Abstract
Eucaryotic ribosome biogenesis involves many cis-acting sequences and trans-acting factors, including snoRNAs. We have used directed mutagenesis of rDNA plasmids in yeast to identify critical sequence and structural elements within and flanking the ITS2-proximal stem. This base paired structure, present in the mature ribosome, is formed between the 5′-end of 25S and the 3′-end of 5.8S rRNAs. Previously we demonstrated that formation of this structure was critical for pre-rRNA processing in yeast. Here we show that there are no sequence-specific recognition elements within the ITS2-proximal stem, rather the structure of this stem is critical for processing. This stem cannot exceed a specific length, but there are different length restrictions for different regions within this tripartite stem. Neither the conserved unpaired nucleotides within the stem nor the sequence of the mature rRNA at the processing sites are required for processing. Collectively, these results suggest a measuring model whereby initial cleavage within ITS2 at the C2 processing site and termination of subsequent exonuclease activity yielding the mature termini are affected by the relative position of sequence and structural elements within the ITS2-proximal stem.
INTRODUCTION
Eucaryotic ribosome biogenesis is a complex process orchestrated by a wide variety of factors acting both in cis and in trans (reviewed in 1,2). Co- or post-transcriptionally, the pre-rRNA undergoes maturation involving processing, modification (including methylation and pseudouridylation), folding and assembly to produce a mature ribosome. Collectively, these various events require assembly, or transient association, with ribosomal and non-ribosomal proteins and ∼100 small nucleolar ribonucleoprotein particles (1,2). Three of the four ribosomal RNAs (rRNAs) are synthesized as a single RNA polymerase I transcript from which two external transcribed spacers (ETS) and two internal transcribed spacers (ITS) are removed via a series of endo- and exonucleolytic cleavages that have been only partially defined (Fig. 1A) (3–9). This research focuses on identification of RNA sequence and structural elements required for ITS2 removal and the formation of the mature ends of 5.8S and 25S rRNAs in yeast.
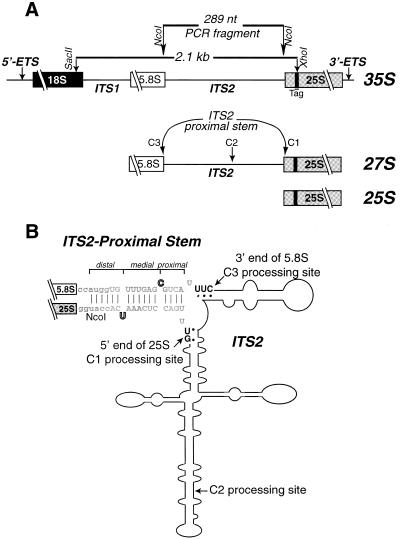
Figure 1.
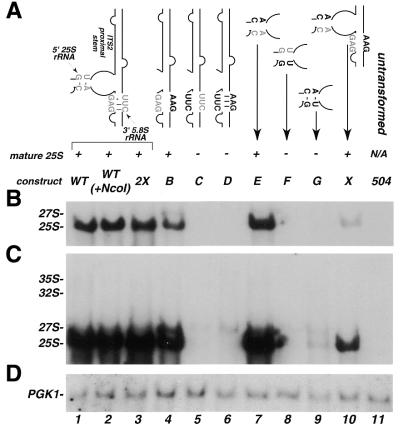
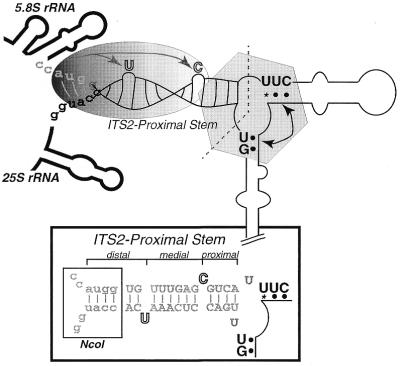
Mutagenesis strategy. (A) Schematic of the 9.1 kb rDNA with positions of relevant restriction sites noted. Mature rRNAs, the 5′ and 3′ ETS and ITS1 and ITS2 are as indicated. Mutations were introduced into the ITS2-proximal stem by PCR of a 289 nt fragment spanning the 3′-end of 5.8S to the 5′-end of 25S. NcoI sites were added via PCR for cloning purposes. The position of the unique 18 nt sequence within 25S rDNA (Tag), specific for the plasmid-borne transcripts, as well as processing sites C1, C2 and C3 are indicated. The composition of pre-rRNA processing intermediates are indicated schematically below. (B) Schematic of the proposed secondary structure of yeast ITS2 [line drawing, based on previous work (10)] showing the sequence of the ITS2-proximal stem formed between the 3′-end of 5.8S and the 5′-end of 25S rRNAs. Three regions were examined by mutagenesis: (i) sequence and length of the ITS2-proximal stem, shown in gray text; (ii) identity and position of the unpaired nucleotides within the ITS2-proximal stem, shown in outlined letters; and (iii) sequence and structure of the mature termini of rRNA, shown in black text. The sequence of the WT(+NcoI) ITS2-proximal stem is indicated here. The NcoI site is shown in lower case letters. The 3′-end of 5.8S (C3 processing site), the 5′-end of 25S (C1 processing site) and C2, the site of initial endonucleolytic cleavage within ITS2, are indicated.
A secondary structure was proposed for yeast ITS2 based upon chemical and enzymatic structure mapping (Fig. 1B) (10). Studies of closely related yeast species indicated their ITS2 sequences could be folded into a similar secondary structure (4) and several regions of high sequence conservation among yeast were identified (4,5). Functional analysis of the processing capacity of plasmids encoding ITS2 deletions or cross-species hybrid ITS2 sequences indicated that there are many sequence and/or structural elements within ITS2 that are required for processing (4,5). Thus, ITS2 plays a direct role in both processing of rRNA and in formation of functional 60S subunits.
The size, sequence and complexity of ITS2 segments vary considerably within eucaryotes. Yet despite the great diversity in ITS2 sequence there is a remarkable conservation of structure at the mature rRNA termini flanking the ITS2 sequence. In the assembled ribosome of all eucaryotes it is predicted that the mature 3′-end of 5.8S is base paired to the 5′-end of 25S/28S rRNA (for examples see 3,11–16; reviewed in 17 and at http://www.rna.icmb.utexas.edu). Together the termini of 5.8S and 25/28S in eucaryotes form an imperfect base paired structure called the ITS2-proximal stem (16). Although both Eubacteria and Archaea lack an ITS2 sequence, the 5′-ends of the respective 23S rRNAs have been proposed to share a common evolutionary origin with eucaryotic 5.8S (18). There exists a stem–loop structure at a position equivalent to the ITS2-proximal stem in Eubacteria and Archaea (review of structures in 17 and at http://www.rna.icmb.utexas.edu). This equivalent stem–loop (stem #10 in Escherichia coli nomenclature; 19) was demonstrated to be present in the mature ribosome of Archaea (20).
