ABSTRACT
The trans-activator Tat protein of HIV-1 belongs to the large family of intrinsically disordered proteins (IDPs), and is known to recruit various host proteins for the transactivation of viral RNA synthesis. Tat protein interacts with the transactivator response RNA (TAR RNA), exhibiting RNA chaperone activities for structural rearrangement of interacting RNAs. Here, considering that Tat-TAR RNA interaction is mutually cooperative, we examined the potential role of TAR RNA as Chaperna – RNA that provides chaperone function to proteins - for the folding of HIV-1 Tat. Using EGFP fusion as an indirect indicator for folding status, we monitored Tat-EGFP folding in HeLa cells via time-lapse fluorescence microscopy. The live cell imaging showed that the rate and the extent of folding of Tat-EGFP were stimulated by TAR RNA. The purified Tat-EGFP was denatured and the fluorescence was monitored in vitro under renaturation condition. The fluorescence was significantly increased by TAR RNA, and the mutations in TAR RNA that affected the interaction with Tat protein failed to promote Tat refolding. The results suggest that TAR RNA stabilizes Tat as unfolded, but prevents it from misfolding, and maintaining its folding competence for interaction with multiple host factors toward its transactivation. The Chaperna function of virally encoded RNA in establishing proteome link at the viral-host interface provides new insights to as yet largely unexplored RNA mediated protein folding in normal and dysregulated cellular metabolism.
KEYWORDS: HIV Tat, molecular chaperone, protein folding, RNA, TAR RNA
Abbreviations
- DNA
DNA
- EDTA
EthyleneDiamineteTraacetic Acid
- EGFP
Enhanced Green Fluorescent Protein
- E. coli
Escherichia coli
- FBS
Fetal Bovine Serum
- GFP
Green Fluorescent Protein
- HIV
Human Immunodeficiency Viruses
- HPLC
High-Performance Liquid Chromatography
- IPTG
IsoPropyl-β-D-ThioGalactopyranoside
- IUPs
Intrinsically Unstructured Proteins
- MEM
Minimum Essential Media
- NLS
Nuclear Localization Signal or Sequence
- PBS
Phosphate-Buffered Saline
- PCR
Polymerase Chain Reaction
- P-TEFb
Positive Transcription Elongation Factor
- RBD
RNA-Binding Domain
- RNA
RiboNucleic Acid (also, mRNA, nRNA, rRNA, tRNA)
- RNase
RiboNuclease
- RNP
Ribonucleoprotein
- rNTPs
riboNucleoside Tri-Phosphate
- ROI
Region Of Interest
- RRE
Rev Responsive Element
- SDS-PAGE
Sodium Dodecyl Sulfate-PolyAcrylamide Gel Electrophoresis
- ssDNA
single-stranded DNA molecules
- TAR
Trans-Activation Response Element
- Tat
Trans-activating Protein
Introduction
RNA molecules play a central role and support functions in cellular processes. Many cellular RNAs have functions that extend well beyond the central dogma.1–5 Though versatile roles of RNA have been newly discovered, the functions of many non-coding RNAs transcribed from the genome are not well characterized yet.6–8 Varieties of IDPs of human and viral origins are known to interact with RNA molecules for establishing protein networks.9 IDPs, due to intrinsically unstable nature, are prone to misfolding, and possibly are stabilized by interaction with RNAs. And yet, the potential role of RNAs in assisting the folding of IUPs, either in normal cellular environment or in pathogenic conditions, remains largely unexplored.
Proteins frequently encounter off-pathways in the folding process such as misfolding and consequent aggregation in the folding process. Productive folding into native conformation often requires the assistance from molecular chaperones.10 Chaperones and chaperonins interact with nascent polypeptide and, by shielding exposed hydrophobic residues, influence its kinetic network in favor of folding into native conformation11–16 However, genetic and biochemical analyses showed that only a limited number of proteins are folded by the assistance of molecular chaperones17–20 Limited role of molecular chaperones implies that other factors may involve, directly or indirectly, to ensure the folding of proteins in the crowded cellular environment. Experimental evidences are being accumulated that RNAs provide potential chaperone function.21 Polyanionic macromolecules have been suggested as a candidate for another chaperone type. For instance, refolding of Arc repressor can be accelerated by binding nucleic acids and other polyanions.22 RNAs strongly potentiates the chaperone function of DnaK in vitro.23 In addition, 23S rRNA, the component of 50S ribosomal subunit in E. coli, has shown to stimulate refolding of proteins in vitro.24 Moreover, it was demonstrated that RNAs have a chaperoning effect on the protein linked to RNA-binding domain (RBD).25,26 The results suggest that RNAs can function as a stabilizer of highly dynamic folding intermediates, which, otherwise, would yield non-functional aggregates or even toxic consequences. It has been reported that the natively unfolded or partially disordered proteins or domains turn into an ordered structure upon binding to their cognate RNA.27,28 For instance, the α-helical conformation of an HIV-1 RRE peptide become stable upon interaction with RRE RNA.29 In the phenylalanine tRNA synthetase, a disordered N-terminus of the protein shapes into a long coiled-coil helical domain upon binding with cognate RNA.30 The HIV Tat, as intrinsically disordered protein,31 is able to interact with multiple cellular proteins. This ability plays a pivotal role in HIV transactivation by recruiting the kinase activity of the P-TEFb complex to the viral mRNA's stem-bulge-loop structure of TAR into stabilized transcriptional elongation complex.32 Previously, Tat was known to exhibit RNA chaperoning activity assisting the folding and structural rearrangement of RNA molecules,33 possibly mediated by interaction with intrinsically disordered region.34 These results prompted us to investigate if HIV TAR RNA in turn provides a chaperoning function to its interacting Tat protein. Wealth of information is accumulated on specific interaction between Tat and TAR RNA35–37 Tat acts by binding to TAR RNA of a stem-loop structure located at the 5’ end of the HIV-1 transcript,38,39 and this specific interaction is required for efficient transcription elongation of the HIV-1 genome40–42 Despite detailed studies on the structure and function of Tat-TAR RNA complexes, a direct role of TAR RNA on the folding status of Tat protein has not been studied yet. In fact, the majority of Tat exists as extended random coil where the structure is highly dictated by an extensive contact with binding partners.43 As such, highly dynamic nature of folding intermediates or intrinsically disordered region (IDR) of IDPs precludes detailed characterization of their structures,44 and could be inferred from various in vitro and in vivo evidences. Here, the potential role of TAR RNA on the folding status of the HIV-1 Tat was addressed by the time-lapse fluorescence microscopy in live cells and in vitro refolding condition using EGFP fusion as reporter for folding.45–47 Our data suggests that the virally encoded TAR RNA keeps the Tat protein as ‘folding-competent’, which enables its interaction with multiple cellular factors for transactivation, underlying the pathogenesis associated with HIV-infection.
