
Fig. S3. In vivo delivery of siRNA to the mammary ductal tree.
Fig. S4. Monitoring systemic response to in vivo RNAi.