In the pre-rRNA of eucaryotes, the ITS2-proximal stem consists of the mature termini adjacent to ITS2, which is removed in the maturation process. The ITS2-proximal stem in eucaryotes can be viewed as tripartite, with two unpaired nucleotides dividing it into proximal (i.e. adjacent to the processing sites), medial and distal segments (Fig. 1B). In vivo competition studies using 2′-O-methyl RNA oligonucleotides in Xenopus oocytes provided indirect evidence that formation of the ITS2-proximal stem was a prerequisite for later stages in processing of the large subunit rRNAs, including removal of ITS2 (21). Direct evidence that the formation of the ITS2-proximal stem was critical for processing was obtained using rDNA plasmids in yeast. Mutations that inhibited formation of this stem resulted in an absence of mature 25S rRNA and destabilization of the 25S precursors (16). While compensatory mutations in this structure restored accurate processing (16), it was not clear what elements (sequence or structural) govern the ability to form mature rRNA (Fig. 1A and B).
In this study, we have undertaken a systematic analysis of this region of the rRNA to identify sequences and/or structural elements within (or flanking) the ITS2-proximal stem that are necessary for processing. We have identified a number of cis-elements within the mature rRNA sequences that define structural constraints required for accurate and efficient processing of the large subunit rRNAs. Our analyses have identified several mutations that not only affect pre-rRNA processing, but also lend insight into possible recognition mechanisms used by the pre-rRNA processing machinery. This is the first description of a systematic functional analysis to identify sequences within the mature rRNAs required for accurate processing of pre-rRNA.
MATERIALS AND METHODS
Plasmid construction
The tagged WT, 5.8S, 25S and 2X constructs have been described previously (16). The WT(+NcoI) construct was created through a multistep cloning process (Fig. 1) in which a 289 nt fragment corresponding to a region near the 3′-end of 5.8S to a point just within the 5′-end of 25S rRNA was PCR amplified from the WT plasmid (16) using the oligonucleotide pair 1s and 2a (Table 1). The PCR fragment was gel purified (Qiagen) and subcloned using the PCR-Script Amp Cloning Kit (Stratagene) for sequence verification. The NcoI fragment from this construct was inserted into the NcoI site of ystSXpBS, which consisted of pBluescript (Stratagene) containing the 2.1 kb SacII/XhoI fragment from 2X (Fig. 1A). The final construct was generated by replacement of the WT SacII/XhoI fragment in pNOY102(tag) (16) with the mutations encoded in a SacII/XhoI cassette. All additional mutations were cloned essentially as described for WT(+NcoI) with PCR fragments amplified using the primer pairs shown in Table 1. The WT and WT(+NcoI) constructs served as templates for PCR-mediated mutagenesis. AllAU was amplified using primers 3s and 4a; allGC from primers 5s and 6a; +AU/AU from primers 7s and 8a; +GC/GC from primers 9s and 10a; +AU/GC from primers 7s and 10a; +GC/AU from primers 8a and 9s; +AU/AU:SB from primers 11s and 12a; +2M from primers 13s and 14a; +2P from primers 15s and 16a; +2D from primers 17s and 18a; ΔB from primers 19s and 20a; CB from primers 21s and 22a; RB from primers 23s and 24a; RB/– from primers 23s and 2a; –/RB from primers 1s and 24a; B from primers 25s and 2a; C from primers 26s and 2a; D from primers 27s and 2a; E from primers 1s and 28a; F from primers 29s and 2a; G from primers 28a and 29s; and X from primers 25s and 28a.
Table 1. Oligonucleotides used in this study.
| Name |
Sequence |
| 1s |
5′-gggggccatggTGTTTGAGCGTCATTTCCTTCTC-3′ |
| 2a |
5′-gagagatccatggTGATTTGAGGTCAAACTTTAAG-3′ |
| 3s |
5′-ggggccatggTGTTTATACATAATTTCCTTCTCAAACATTCTG-3′ |
| 4a |
5′-cccccatggTAATTTATAATAAAACTTTAAGAACATTGTTCG-3′ |
| 5s |
5′-ggggccatggCGCCCGCGCGCATTTCCTTCTCAAACATTCTG-3′ |
| 6a |
5′-cccccatggCGACCCGCGGCCAAACTTTAAGAACATTGTTCG-3′ |
| 7s |
5′-ggggccatggTGTTTGAGCGTCATATTATTTCCTTCTCAAACATTTG-3′ |
| 8a |
5′-cccccatggTAATTTGAGGTCATATTAAACTTTAAGAACATTGTTCG-3′ |
| 9s |
5′-ggggccatggTGTTTGAGCGTCGGGCCATTTCCTTCTCAAACATTCTG-3′ |
| 10a |
5′-cccccatggTAATTTGAGGTCGGGCCAAACTTTAAGAACATTGTTCG-3′ |
| 11s |
5′-aaaaccatggTGTTTGAG*GTCATCATTATTTCCTTCTCAAAC-3′ |
| 12a |
5′-ttttccatggTG*TTTGAAGGTCATATTAAACTTTAAGAAC-3′ |
| 13s |
5′-aaaaccatggTGTTTGAGAGCGTCATTTCC-3′ |
| 14a |
5′-aaaaccatggTGATTTGAGAGGTCAAACTT-3′ |
| 15s |
5′-aaaaccatggTGTTTGAGCGTGACATTTCC-3′ |
| 16a |
5′-aaaaccatggTGATTTGAGGTGACAAACTT-3′ |
| 17s |
5′-aaaaccatggGATGTTTGAGCGTCATTTCC-3′ |
| 18a |
5′-aaaaccatggGATGATTTGAGGTCAAACTT-3′ |
| 19s |
5′-aaaaccatggTGTTTGAGGGTCATTTCCTTC-3′ |
| 20a |
5′-ttttccatggTGTTTTGAGGTCAAAC-3′ |
| 21s |
5′-ggggccatggTGATTTGAGCGTCATTTCCTTCTCAAACATTCTG-3′ |
| 22a |
5′-cccccatggTAATTTGAGCGTGCAAACGTTTAAGAACATTGTTCG-3′ |
| 23s |
5′-aaaaccatggTGTTTGAG*GTCATTTCCTTC-3′ |
| 24a |
5′-ttttccatggTG*TTTGAGGTCAAAC-3′ |
| 25s |
5′-aaaaccatggTGTTTGAGCGTCATAAGCTTCTCAAACATTC-3′ |
| 26s |
5′-aaaaccatggTGTTTGAGCGTCATTTCCTTCTCAAACATTCTGTTTGGTAGTCTTTGATACTCTTTGG-3′ |
| 27s |
5′-aaaaccatggTGTTTGAGCGTCATAAGCTTCTCAAACATTCTGTTTGTAGTCTTTGATACTCTTTGG-3′ |
| 28a |
5′-cccccatggTGATTTGAGGTCAATGTTTAAGAACATTGTTCG-3′ |
| 29s |
5′-aaaaccatggTGTTTGAGCGTCATTTCCTTCTCAAACATTCTGTTTGGTAGTGAGTGATTGTCTTTGGAGTTAAC-3′ |
| 25S400 |
5′-CTCGCATAGACGTTAGACTC-3′ |
| TagA |
5′-GTACTGAAGCTCTCGAGT-3′ |
| TagB |
5′-ACTCGAGAGCTTCAGTAC-3′ |
| PGK1 | 5′-CGAAGGCATCGTTGTAGTAAACATCAGCC-3′ |
Lower case letters include the NcoI site used for cloning purposes. Underlined regions denote areas of mutagenesis and asterisks represent positions of nucleotides that have been removed.