Materials and methods
Construction of protein and RNA expression vectors
The HIV Tat plasmid was constructed as follows: the HIV-1 tat gene was chemically synthesized (GenScript). The sequence of the gene was selected from the HIV-1 complete genome (GenBank ID: NC_001802) and used as a template DNA for PCR. The plasmid pGE-LysRS, a derivative of pGEMEX-1 (Promega), was used for the construction of plasmids for Tat and Tat-EGFP expression. The tat gene was ligated into the NdeΙ and SalΙ sites of pGE-LysRS, yielding pGE-Tat. Tat mutants replacing the residues R52, R53 by alanine were produced by site-directed mutagenesis. BamH1 and EcoRI restriction sites were generated by insertion into pcDNA3.1+ (Invitrogen) using standard restriction cloning methods. The oligonucleotide forward primer was: 5′-GGC ACA AGC TGG AGT ACA AC-3′and reverse primer was 5′-ATG CCG TTC TTC TGC TTG TC-3′for EGFP. The vector modified from pcDNA3.1+ (Invitrogen) was used for the construction of the expression vectors for the human wild type HIV Tat-EGFP protein using standard restriction cloning methods. The EGFP gene was amplified from pEGFP-N1 (Clontech), and inserted, using KpnΙ and SalΙ sites, into the 3′ end of the tat gene that was ligated into the NdeΙ and KpnΙ sites of pGE-LysRS, yielding pGE-Tat-EGFP. For in vitro transcription, a template including T7 promoter-TAR RNA gene-T7terminator was ligated into the NdeΙ and SalΙ sites of pGE-LysRS, yielding pGE-TAR. Similarly, TAR RNA mutant were produced by site-directed mutagenesis and used to generate pGE-TAR derivatives. To construct a TAR RNA co-expression vector, a DNA fragment that contained the arabinose promoter, the TAR RNA gene, and the rrnB terminator was inserted into the SphΙ and SalΙ sites of pLysE (Novagen), yielding pLysE-TAR. E. coli strain BL21 Star™ (DE3) (Invitrogen), 1 mM IPTG, and 0.04% L-arabinose were added to induce the expression of protein and RNA simultaneously. After culture for 3 h, the cells were harvested. For HeLa cells, TAR RNA oligos (57mers) were synthesized with 100 nmol from M-biotech Inc. (the Korean branch of IDT Inc.) (5′- GGU CUC UCU GGU UAG ACC AGA UCU GAG CCU GGG AGC UCU CUG GCU AAC UAG GGA ACC -3′ in RNase Free water by HPLC purification).
Cell culture and Tat protein & TAR RNA co-transfection
HeLa cells were grown in MEM (Welgene Inc., Korea) supplemented with 10% FBS (v/v) and 1% penicillin/streptomycin (v/v) in a 5% CO2 atmosphere at 37°C. Cells grown at 60–90% confluence were transfected with 500 ng plasmid per 2 × 106 cells using 2 µL Lipofectamine 2000 (Invitrogen). Cells were washed once with growth medium, twice with 10X PBS at pH 7.4 and finally with PBS alone. Co-expression of HIV Tat and TAR RNA was performed by co-transfection of HIV Tat-EGFP fusion pCDNA3.1+ (Invitrogen) and TAR RNA oligos (57mers).
Time-lapse live cell fluorescence microscopy
Time-lapse, multicolor images in living cells as well as direct EGFP fluorescence images in HeLa cells were obtained using the A1Rsi confocal microscope system (Nikon). To maintain physiologic temperature during live observation, the microscope was kept at 37°C in a temperature-controlled chamber. The light intensity was set to 7 at the sample plane. Transfections with EGFP or Tat -EGFP fusion plasmid were performed using Lipofectamine 2000 at 37°C for 24 h. Cells were grown in a 12-well culture dish (Nunc). The EGFP fusion plasmids (0.5–0.8 μg) were transfected into cells with Lipofectamine 2000 according to the manufacturer's protocol. A 20X phase-contrast objective was used with a Nikon perfect focus system (PFS) was used to maintain focus throughout the time-lapse imaging every 10 min for 2 h. PFS requires cells plated on 35 mm Petri dish containing a 14 mm glass microwell (MatTek). The system was equipped with a temperature-controlled 37°C chamber and is provided with an oxygen supply; images were acquired using a 40X short distance 0.75 NA air objective. Analysis was performed with NIS-Elements Viewer 4.20 software (Nikon), which compensates for stage shift, vibration, or similar small whole field movement that can occur during time-lapse acquisition. Whole HeLa cell extracts were made using EzRIPA Lysis kit (Atto Co.). The concentration of the isolated proteins was determined using BCA Protein Assay Reagent (Pierce).Western blot was performed according to standard methods, using an anti-GFP antibody (1:100 in TBST) (Clontech) and β-Actin antibody (1:2000 in TBST) (Cell signaling) for the detection of Tat-EGFP protein.