Yeast transformation and RNA preparation
Plasmids were transformed into Saccharomyces cerevisiae strain NOY504 MATα rrn4::LEU2 ade2-101 trp1-1 leu2-3, 112 his3-11 can1-100 (a generous gift of Dr M.Nomura, University of California, Irvine) (22) by lithium acetate (23), plated on synthetic defined (SD) media plus 2% glucose supplemented with essential nutrients and grown at 25°C. Transformants were maintained on SD + 2% galactose with essential nutrients. The presence of the plasmid was confirmed in transformants by PCR analysis using total DNA isolated (23) from each transformant and primers specific for the tag sequence within 25S rDNA (TagA) and a sequence located ∼400 bp downstream (25S400) in mature 25S rRNA.
For RNA isolation, cells were grown at 25°C in liquid media (SD + 2% galactose + essential nutrients) to OD600 of ∼0.1 then shifted to 37°C for 6 h. Cells were harvested and total RNA was isolated as previously described (24).
RNA analysis
For northern blot analysis, total RNA (∼8 µg, normalized to PGK1, see below) was resolved on a 1.2% agarose–formaldehyde gel and transferred to nylon membrane (ZetaProbeGT, Bio-Rad) by capillary transfer in 20× SSC. After transfer, RNA was crosslinked to the membrane (UV Stratalinker 1800, Stratagene) which was then hybridized with the appropriate probe. Radioactive probes were generated by 5′-end-labeling deoxyoligonucleotides in the presence of [γ-32P]ATP (NEN DuPont) with T4 polynucleotide kinase (USB/Amersham). Hybridizations were in 6× SSC and 10× Denhardt’s for 16 h at 42°C. Membranes were washed twice for 5 min in 6× SSC and once for 5 min in 1× SSC then exposed to BioMax MR film (Eastman Kodak). To analyze pre-rRNA processing, probes were complementary to the unique tag sequence in the plasmid-encoded 25S rRNA (TagB). As a control for equal loading, a probe complementary to S.cerevisiae 3-phosphoglycerate kinase (PGK1; SGDID #S0000605) was hybridized to the same membrane. Hybridization signal intensities were quantitated by phosphoimager analysis (Image Gauge v. 3.3, Fuji) and RNA levels were normalized to PGK1 of WT(+NcoI).
To analyze the accuracy of processing, the 5′-end of 25S rRNA transcript derived from the plasmid-borne template was mapped as described previously (16). Briefly, total RNA (2 µg) was used as a substrate for primer extension using 20 ng of 5′-end-labeled oligonucleotide (TagB) and AMV reverse transcriptase (NEB) (25). After extraction with phenol and precipitation with ethanol, samples were resolved on a 10% denaturing polyacrylamide gel and visualized by autoradiography. The sequences at the termini were determined by comparison with a DNA sequencing ladder generated by dideoxy chain termination (T7 Sequenase Quick-Denature Plasmid Sequencing Kit, USB) using WT(+NcoI) plasmid DNA and the TagB primer in the presence of [35S]dATP (Amersham).
RESULTS
To conduct a systematic analysis of the role of the ITS2-proximal stem in pre-rRNA processing, we have taken advantage of a rDNA expression system developed in yeast (22). The yeast plasmid, pNOY102, encodes a complete repeat of the rDNA gene under the control of a galactose-inducible polymerase II promoter. A unique 18 nt tag sequence (26) was inserted into the 25S coding region to facilitate detection of the plasmid-borne transcripts over the background of the endogenous rDNA genes (16). Each mutagenized construct was transformed into a yeast strain (NOY504) conditional for RNA polymerase I activity (22). This allowed analysis of the plasmid-derived pre-rRNA under conditions of reduced competition with endogenous rRNA. The efficiency and accuracy of processing of plasmid-borne pre-rRNA transcripts was determined by northern blot and primer extension analyses using a probe complementary to the unique 18 nt tag sequence within mature 25S rRNA (Fig. 1A).
rDNA plasmids were generated that encoded mutations to identify structural and sequence requirements within three specific areas (Fig. 1B). These three regions consisted of: (i) sequences within the ITS2-proximal stem formed by the interaction between the 3′-end of 5.8S and the 5′-end of 25S rRNAs; (ii) the sequence and position of the conserved unpaired nucleotides located within the ITS2-proximal stem; and (iii) sequence and structure of the mature termini of 5.8S (3′-end) and 25S (5′-end) rRNAs, defined as the C3 and C1 processing sites, respectively.
Formation of a stable ITS2-proximal stem is essential for pre-rRNA processing
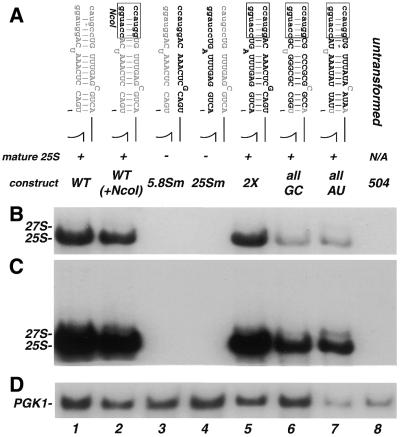
Our previous results demonstrated that formation of a base paired ITS2-proximal stem structure is required for processing (16). Mutations that alter the sequence in either 5.8S or 25S to prevent formation of a base paired structure inhibit processing (Fig. 2, lanes 3 and 4). A compensatory double mutation that restores the base paired structure allows mature 25S rRNA to accumulate (Fig. 2, lane 5). Global alteration of the sequence (but preservation of the structure) of this region reduces the efficiency, but not the accuracy of processing (2X mutation; 16) (Fig. 2A, B and the darker exposure in C, compare lane 1 with lane 5).
Figure 2.
Analysis of plasmid-encoded rRNA containing mutations within the ITS2-proximal stem. (A) Schematic representation of the mutations and the name by which they are referred to in the text. Wild-type sequences are indicated by gray lettering, mutated sequences are black and the NcoI site is denoted by lower case black letters. Maturation of rRNA scored as (+) normal processing with accumulation of mature 25S; and (–) low or no detectable levels of mature 25S. Below, northern blot analysis of mutations illustrated in (A). Total RNA (8 µg) from each transformant was resolved on a denaturing 1.2% agarose–formaldehyde gel and transferred to nylon membrane. The membrane was divided in half and probed with 5′-end-labeled oligonucleotides complementary to either the tag sequence within 25S (B and C) or PGK1 (D) as a control for loading. (C) Longer exposure of (B) presented to visualize the lower amounts of precursors and mature 25S present in some of the mutants. The positions of mature 25S rRNA precursors are noted on the left. 504 has no plasmid and indicates background hybridization for the tag probe.
To further examine the effects upon processing that could be attributed to sequence in this region, we generated three constructs. In each, the sequence of the stem was altered while maintaining the ability to form a base paired structure. The WT(+NcoI) construct was similar to WT except that 5 bp at the distal end (relative to ITS2) of the ITS2-proximal stem were changed to introduce the NcoI sites necessary for cloning [Fig. 2A, WT(+NcoI)]. The WT(+NcoI) mutation served as the parent construct for all other mutations described here. Two other constructs more completely altered the sequence identity within the stem by either maximizing (allGC) or minimizing (allAU) the G/C content (Fig. 2A).
Figure 2B and C shows that all of the mutations that allowed formation of the stem structure allowed accumulation of mature 25S (Fig. 2, lanes 5–7). However, the hybridization signal intensities of mature 25S rRNA from both the allGC (lane 6) and allAU (lane 7) transformants were reduced compared to WT(+NcoI) (lane 2) and 2X (lane 5) by an amount that could not be accounted for by loading (Fig. 2D, compare lanes 5, 6 and 7). Furthermore, as was first observed with the 2X mutant, a slightly elevated level of 27S precursor was detected in the allGC lane and was considerably more prominent in the allAU lane (Fig. 2, compare lanes 5, 6 and 7). Here, we scored any significant accumulation of 25S rRNA with correct termini as ‘accurate processing’. When a construct accumulated a decreased amount of mature 25S [compared to WT(+NcoI), the parental construct] that could not be accounted for by loading (see Materials and Methods), we referred to this construct as displaying a decreased efficiency of processing. Thus, these results demonstrate that while the sequence composition of the ITS2-proximal stem plays some role in the efficiency of processing, no sequence-specific recognition elements in this stem are required for the accuracy of processing of mature 25S.
The structure of the ITS2-proximal stem affects processing
The ITS2-proximal stem is an imperfect tripartite base paired structure. The length of the stem and the placement of unpaired nucleotides that define the tripartite nature are well conserved among eucaryotes (16,17; http://www.rna.icmb.utexas.edu). This conservation of structure raised the possibility that the position of the unpaired (bulged) nucleotides and the length of the proximal, medial and distal stem segments (Fig. 1B) might be critical for recognition by the processing apparatus. A series of mutations were constructed to test these possibilities.
Lengthening the stem by 5 bp. Constructs were generated that would test whether the absolute length of the ITS2-proximal stem affects pre-rRNA processing. Four constructs were generated that inserted 5 nt pairs (+5) at the proximal end (nearest ITS2) of this stem, closest to the mature termini (Fig. 3A). In two constructs (+GC/GC and +AU/AU) the potential for the added sequence to base pair was maintained and would lengthen the ITS2-proximal stem by 5 bp. Two other constructs (+AU/GC and +GC/AU) contained the same number of added bases, but no stable base pairing potential was predicted for the additional sequence (Fig. 3A).
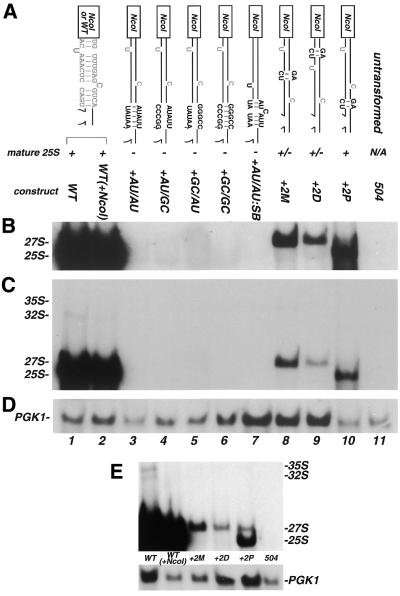
Figure 3.
Analysis of plasmid-encoded rRNA containing mutations that shifted the relative positions of the bulged nucleotides. (A) Schematic representation of the mutations and the name by which they are referred to in the text. Schematics coded by color and text as indicated in Figure 2. Maturation of rRNA scored as (+) normal processing with accumulation of mature 25S; (+/–) no mature 25S but significant accumulation of stable precursors; (–) low or no detectable levels of mature 25S. Below, northern blot analysis of mutations illustrated in (A). Gel was manipulated as indicated in Figure 2. (B) Longer exposure of (C) presented to visualize the lower amounts of precursors and mature 25S present in some of the mutants; (D) probe for PGK1. (E) Separate experiment where the PGK1 levels for +2P are much more equivalent to that in WT. Processing is less efficient, but mature 25S clearly accumulates.
Yeast were transformed with these four constructs and processing was examined by northern blot analysis of total RNA using a probe complementary to the tag sequence (Fig. 3B and the shorter exposure in C). All of the +5 mutations resulted in a failure to accumulate mature 25S rRNA. Presumably the 25S precursors were unstable; visible amounts of precursors were not detected, even on much longer exposures (Fig. 3B, lanes 3–6). However, we cannot rule out instability of the mature product. Regardless, increasing the length of the stem by 5 base insertions prevented the accumulation of mature 25S and all its precursors.
Shifting the unpaired positions by 5 bp. Addition of 5 bp in the above constructs not only lengthened the stem, but also displaced the two conserved unpaired nucleotides further away from the mature rRNA termini [Fig. 3A, compare +AU/AU with WT(+NcoI)]. An additional mutation was generated to examine whether the lack of mature 25S rRNA observed for the +5 mutations could be attributed to repositioning the unpaired nucleotides within the stem.
In the ‘shift bulge’ mutation (Fig. 3A, +AU/AU:SB), both unpaired nucleotides were restored to their original positions relative to the mature termini in the context of a +5 mutation. The overall structure of the proximal end (closest to the mature termini) of the stem in this construct most closely resembles WT(+NcoI), but the ‘shift bulge’ construct now contains a longer, perfect base paired region at its distal end [Fig. 3A, compare WT(+NcoI) with +AU/AU:SB]. Northern blot analysis (Fig. 3B, lane 7) indicated that restoring these unpaired nucleotides to their original positions relative to the mature termini neither rescued processing nor stabilized the precursors. Thus, the negative effects of the +5 mutations on processing are not due to simply repositioning the unpaired nucleotides along the stem.
Shifting the relative positions of the bulges by 2 bp. To further examine how the length of the ITS2-proximal stem affects processing and/or stability, a series of 2 bp insertions were made within the three different segments of the tripartite stem. Insertion of 2 adjacent bp between the bulged positions (Fig. 3A, +2M) separated these unpaired nucleotides by 2 additional bp, and also moved the unpaired nucleotide in 25S further from ITS2. Insertion of 2 adjacent bp distal (+2D) to the unpaired nucleotides (relative to the mature rRNA termini) maintained the original position and spacing of both bulged nucleotides relative to the termini, but moved these structures further from the rest of the mature rRNA. Finally, addition of 2 adjacent bp at the proximal end of the stem (+2P) shifted both unpaired nucleotides 2 bp further away from ITS2, but their position relative to the rest of the mature rRNA remained unchanged.