Protein expression
The E.coli strain BL21 Star™(DE3) pLysS (Invitrogen) was used as a host for protein expression. pGE-Tat-EGFP and pGE-EGFP were ligated into the restriction sites of pGE-LysRS using standard restriction cloning methods and pLysE-TAR synthesized by in vitro transcription. A single transformation colony was selected and inoculated into 3 mL LB containing 50 μg/mL ampicillin and 30 μg/mL chloramphenicol, and cultured overnight at 37°C. 1 mL culture was diluted into 15 mL fresh LB with the same antibiotics, and cultured until the optical density (OD) reached to 0.5∼0.8 at 600 nm. Proteins were expressed for 3 h after the addition of 1 mM IPTG. The harvested cells from 10 mL culture were suspended in 0.3 mL PBS, then lysed by sonication. The total lysates were centrifuged at 12 000 rpm for 15 min to separate the soluble and pellet fractions from the total lysates. The separated pellet fractions were resuspended with PBS having the same volume as the soluble fractions. Each 50 μL of soluble and pellet fractions was mixed with the same volume of 2X SDS loading buffer. After boiling, the samples were loaded and run on SDS-PAGE. The loading volume of each sample was adjusted to the equivalent amount based on the final cell OD600 nm. The gels were stained with Coomassie brilliant blue and the solubility of proteins was estimated using a gel densitometer. Precipitated His-tagged HIV Tat proteins were further analyzed by SDS-PAGE and western blot according to standard methods, using an anti-His penta antibody (1:1000 in TBST) (Qiagen) for detection.
Protein purification
E.coli cells were harvested from 500 mL culture by centrifugation, suspended, and lysed by addition of 10 mL buffer A (50 mM Tris-Cl (pH 7.5), 300 mM NaCl, 10% glycerol, 10 mM imidazole, 2 mM β-mercaptoethanol, 0.5% NP-40) with 1 mM PMSF followed by sonication. After centrifugation, the soluble fraction of the lysates was loaded on a HisTrap HP column (GE Healthcare). After sufficient washing with buffer A, proteins were eluted under an imidazole gradient from 10 to 300 mM by mixing buffer A and buffer B (50 mM Tris-Cl (pH 7.5), 300 mM NaCl, 10% glycerol, 300 mM imidazole, 2 mM β-mercaptoethanol, 0.5% NP-40). The fractions were analyzed by SDS-PAGE, and then the fractions containing the proteins of interest were pooled and dialyzed against a buffer containing 50 mM Tris-Cl (pH 7.5), 50 mM NaCl, 1 mM DTT, and 0.1 mM EDTA.
In vitro RNA transcription
The linear DNA templates for in vitro transcription were obtained by PCR. Templates containing a T7 promoter upstream of the RNA coding sequence and restriction enzyme sites at the 5′ and 3′ ends were digested with restriction enzymes, and RNA was transcribed using the RiboMAX™ large scale RNA production system-T7 (Promega). Following transcription, the DNA templates were removed by digestion with RNase-free DNase for 15 min at 37° C. One volume of citrate-saturated phenol (pH 4.7): chloroform: isoamyl alcohol (125:24:1) was added to the sample, which was vortexed and centrifuged at 12 000 rpm for 2 min. The upper, aqueous phase was transferred to a new tube, and then 1 volume of chloroform: isoamyl alcohol (24:1) was added. After vortexing and centrifugation, the aqueous phase was carried to a new tube, and 0.1 volume of 3 M sodium acetate (pH 5.2) and 1 volume of isopropanol were added to the sample. The pellet obtained from centrifugation was washed with 1 mL 70% ethanol, dried, and suspended in nuclease-free water. The unincorporated rNTPs were removed by illustra™ MicroSpin G-25 columns (GE Healthcare). The concentration of RNA was measured by the absorbance in 260 nm.
In vitro EGFP refolding assay
Purified EGFP and Tat-EGFP was denatured in 6M guanidine hydrochloride and 1 mM DTT at 40°C for 20 min. The denatured protein was 25-fold diluted in the refolding buffer containing 50 mM Tris-Cl (pH 7.5), 50 mM NaCl, and 5 mM MgCl2, either in the absence or the presence of RNAs. The fluorescence intensity of the refolded EGFP was monitored by a fluorescence spectrophotometer (Varian) with excitation at 489 nm and emission at 509 nm. Alternatively, the fluorescence emission at 491 nm after excitation at 517 nm was also monitored by FlexStation 3 plate reader (Molecular Devices).
Results
The subcellular localization of HIV Tat-EGFP fusion protein
To test the feasibility of EGFP as a folding reporter of Tat in living cells, the HeLa cells were transfected with a Tat-EGFP fusion vector and the cellular localization of the Tat protein was analyzed. Among the 3 major versions of the Tat protein, we used the most widely studied 86-amino acid form where residues 1–72 are encoded by the first exon.48–52 Tat contains multiple functional domains in which the amino acids 49–72 are responsible for the binding of TAR RNA and in particular 3 amino acids (59–61) functions as basic nuclear localization signal (NLS).49,53–57
Fig. 1 compares the location of Tat-EGFP protein in transfected HeLa cells. Each construct is featured in 3 images: GFP filtered images corresponding to the location of Tat-EGFP protein, DAPI stains of the nucleus, and the merged image of both GFP and DAPI staining. As shown in Fig. 1, EGFP was uniformly distributed throughout the cell, whereas Tat-EGFP was mainly localized in the nucleus with or without nuclear speckles of variable sizes and irregular shapes. The morphology and nuclear location of Tat was consistent with its known role as a trans-activator, which colocalized with the spliceosome assembly factors such as nuclear speckles.50,58–60 These results suggest that Tat in its EGFP fusion form is suitable as a folding reporter in living cells.45
Figure 1.