Data from northern blot analyses are shown in Figure 3B. Processing for the +2 mutations differed from that of the +5 mutations described above. Mature 25S rRNA was seen in the +2P construct with little accumulation of precursor (Fig. 3B, lane 10). Thus, insertion of 2 bp at a position proximal to the C1 and C3 processing sites resulted in stable accumulation of mature 25S. Both the +2M and +2D constructs blocked accumulation of mature 25S and showed significant accumulation of 27S precursor. While these 27S precursors were not processed to mature 25S, they were not efficiently degraded; stable accumulation of 27S and some 35S precursor was detected (Fig. 3B, lanes 8 and 9). Thus, insertion of 2 bp within the medial or distal part of this stem resulted in a block of processing distinct from the block observed for the +5 mutations.
Altering the sequence identity of the unpaired nucleotides. The three +2 constructs differed from each other in the position of the bulged nucleotides relative to the mature rRNA termini. The observed differences in processing phenotypes among the +2 mutants suggested that there might be some interplay between the length of the stem and the positions of the bulged nucleotides in determining whether accurate processing occurs. To test this possibility, a series of mutations were constructed that altered the identity of the unpaired nucleotides or their structure (paired or unpaired) within the stem.
The identity of both unpaired nucleotides was altered in the 2X construct examined previously (16). RNA containing this mutation was processed (Fig. 4, lane 3) (16). However, the identity of all bases throughout the stem was altered in the 2X construct. Construct ΔB changed the sequence identity of both bulged positions in the context of a wild-type sequence ITS2-proximal stem (ΔB; Fig. 4, lane 4). Similar to that observed in the 2X mutant, these changes had no significant effect upon accumulation of mature 25S rRNA. Thus, there appears to be no specific sequence requirement for the identity of the unpaired nucleotides in processing.
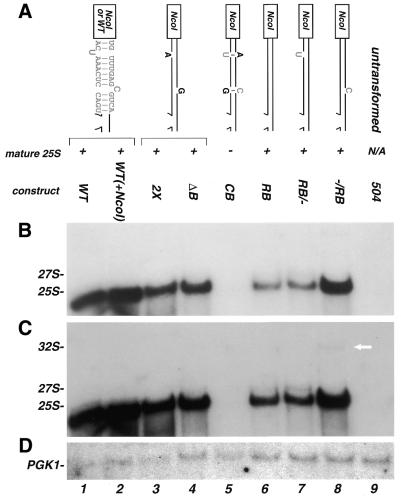
Figure 4.
Analysis of plasmid-encoded rRNA containing mutations which removed the bulged nucleotides. (A) Schematic representation of the mutations and the name by which they are referred to in the text. Schematics coded by color and text as indicated in Figure 2. Maturation of rRNA scored as (+) normal processing with accumulation of mature 25S; and (–) low or no detectable levels of mature 25S. Below, northern blot analysis of mutations illustrated in (A). Gel was manipulated as indicated in Figure 2. (C) Longer exposure of (B) presented to visualize the lower amounts of precursors present in some of the mutants; (D) probe for PGK1. The arrow points to the trace amount of 32S that is reproducibly seen in the –/RB mutant, as described in the text.
Removing the unpaired nucleotides. To test whether the structure generated by the unpaired nucleotides was being recognized by the processing machinery, a set of four mutations was designed as follows. In the first, designated CB (Fig. 4A), complementary bases were inserted into the strand opposite each of the two unpaired positions. Note that this both ‘closed the bulge’ by creating a potential base pair at each site and increased the length of the stem. The second and third constructs removed a single unpaired base, with the proximal (relative to the processing sites) unpaired base removed in construct RB/– and the distal base removed in construct –/RB. Both unpaired bases were removed in the fourth construct, designated RB.
Processing was assayed for each of these constructs by northern hybridization and the results are shown in Figure 4. Removal of both unpaired bases in the RB construct allowed accumulation of 25S rRNA. In contrast, insertion of 2 nt in the CB construct resulted in the absence of detectable 25S mature rRNA and its precursors. This block in processing was similar to that observed for the +5 bp constructs described above. Comparison of the RB and CB constructs suggests that while the unpaired nucleotides are not required for processing, in their absence, changes that increase the length of the double-stranded region can significantly impact processing.
Removal of either unpaired nucleotide in the RB/– and –/RB constructs allowed accumulation of mature 25S rRNA but also resulted in an increased level of 27S or 32S precursors, consistent with a decrease in processing efficiency. These mutations demonstrate that there are underlying structural parameters in this region that affect processing efficiency. When present, these unpaired bases may bulge out of or stack into this helix; removing them may provide excess flexibility or rigidity in this stem, which appears to be detrimental only when there is a concomitant change in overall stem length.
Requirement for sequence and structural elements at the processing sites of mature rRNA
We next examined whether the processing machinery recognized sequences and/or structures at the mature termini of 5.8S and 25S rRNAs (corresponding to the C3 and C1 processing sites, respectively; Fig. 1B). In designing mutations to test these requirements, we used the proposed secondary structure for yeast ITS2 determined by chemical and enzymatic structure probing (Fig. 1B) (10). In this structure, the mature rRNA termini are paired with separate, short segments within ITS2. Thus, to distinguish between sequence and secondary structure requirements it was necessary to create single and compensatory mutations within mature rRNA termini and their respective pairing segments within ITS2.
Three mutations were designed to test requirements at the C3 processing site, corresponding to the mature 3′-end of 5.8S rRNA. Construct B (Fig. 5A) changed three bases at the 3′-end of 5.8S rRNA (UUC in WT) to their Watson–Crick counterparts (i.e. AAG in construct B). Note that this changed both the sequence and predicted structure at the C3 site. Construct C was designed to alter structure by changing only the ITS2 pairing segment, leaving the mature 5.8S sequence intact. Construct D contained compensatory changes at both sites, thereby retaining the predicted secondary structure while altering the 5.8S sequence. Processing for each of these constructs was examined by northern hybridization and the results are shown in Figure 5B and C (lanes 4–6). Surprisingly, alteration of both sequence and predicted structure of the 5.8S segment (construct B) did not inhibit processing. Thus, processing does not seem to include any specific sequence requirements within this region of the mature rRNA. In contrast, changing the sequence of the ITS2 pairing segment, in either the complementary unpaired (construct C) or compensatory paired (construct D) configuration significantly reduced processing. This result is consistent with a sequence-specific requirement within the ITS2 segment predicted to pair with the 5.8S 3′-end.
Figure 5.
Analysis of plasmid-encoded rRNA containing mutations that altered the mature termini. (A) Schematic representation of the mutations and the name by which they are referred to in the text. Schematics coded by color and text as indicated in Figure 2. Maturation of rRNA scored as (+) normal processing with accumulation of mature 25S; and (–) low or no detectable levels of mature 25S. Below, northern blot analysis of mutations illustrated in (A). Gel was manipulated as indicated in Figure 2. (C) Longer exposure of (B) presented to visualize the lower amounts of precursors and mature 25S present in some of the mutants; (D) probe for PGK1.
This same strategy was applied to an examination of the requirements for sequence and/or structure at the C1 processing site, corresponding to the mature end of 25S rRNA. Similar results were obtained (Fig. 5, lanes 7–9). Alteration of both sequence and predicted structure of the first two bases in 25S (construct E) had little or no effect on processing. Thus, formation of the mature termini does not seem to include any specific sequence requirements within this segment of mature 25S rRNA. In contrast, alterations in the predicted ITS2 partner, whether in the unpaired (construct F) or paired (construct G) configuration, significantly reduced processing (the observed reduction was greater for F than for G). This result is consistent with a sequence-specific requirement for the ITS2 segment predicted to pair with the 25S 5′-end.
These results are extended by a double mutation (construct X; Fig. 5, lane 10) that altered sequence and predicted structure at both the 5.8S and 25S mature ends, while retaining the sequence of the ITS2 segments. This double mutation had no significant effect on the accumulation of mature 25S. Collectively, these results suggest that processing involves no specific requirements for mature rRNA sequence or structure at the C3 and C1 processing sites. Instead, these processing events may involve sequence-specific recognition of distinct regions within ITS2.
Removal of ITS2 is accurate when sequences and/or structures near the mature termini are mutated
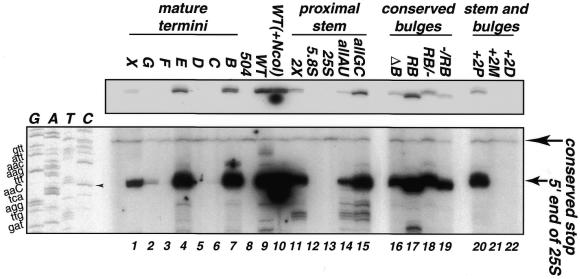
The northern hybridization analyses described in each of the preceding sections provided an effective means for measuring the relative levels of precursors and mature rRNA. However, this assay might not detect small but significant changes in the accuracy of processing. For this purpose, we used primer extension analyses taking advantage of the unique tag sequence present near the 5′-end of the plasmid-encoded 25S sequence (Fig. 1). Use of the TagB oligonucleotide for primer extensions allowed mapping of the 5′-end of plasmid-encoded 25S rRNA processing products without interference from the endogenous rRNA present in the yeast transformants. The accuracy of processing was determined by comparison to a DNA sequencing ladder (refer to Materials and Methods and Fig. 6, lanes G, A, T and C). The data indicated that all transformants that gave a hybridization signal for mature 25S rRNA by northern blot analysis had mature termini that mapped to the same position as endogenous rRNA (Fig. 6, bottom panel). Insertion or removal of 1 or 2 nt resulted in a shift of the primer extension stop by the corresponding number of nucleotides (Fig. 6, lanes 17, 19 and 20). Lanes devoid of a primer extension stop corresponding to the 5′-end of 25S reflect processing deficiencies; these constructs did not generate mature 25S rRNA (Fig. 6, lanes 3, 5, 6, 12, 13, 21 and 22). The doublets (Fig. 6, top panel) observed for many of the mutants presumably correspond to the reported heterogeneity at the terminal nucleotides seen in S.cerevisiae (1). Thus, we have no evidence that any of the mutations analyzed in this work affect the accuracy of processing at the C1 site. Note that this includes mutations that alter the mature 25S sequence at the C1 site (Fig. 6, construct E, lane 7). Instead, processing site selection (i.e. accuracy) must be determined by features of the substrate other than those analyzed in the present experiments (i.e. tertiary structural conformations).
Figure 6.
Mapping the 5′-end of the mutated 25S rRNA. Primer extension analysis was performed on 2 µg of total RNA from each transformant using a 5′-end-labeled oligonucleotide complementary to the unique sequence (tag) within 25S rRNA. The positions of the 5′-end of 25S and an unrelated conserved stop (used here as a loading control and is not plasmid-dependent; see 504, lane 8) are noted on the right. The corresponding DNA sequencing ladder is shown on the left. The upper case bold C (and arrowhead) indicate the expected 5′ terminal nucleotide of 25S rRNA. The upper panel is a lighter exposure of the lower panel.
DISCUSSION
Ribosome biogenesis is a complex process involving trans-acting factors and a very large number of cis-acting sequences within pre-rRNA. The cis-acting sequences are the basis for the intramolecular and intermolecular interactions that fold the mature ribosomal subunit RNAs and form the binding sites for ribosomal proteins. Given that the ribosome is essentially an RNA-based machine with proteins decorating its surface (20,27), it is easy to understand why correct folding of the mature ribosomal RNAs would be critical for ribosome function. Here we have taken a closer look at the ITS2-proximal stem, a structure in the mature ribosome whose formation was previously demonstrated to be essential for maturation of both 5.8S and 25S rRNA in yeast (16). Our results would indicate that formation of correctly folded structures is critical in the early steps of ribosome biogenesis, during folding and assembly, which in turn affects subunit stability.
Processing phenotypes
Two distinct pre-rRNA processing phenotypes were observed for mutations that blocked processing. Many of the mutations resulted in the absence of stable processed products and precursors. This might result from complete degradation of the precursor soon after transcription due to incomplete or inaccurate assembly with the processing/stabilizing machinery. Alternatively the precursor might be stable and rapidly processed (i.e. undergo accurate cleavage at site C2 within ITS2) with subsequent complete degradation of the mature product by exonucleases (6–9) because the processing termination or stabilization apparatus was not correctly assembled on the ITS2-proximal stem. These in vivo assays do not allow us to distinguish between these scenarios. In other constructs, larger amounts of precursor (but no mature 25S) accumulated. Stable binding of processing components and/or ribosomal proteins might have protected and stabilized these pre-rRNAs somewhat, but clearly there has been some degradation of precursors. However, no cleavage occurred within ITS2 at C2 (presumably due to the mutations within the ITS2-proximal stem) so it is the full-length precursors (primarily 27S) that accumulate.
To date, very little is known about the identity of the cleavage machinery at site C2. While specific exonucleases have been demonstrated to be capable of removing the ITS2 segments (6–9) the sequences/structures/complexes regulating or terminating exonuclease activity have not been identified. While a specific complex is not characterized here, structural constraints within the mature rRNA that play a role in maturation have been identified in this study and occur at a site distant to the cleavage at C2. Identification of these critical cis-acting structural elements within mature rRNA is a first real step toward identifying the machinery and understanding the mechanism of pre-rRNA processing of the large subunit RNAs.
Critical structural parameters
Our analysis of mutations that altered primary sequence and/or disrupted secondary structures has identified a number of important structural parameters within the ITS2-proximal stem that are critical for 25S rRNA accumulation in S.cerevisiae and which implicate specific recognition mechanisms for the pre-rRNA processing machinery.
ITS2-proximal stem is critical. A requirement for formation of a base paired ITS2-proximal stem was demonstrated by this and other work (16,21). The fully unpaired constructs (i.e. 5.8Sm and 25Sm) fail to accumulate mature 25S rRNA, but the fully paired, compensatory constructs (i.e. 2X, allGC, allAU) result in the accumulation of stable mature 25S rRNA.