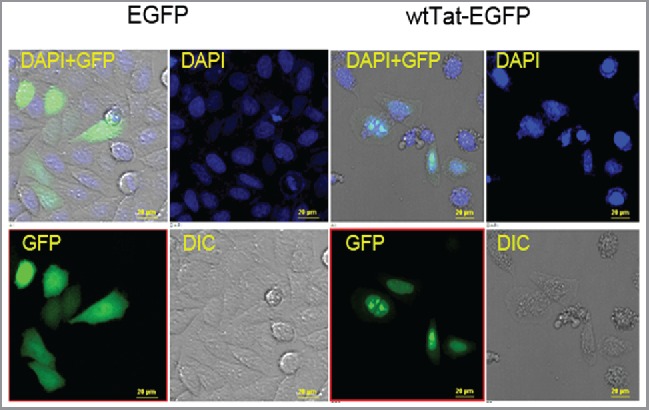
Subcellular localization of Tat-EGFP fusion protein folding in HeLa cells. HeLa cells were transfected with EGFP or wild type (WT) EGFP-Tat fusion constructs, and subcellular localization was determined by imaging EGFP using a fluorescence microscope (40X). The EGFP is present in the cytosol as evident in the merged (DAPI + GFP) image. Tat-EGFP localizes predominantly in the nucleus, frequently as nuclear speckles. GFP filtered images correspond to the location of EGFP or Tat-EGFP protein expression. DAPI stains the nucleus of all fixed cells. The merged image contains the combined images of GFP, DIC (Differential Interference Contrast), and DAPI staining, and shows a representative area under microscopic observation. Scale bar, 20 μm.
Tat-EGFP folding enhancement by TAR RNA in HeLa cells
A ‘folding reporter’ in which a test protein is expressed as an N-terminal fusion with GFP, gives a fluorescence signal directly proportional to the amount of correctly folded protein, and requires no functional assay for the protein of interest or knowledges on its structure or biologic function45–47 The rationale behind the reporter system is that the test protein, if folded properly, would not interfere with the folding of EGFP, hence emitting fluorescence. It should be noted, however, that Tat in itself is prone to misfolding due to the disordered nature, and consequently misfolds its linked EGFP domain. Thus, any factor that would stabilize Tat from misfolding, TAR RNA in this case, increases the fluorescence in the EGFP fusion construct. Thus, fluorescence reports not so much a stable folding of Tat, but rather its inability to misfolding.
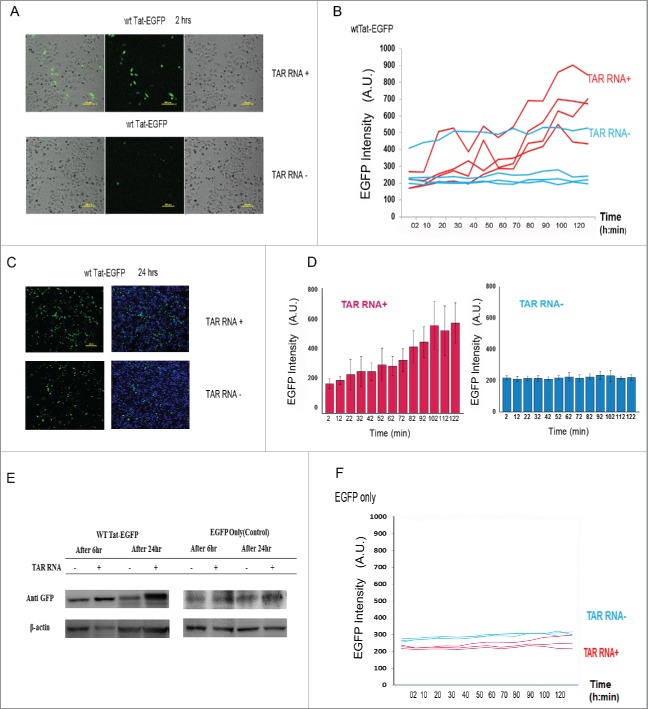
Cells were co-transfected with Tat-EGFP and TAR RNA, and the fluorescent cells were traced and monitored by live cell time-lapse fluorescence microscopy. The time-lapse imaging monitored the formation of Tat-EGFP and the cell migration in real time (Fig. 2 and Fig. S1, Video S1- S3). After transfection and incubation for 5 hours, the initial rate and the yield of wild type Tat-EGFP folding were estimated by monitoring the increase in intrinsic EGFP fluorescence. The temporal change of Tat-EGFP fluorescence was monitored at every 10 minutes for 2 hours by tracking ROIs (region of interest; matched to the fluorescence of the cell) in the videos and graphically represented in Fig. 2B. ROIs were traced using DIC (Differential Interference Contrast) by which the location of the Tat-EGFP fluorescence intensity was clearly identified. The TAR RNA co-expression (TAR RNA + in blue) clearly stimulated the fluorescence as compared with the control without TAR RNA co-expression (TAR RNA – in red) (Fig. 2B). As a control, the fluorescence of EGFP only was not affected by TAR RNA co-expression (Fig. 2F). The level of fluctuation in the measurement of fluorescence signal reflects the degree of difficulties in tracking live, migrating single cells before reaching confluence (Fig. 2B). And yet, the average intensity, taking statistical significance of signals from individual cells into account, reflects notable stimulation of signals in the presence of TAR RNA (Fig. 2D). Of note, the level of Tat-EGFP protein in the transfected cells was increased by co-expression of TAR RNA by western blot analysis (Fig. 2E). The EGFP only as a control, however, was similar regardless of TAR co-expression. As monitored by the movie, there was a marked cytoplasmic EGFP fluorescence representing properly folded Tat (Video S1-S3), which was quantified by the measured mean fluorescence of the ROI. Indicative of the localization/abundance of the Tat-EGFP in the cytoplasm and nucleus at particular time points, the fluorescence by EGFP expression represents Tat-EGFP folding as affected by TAR RNA. Moreover, after co-transfection of Tat-EGFP and TAR RNA plasmids, the number of fluorescent cells was increased over 2–24 h as compared with the Tat-EGFP transfection only controls (Fig. 2A and 2C). The enhancement of both the initial rate and intensity of fluorescence is consistent with potential chaperoning role of TAR RNA for the folding of Tat protein inside cells (Fig. 2D). The results are consistent with the stabilization of Tat by interaction with TAR RNA from misfolding into unstable conformation vulnerable to degradation in the cellular environment.