Lack of sequence requirements in the ITS2-proximal stem. There are no sequence-specific recognition elements within the ITS2-proximal stem. Completely changing the sequence identity of the nucleotides in the ITS2-proximal stem (i.e. 2X, allGC, allAU) did not prevent correct processing. In addition, the sequence identity of the unpaired positions did not affect processing (ΔB and 2X).
Length of stem. The length of the ITS2-proximal stem cannot exceed some critical length. There are different length restrictions for different regions within the ITS2-proximal stem. Within the distal and medial parts of this stem the lack of mature 25S demonstrated that two base insertions are not tolerated (CB, +2M and +2D). However, increased steady-state levels of the 27S precursor were apparent in the +2M and +2D constructs.
Within the proximal part of this stem, small insertions are tolerated. Extending this stem by 5 adjacent bp prevented processing (+GC/GC and +AU/AU). However, insertion of just 2 adjacent bp at the same site (+2P) did not prevent accumulation of mature 25S rRNA. These results indicate that the processing machinery recognizes this stem within specific length parameters. Perhaps the processing machinery could not recognize, or could no longer communicate across the increased distance of 5 bp. Addition of 5 bp to the ITS2-proximal stem also destabilized the precursors; no 25S precursors were detected.
Position of unpaired nucleotides. When correctly positioned within the stem [WT(+NcoI), 2X, +2P] the unpaired nucleotides appear to have a neutral effect upon processing. Either one or both of the unpaired nucleotides could be removed without significantly inhibiting 25S accumulation (RB/–, –/RB and RB). In each of these constructs, the length of the base paired segments was not altered.
Shifting the positions of these unpaired nucleotides within the ITS2-proximal stem affected processing, depending upon the site of the 2 bp insertion. Since the +2P construct processed normally, the lack of processing in +2M and +2D may be due entirely to the increase in length in the distal or medial parts of the stem. Still, it is possible that some of the inhibitory effects upon processing could be attributed to the altered position of the unpaired nucleotide within the stem.
The two unpaired nucleotides, being positioned 6 bp apart on opposite sides of a double-stranded RNA, would lie on the same face of the RNA helix (Fig. 7). Insertions into the stem could rotate these unpaired positions relative to each other or to the ribosome body. These unpaired bases would be an efficient means of inducing local but dramatic conformational changes in the stem depending on whether they stacked into or bulged out of the helix. Unfortunately, the in vivo processing assays used here cannot adequately address the precise structural changes attributable to the precise placement of these unpaired nucleotides.
Figure 7.
Model for recognition of the ITS2-proximal stem by the pre-rRNA processing machinery. The ITS2-proximal stem in yeast is an imperfect 15 bp helix (inset) long enough to produce ∼1.5 helical turns. This would place the unpaired U nucleotide in 25S on the same face of the helix as the C in 5.8S. The distal/medial machinery (shaded oval) recognizes this region of the stem within very strict length and structural parameters. The proximal recognition machinery (hexagon) is more flexible to small increases in length. This machinery may or may not be the same complex that is responsible for the sequence-specific recognition of nucleotides that reside structurally opposite of the mature termini (upper case black nucleotides) that are required to promote accurate and efficient processing of the large subunit rRNAs.
The relative positions of the unpaired nucleotides within the ITS2-proximal stem are well-conserved across evolution (16,17; http://www.rna.icmb.utexas.edu). While previous analyses utilizing sequence comparison and alignments suggested that there was probably no specific sequence requirement in this region, these data demonstrate this directly at a functional level. Clearly these unpaired nucleotides are not required for processing. These well-conserved unpaired nucleotides may instead be required for ribosome function. Most eucaryotes for which a rRNA structure has been determined have an ITS2-proximal stem at this position with the same tripartite composition, with borders demarcated by the unpaired nucleotides (17; http://www.rna.icmb.utexas.edu). The only significant length heterogeneity within the stem is at the ITS2-proximal (the ribosome-distal) part of this stem, where the +2P mutation falls. Curiously, there are two organisms where only one of these two unpaired nucleotides is present in the ITS2-proximal stem. Giardia lacks the unpaired nucleotide in 25S and Plasmodium lacks the unpaired nucleotide in 5.8S (http://www.rna.icmb.utexas.edu). These two organisms represent ‘naturally occurring’ variations of the –/RB and the RB/– mutations.
Sequence requirements at the mature termini
There are no apparent sequence-specific requirements at the mature ends of 5.8S and 25S rRNA (corresponding to C3 and C1, respectively). Alteration of the three furthest 3′ nucleotides in 5.8S and the two furthest 5′ nucleotides in 25S (constructs B and E, respectively) had no dramatic effect upon processing, although these mutations were predicted to alter both sequence and proposed structure in this region.
Sequence requirements within ITS2
While there are no sequence requirements within the ITS2-proximal stem or at the processing borders, the controls we designed demonstrated a requirement for sequence-specific recognition at more internal regions within ITS2. The complementary mutations (constructs C and F) identified sequences which, when altered, prevented mature 25S accumulation. Compensatory mutations (constructs D and G) altered these same sequences and restored the proposed structure in this region but did not efficiently rescue processing. The small but detectable levels of mature 25S in construct G seen both by northern analysis (Fig. 5, lane 9) and primer extension (Fig. 6, lane 2) indicate there may be some rescue of processing due to restoration of structure. There are two possible implications for these results. (i) There is a very strong sequence requirement at more internal regions of ITS2, perhaps related to structural constraints within ITS2 (10). The compensatory mutations, which restored structure, none-the-less altered what might be critical sequence within ITS2. (ii) Alternatively, the predicted structure in this region of ITS2 is incorrect.
One interpretation of the complementary and compensatory data could be that the previously proposed secondary structure for yeast ITS2 (10) is not accurate. The data presented here for the compensatory and complementary mutations cannot completely distinguish between the two different structural models that now exist for yeast ITS2 (10,28). Collectively, these data (particularly construct G) support a sequence-specific recognition mechanism of the structure proposed by Yeh and Lee (10) (Fig. 7). However, the data are also consistent with a mechanism that requires both sequence and structural elements in the model proposed by Joseph et al. (28 and data not shown). Previous mutagenesis studies of sequences and structures within ITS2 required for processing do identify critical regions within ITS2, but also fail to discriminate between these two models (4,5). Additional functional and structural analyses will be required to distinguish between these various models.
Structural parameters for recognition of ITS2-proximal stem by the processing machinery
Together these data implicate a processing apparatus with three distinct features, as indicated in Figure 7.
Dual specificity. This processing system demonstrates dual specificity (or a bipartite recognition mechanism) for different regions within the ITS2-proximal stem. For simplicity, we will refer to these as two distinct processing complexes, each with its own specificity, but this may be one complex with two distinct recognition mechanisms for these different domains. Figure 7 depicts the regions within the ITS2-proximal stem recognized by this machinery. One complex (the ‘distal/medial machinery’, shown as an oval in Fig. 7) recognizes both the distal and medial parts of the ITS2-proximal stem (relative to ITS2). This machinery either has limited tolerance for length increases in this region or requires the two unpaired nucleotides, when present, to be located at specific positions within the helix. The RB set of constructs would have a more rigid structure in this region of the stem but are the correct length and are correctly processed. This indicates that structural rigidity in this stem does not inhibit processing. In fact, one role of the distal/medial machinery may be to add rigidity to this helix, in which case these unpaired nucleotides may stack into the helix, rather than protrude from it.