Figure 2.
Time course Tat folding by TAR RNA in HeLa cells. After transfection and 5 h of incubation, HeLa cells were co-transfected with plasmids encoding Tat-EGFP and TAR RNA and observed under time-lapse fluorescence microscopy at 10 min intervals for 2 h. (A) Merged images (40X, left) of the Differential Interference Contrast (DIC) and the fluorescence (GFP) after 3 h after transfection. (B) Time-dependent increase of EGFP based on a scatterplot of region of interest (ROI) corresponding to Videos S1–S3 of EGFP fluorescence in the control (blue) and TAR RNA co-expressed (red) cells. Time-lapse images of Tat-EGFP expression in HeLa cells at 4 independent positions were captured. Mean fluorescence of ROIs were traced using DIC (Differential Interference Contrast) covered the entire numbering field. (C) Overlaid image (10X) used to generate numbers of Tat-EGFP expressing cells 24 h after transfection. Scale bar, 200 μm. (D) Graphical presentation of Fig. 2B for the time dependent increases of Tat-EGFP expression in HeLa cells. (E) SDS-PAGE and western blot analysis of HeLa extracts for EGFP proteins. (F) Time-lapse fluorescence observations of EGFP expression with (red) and without TAR RNA co-expression (blue).
TAR RNA-mediated solubility enhancement of Tat
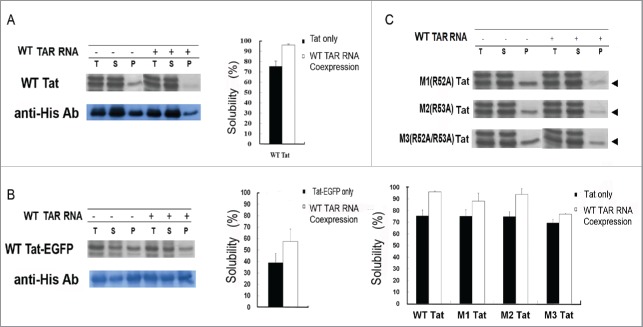
Maintaining the solubility of a protein by preventing it from misfolding is necessary for keeping it as folding competent inside the cells. The potential effect of TAR RNA on the solubility of Tat was studied in E.coli system. We constructed 2 independent plasmids, expressing either Tat-EGFP under the T7 promoter or TAR RNA under the arabinose promoter, which could be induced by IPTG and arabinose, respectively. After expression, the total extract [T] were separated into soluble[S] and pellet [P] fractions by centrifugation and analyzed by SDS-PAGE, and representative data are shown in Fig. 3. The band intensity of Coomassie staining (Fig. 3A, upper panel) was estimated by densitometric scanning of the band corresponding to that identified by Western blot with anti-his-antibody (Fig. 3A, lower panel). The relative solubility was calculated by comparing the band intensities of [S] and [P] fractions. The solubility of wild type Tat (wtTat) was greatly increased upon co-expression of TAR RNA (96%) as compared with the control without TAR RNA expression (75%) (Fig. 3A). Consistent with SDS-PAGE Coomassie staining, western blot analysis also showed a notable decrease of Tat protein in the pellet fraction upon co-expression of TAR RNA (50%), as compared with TAR RNA(-) control (36%). A similar observation was also made in Tat-EGFP, where modest increase in solubility was observed by TAR expression (57%) as compared with the control (38%) in Coomassie staining (Fig. 3B), as well as in western blot (46% and 42%, respectively). The overall solubility of Tat-EGFP was relatively lower than Tat, both with and without TAR RNA co-expression (Fig. 3A and 3B), probably reflecting additional requirements for folding of GFP, including auto-catalytic cyclization and oxidation reactions for chromophore formation.61 The arginine-rich motif of Tat has been shown to be important for the recognition and binding of TAR RNA,35 and the relative importance of specific amino acids within the motif have been determined.62
Figure 3.
In- cell solubility of Tat protein in E.coli by TAR RNA. The effect of TAR RNA enhances Tat protein co-expression on the solubility in vivo. (A) The expression of wild type Tat was controlled by T7 promoter and induced by IPTG, whereas the expression of wt TAR RNA was under the control of arabinose promoter and induced by L-arabinose. The band intensity of each Tat protein fraction on the SDS-PAGE gel was estimated by densitometric scanning. T, S, and P represent total lysate, soluble fraction, and pellet fraction, respectively. (B) Increase of Tat-EGFP solubility by TAR co-expression as analyzed by Coomassie staining and western blot using the anti-His antibody. (C) SDS-PAGE gel analysis of Tat mutants. The relative solubility of Tat mutants based on the Coomassie stained band intensities is shown in bars: with (white) and without (black) TAR RNA coexpression. The band intensity of each Tat protein fraction was measured by a densitometer. The mutants M1, M2, and M3 Tat carry amino acid substitutions, R52A, R53A, and both R52A and R53A, respectively. T, S, and P represent total lysate, soluble fraction, and pellet fraction.
We therefore constructed Tat mutants by site-directed mutagenesis, to evaluate the effect of TAR RNA binding to Tat on the solubility in vivo. We replaced the arginines at positions 52 and 53 (R52 and R53), both of which are crucial for binding, yielding 2 single point mutants, R52A (M1) and R53A (M2), respectively, and a double mutant R52A/R53A (M3) (Fig. 3C). Co-expression of TAR RNA only partially rescued M1 and M2 solubility, whereas the combination of the 2 mutations (M3) strongly abolished the rescue. These results show that the arginine residue in the conserved basic region of Tat crucial for binding with TAR RNA is required for maintaining the solubility of Tat protein. Besides the affinity, the size of RNA may also influence the solubility and folding of interacting proteins.63 Thus, to examine if TAR RNA size influenced the Tat solubility, RNAs of varying sizes were tested, including the 5′-end proximal TAR and downstream secondary stem-loop structures. Thus, TAR (80 nt) and TAR (104 nt), corresponding to +1 to +80 and +1 to +104 of the HIV-1 transcript, respectively, were compared with TAR (57 nt) (Fig. 4A). The solubility of co-expressed Tat-EGFP, was similar regardless of the size of the TAR RNAs tested (Fig. 4B). The results show that direct interaction at specific residues is more important than RNA size for preventing Ta from misfolding into insoluble aggregates.
Figure 4.
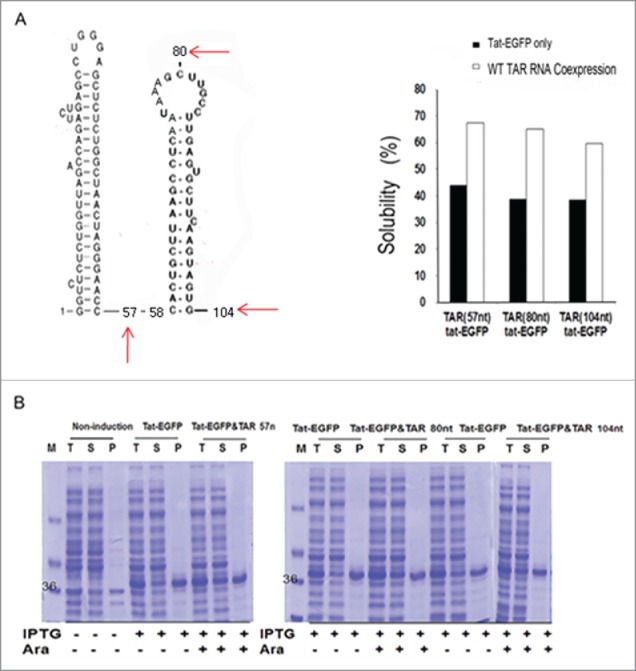
The effect of the length and the structure of TAR RNA on the solubility of Tat-EGFP. (A) The predicted secondary structure of the 5’-end of the HIV-1 transcript including the 5-proximal TAR sequence84 and the relative solubility of Tat protein upon co-expression with TAR RNAs of varying size. (B) SDS-PAGE gel shows the wild type Tat protein co-expressed with TAR RNAs of various size: 57, 80, or 104 nt in length.
In vitro refolding of Tat-EGFP in the presence of TAR RNA
The influence of TAR RNA on Tat protein folding was further investigated in vitro. The sites of mutations on TAR and Tat were guided by previous reports.64–68 Tat-EGFP containing a C-terminus His-tag was expressed from E. coli and purified by Ni-affinity chromatography (Fig. S2). The protein was solubilized in 6M guanidium chloride, and diluted 25 fold in a refolding buffer in the presence or absence of TAR RNAs and the extent of folding was monitored by the fluorescence. The extent of refolding was enhanced by the presence of TAR RNAs, and the degree of enhancement was dependent on its concentration (Fig. 5A). Based on the fluorescence of native EGFP of equal amount, the refolding yield was 31% and 40% for 0 and 0.14 μM TAR RNA, respectively, and appeared saturated at higher RNA concentration (0.28 μM) (Fig. 5A). The experiment was repeated 3 times and the average refolding yield shown in Fig. 5B. The data showed that maximum stimulation (∼40%) was achieved at about equimolar concentration, e.g., 0.14 μM TAR RNA and 0.14 μM Tat-EGFP, suggesting that folding is enhanced by a direct 1:1 interaction. Therefore, in subsequent experiments, the final concentration of protein in the refolding buffer was kept at 0.14 μM. As a negative control, the refolding of EGFP was insensitive to the presence of TAR RNA, even at the highest concentration tested (12 uM) (Fig. 5G). As another control, the native Tat-EGFP was treated with TAR RNA in the refolding buffer. At varying concentrations of TAR RNA and incubation times, the fluorescence remained similar without appreciable changes (Fig. S3). The results suggest that the increases in fluorescence by TAR RNA in refolding condition is actually due to a stimulation of folding rather than a post-folding event such as stabilization of Tat structure by RNA binding. When TAR RNA was replaced with tRNALys in the refolding mixture, we failed to observe a similar increase in fluorescence (Fig. 5C). Alternatively, RNAase treatment during refolding quenched the stimulation of Tat-EGFP refolding (Fig. 5D), demonstrating that the refolding of Tat-EGFP was mediated by TAR RNA interaction. A low level stimulation even after the RNAse treatment may be due to the presence of short RNA fragment, which was investigated no further. As an analog of TAR RNA, TAR ssDNA (0.14 μM) was also shown to stimulate Tat-EGFP refolding similar to that of TAR RNA at the same concentration (Fig. 5E). It has been shown that the 3-nucleotide bulge in the TAR RNA sequence is important for Tat protein recognition.68 This bulge was either deleted (M1 TAR) or substituted (M2 TAR) by site-specific mutagenesis. As shown in Fig. 5F, the refolding yields of Tat-EGFP in the presence of TAR RNA mutants at the equimolar concentration (0.14 μM) were either decreased (M1 TAR) or as low as in the absence of wt TAR RNA (M2 TAR). Together, our data support that the refolding of Tat-EGFP is mediated by direct interaction with TAR RNA through its specific recognition site.