The machinery recognizing the proximal end of this stem (Fig. 7, hexagon) is more flexible with regard to length constraints. While insertion of 5 bp was not tolerated, insertion of 2 bp in this region was correctly processed. In part, this may be due to communication required between the distal/medial and the proximal recognition complexes. The proximal recognition machinery may (or may not) be the same components recognizing sequences within ITS2 (see below).
Indirect recognition of mature termini. Together this processing system comprises an integral measuring device analyzing the ITS2-proximal stem. The substrate must be within specific structural and length parameters for initial binding/recognition, but neither the structure nor the sequences at the mature termini are recognized directly by the processing machinery. Once binding has occurred, the efficiency of processing is determined by other factors, including the stability of the ITS2-proximal stem.
Possible recognition of RNA conformational changes. Processing is much less efficient in the allGC and allAU constructs where the base composition was significantly altered, and thus the free energy or stability of the structure was also altered. One possible explanation is that the allGC and allAU have very different free energies, so the inherent stability of these stems differs as well. The ITS2-proximal stem is not a perfect helix and perhaps flexibility is a critical part of assembling the recognition/processing machinery. If the helix is too stable, as in allGC or RB, then the machinery may not be able to effectively distort this helix and the processing efficiency drops. Likewise, the allAU stem may be too flexible and may breathe, preventing efficient assembly/processing.
The model for the recognition/processing machinery proposed in Figure 7 makes several very important testable predictions for pre-rRNA processing. While the ITS2-proximal stem must form as a prerequisite to processing and is predicted to exist in the mature ribosome, these data question whether a conformational transition must occur in the RNA after initial (and possibly co-transcriptional) binding/recognition by the processing machinery. Are there additional primary recognition elements (sequence or structure) located within ITS2 or mature rRNA that are required for processing? Are the processing sites defined by their position relative to these primary recognition elements? Gaining a better understanding of the recognition elements involved in processing will allow us to identify and characterize the processing machinery and its mechanism of action.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank George Poy at NIH for the deoxyoligonucleotide synthesis and sequencing that made these analyses possible and Masayasu Nomura for his generous gift of plasmid pNOY102 and yeast strain NOY504. Also, we thank Chris Greer and members of the Peculis lab for helpful discussions, and anonymous reviewers for identifying confusing passages in the manuscript. We thank Chris Greer and Vera Nikodem for critical reading of this manuscript.
References
- 1.Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- 2.Kressler D., Linder,P. and de la Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veldman G.M., Klootwijk,J., van Heerikhuizen,H. and Planta,R.J. (1981) The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA and the processing of the primary transcript. Nucleic Acids Res., 9, 4847–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Sande C.A., Kwa,M., van Nues,R.W., van Heerikhuizen,H., Raue,H.A. and Planta,R.J. (1992) Functional analysis of internal transcribed spacer 2 of Saccharomyces cerevisiae ribosomal DNA. J. Mol. Biol., 223, 899–910. [DOI] [PubMed] [Google Scholar]
- 5.van Nues R.W., Rientjes,J.M., Morre,S.A., Mollee,E., Planta,R.J., Venema,J. and Raue,H.A. (1995) Evolutionarily conserved structural elements are critical for processing of internal transcribed spacer 2 from Saccharomyces cerevisiae precursor ribosomal RNA. J. Mol. Biol., 250, 24–36. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell P., Petfalski,E. and Tollervey,D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 8.Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geerlings T.H., Vos,J.C. and Raue,H.A. (2000) The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA, 12, 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh L.C.C. and Lee,J.C. (1990) Structural analysis of the internal transcribed spacer 2 of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol., 211, 699–712. [DOI] [PubMed] [Google Scholar]
- 11.Gasser R.B., Zhu,X., Chilton,N.B., Newton,L.A., Nedergaard,T. and Guldberg,P. (1998) Analysis of sequence homogenisation in rDNA arrays of Haemonchus contortus by denaturing gradient gel electrophoresis. Electrophoresis, 19, 2391–2395. [DOI] [PubMed] [Google Scholar]
- 12.Hershkovitz M.A. and Zimmer,E.A. (1996) Conservation patterns in angiosperm rDNA ITS2 sequences. Nucleic Acids Res., 24, 2857–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakker F.T., Olsen,J.L. and Stam,W.T. (1995) Evolution of nuclear rDNA ITS sequences in the Cladophora albida/sericea clade (Chlorophyta). J. Mol. Evol., 40, 640–651. [DOI] [PubMed] [Google Scholar]
- 14.Chilton N.B., Hoste,H., Newton,L.A., Beveridge,I. and Gasser,R.B. (1998) Common secondary structures for the second internal transcribed spacer pre-rRNA of two subfamilies of trichostrongylid nematodes. Int. J. Parasitol., 28, 1765–1773. [DOI] [PubMed]
- 15.Michot B., Bachellerie,J.P. and Raynal,F. (1982) Sequence and secondary structure of mouse 28S rRNA 5′ terminal domain. Organisation of the 5.8S–28S rRNA complex. Nucleic Acids Res., 10, 5273–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peculis B.A. and Greer,C.L. (1998) The structure of the ITS2-proximal stem is required for pre-RNA processing in yeast. RNA, 4, 1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutell R.R., Gray,M.W. and Schnare,M.N. (1993) A compilation of large subunit (23S and 23S-like) ribosomal RNA structures. Nucleic Acids Res., 21, 3055–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacq B. (1981) Sequence homologies between eukaryotic 5.8S rRNA and the 5′ end of prokaryotic 23S rRNA: evidences for a common evolutionary origin. Nucleic Acids Res., 9, 2913–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brimacombe R. and Stiege,W. (1985) Structure and function of ribosomal RNA. Biochem. J., 229, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 21.Peculis B.A. (1997) The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol., 17, 3702–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogi Y., Yano,R. and Nomura,M. (1991) Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl Acad. Sci. USA, 88, 3962–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- 24.Lin R.-J., Kim,D.-H., Castanotto,D., Westaway,S. and Rossi,J.J. (1996) RNA preparations from yeast cells. In Krieg,P.A. (ed.), A Laboratory Guide to RNA: Isolation, Analysis and Synthesis. Wiley-Liss, Inc., New York, NY, pp. 43–50.
- 25.Peculis B.A. and Steitz,J.A. (1993) Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell, 73, 1233–1245. [DOI] [PubMed] [Google Scholar]
- 26.Musters W., Boon,K., van der Sande,C.A.F.M., van Heerikhuizen,H. and Planta,R.J. (1990) Functional analysis of transcribed spacers of yeast ribosomal DNA. EMBO J., 9, 3989–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schluenzen F., Tocilj,A., Zarivach,R., Harms,J., Gluehmann,M., Janell,D., Bashan,A., Bartels,H., Agmon,I., Franceschi,F. and Yonath,A. (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 28.Joseph N., Krauskopf,E., Vera,M.I. and Michot,B. (1999) Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res., 27, 4533–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]