Figure 5.
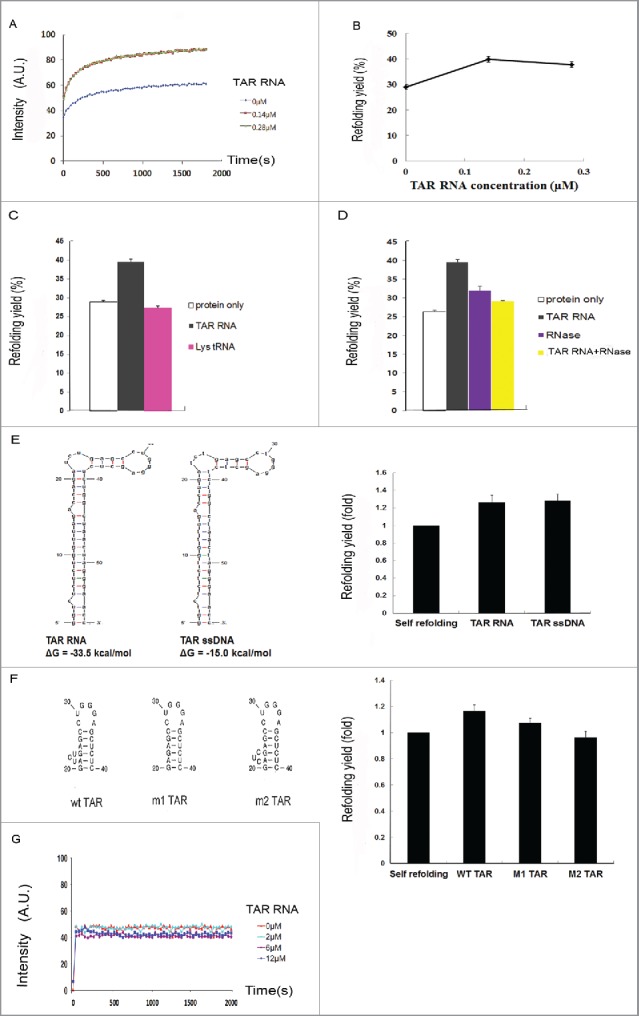
In vitro refolding of Tat-EGFP. (A) Time course of fluorescence intensity and (B) refolding yield as measured by EGFP fluorescence at different TAR RNA concentrations. Error bars represent the standard error of the means. The denatured Tat-EGFP in 6M guanidine-HCl was diluted to a final concentration of 0.14 μM in the refolding buffer containing different concentrations of TAR RNA. Refolding of Tat-EGFP (C) in the presence of cognate TAR RNA or irrelevant Lys tRNA, or (D) after RNase treatment (25 μg/mL RNase A at 37°C for 30 min). The concentration ratio of Tat-EGFP to RNA was adjusted to 1:1. Error bars represent the standard error of the means. To verify it TAR RNA binding is an important factor for HIV-1 Tat refolding, a mutation of the TAR RNA binding site was constructed. (E) Secondary structures and free energies of TAR RNA and single-stranded (ssDNA) were predicted by the Mfold program68. The effects on Tat-EGFP refolding are compared. The concentration of Tat protein, ssDNA and TAR RNA was 0.14 μM. The folding yield is shown as relative to the self-refolding of Tat-EGFP only, which was taken as 1. (F) The refolding of Tat-EGFP in the presence of TAR mutants. (G) Time course of refolding of EGFP as control at various concentrations of TAR RNA.
Discussion
The present results show that TAR RNA influences the folding status of its bound Tat protein preventing it from misfolding both in vivo and in vitro. As a typical example of IDPs,44 Tat protein exists as extended random coil where the structure is highly dictated by an extensive contact with binding partners.43 As such, addressing to the structure of its folding intermediates is even more difficult and could be inferred from various in vitro and in vivo evidences. As a means to approach to this problem, here we adopted EGFP fluorescence as an indirect reporter for the folding status as EGFP-Tat fusion. As such, the system does not report the stable folding of Tat, which in itself is unfolded, but rather its inability to misfold and interfere with folding of interacting partners. Thus, the assay is relevant for the chaperone activity, considering that the major function of molecular chaperones is the prevention of aggregation of unfolded proteins rather than active promotion of folding.16
The time-lapse fluorescence microscopy was used for direct visualization of live cells for EGFP fluorescence as a tracer and an indirect indicator for Tat folding in real time in migrating HeLa cells. HeLa cells co-transfected with TAR RNA and the Tat-EGFP indicator displayed a gradual, but distinct increase of the in - cell fluorescence intensities (Fig. 2). Of note, the amount of Tat-EGFP was increased by the coexpression of TAR RNA (Fig. 2E). In vitro refolding experiment indeed showed that TAR RNA stimulates Tat folding (Fig. 5), whereas TAR itself exerts little effects on the stability of Tat (Fig. S3). Overall, the results are interpreted to mean that Tat protein acquired protein stability by TAR RNA binding, which kept Tat form misfolding and subsequent degradation in the cellular environment. The results are reminiscent of the proteostasis function of molecular chaperones that provide quality control over their cargo proteins from folding to expedited clearance of misfolded proteins.69,70 The solubility of the Tat protein was increased upon TAR RNA co-expression as tested in the E.coli recombinant system (Fig. 3). Misfolding usually results in insoluble aggregates, and therefore, the increased solubility is an important indicator for proper folding of Tat in the presence of TAR RNA. Mutations in either Tat or TAR RNA that affected their mutual interactions decreased the solubility and folding of Tat in vivo (Fig. 3C) and in vitro refolding (Fig. 5F). Studies with TAR RNA size variants showed that the specific Tat-TAR interaction, rather than the size of the TAR RNA, was important in the enhancement of Tat protein solubility (Fig. 4). Consistent with the co-expression study, the refolding yield of Tat-EGFP in vitro was increased in the presence of TAR RNA, whereas the RNAase treatment abolished the stimulation (Fig. 5D). Non-cognate RNA (Lys tRNA) or mutants of TAR RNAs that affected Tat interaction failed to promote Tat refolding (Fig. 5C and 5F). Thus, the analyses in vivo both in HeLa and E. coli cells, as well as refolding in vitro, suggest that TAR RNA binding stabilizes Tat from misfolding into non-functional form vulnerable to degradation.
With respect to the mechanism by which RNA prevents misfolding and promotes the solubility of its interacting protein, various possibilities could be advanced. Primarily, the chaperoning effect of RNA can be explained by its charge and steric hindrance.71,72 These characteristics could stabilize the folding intermediates and interfere with protein aggregation. RNA carries a highly negative charge and, the negatively charged ribonucleoprotein (RNP) complex would remain in a monomeric state due to charge-charge repulsion, resisting intermolecular interactions into misfolded aggregation. It has been reported that the net charge of a protein is an important factor for its rate of aggregation, and that intermolecular electrostatic repulsion among charged residues of proteins influences their solubility.63,72 In addition, RNA as a macromolecule with bulky size could exert steric hindrance, minimizing interactions among folding intermediates of highly dynamic and aggregation-prone nature. Another view is that RNA could function as a specific ligand to its binding proteins. In this regard, meta-stable folding intermediates might be induced to fold into stable conformations upon binding with RNA ligand.73 Thus, TAR RNA might dictate the folding status of Tat as its specific ligand through a mechanism similar to the RNA binding-induced protein folding that occurs in the RNP complex. Thus, the conformation of Tat is stabilized or dictated in space by the overall conformation and specific interaction with TAR.68,74 As such, the bulge region of TAR RNA involved in its interaction with Tat 64,75 plays a role in Tat-EGFP refolding (Fig. 5F). This result suggests that the overall charge and structure of RNA as polyanionic macromolecules are influential factors for the folding status of Tat protein.
The present results are intriguing, especially considering previous reports that Tat itself was shown to assist the folding and structural rearrangement of RNA molecules,33 although there was no direct evidence for its role in the folding of TAR RNA. Of note, RNA chaperone activity is mediated by interaction between Tat and TAR, at the similar sites in the present analyses. It could therefore be speculated that the same Tat-TAR interactions operate mutually and synergistically for the folding of their binding partners. Distinct from RNA chaperone- a protein that assists the folding of interacting RNA, here we suggest the chaperna activity - RNA based chaperone that assists folding of interacting proteins. Of note, the M1 RNA, a prototype ribozyme, was recently shown to exert potent chaperna activity to its interacting C5 protein in E. coli RNase P complex.73 Both activities, RNA chaperone and chaperna, may not be mutually exclusive, but function in a cooperative manner where the intermolecular contact mediates intra-molecular stabilization of both RNA and protein.76 This possibly also operates in the RNA-directed remodeling of HIV Rev protein by Rev Responsive Element (RRE) RNA.29,77,78
The RNA-mediated stabilization of folding intermediates has been suggested in ribozymes73 and in RNA binding mediated protein folding vehicles.79 Here, the TAR RNA ligand mediated Tat folding further addresses to molecular pathogenesis associated with HIV infection. It has been reported that, for a full-fledged transcriptional activation, Tat forms a multiple complex with host proteins including cyclin T1 and P-TEFb,42,80–83 and a dynamic role of TAR RNA has been suggested in this process.43,84 How the TAR acquired folding competence plays in this process remains to be further elucidated, although the basic and intrinsically disordered nature of Tat may well account for its ability to interact with RNAs and multiple cellular proteins.31 It should also be mentioned that there is no established assays so far available for the of folding of IDPs. The reporter folding assay used in the present report 45 directly reports on the folding of the EGFP portion, but does not precisely describe the folding status of Tat. The folding enhancement of the EGFP by TAR RNA is an outcome of inability of Tat from interfering with EGFP folding. The results are interpreted to mean that the TAR interaction with Tat at the TAR interaction domain would maintain the rest of Tat as ‘folding competent’ (Fig. 6), escaping from a kinetic trap into misfolding. This may enable its transition into a stabilized structure upon binding with cellular proteins.28,76 The proposed role of TAR on Tat is therefore reminiscent of molecular chaperones, which does not actively promote folding, but prevents aggregation of unfolded proteins.16 It is likely that the ‘entropy transfer’ between the IDR of Tat and TAR 28 could stabilize Tat from misfolding, and maintains its folding competence.
Figure 6.
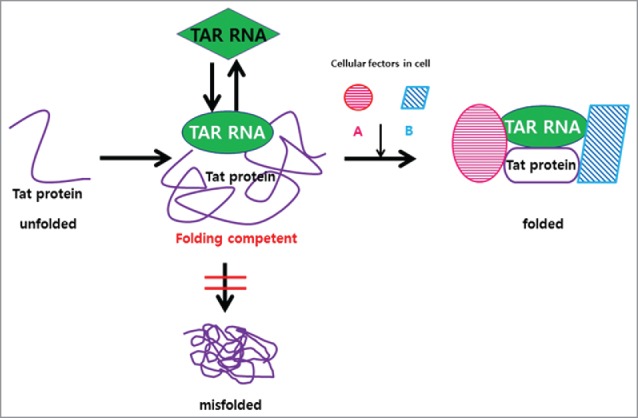
TAR RNA as molecular chaperone for Tat protein. The folding of the HIV virulence factor Tat protein is regulated by its interacting TAR RNA. The TAR–Tat interaction at the TAR interaction domain prevents Tat from misfolding and keeps it as ‘folding competent’ state, enabling Tat to further interact with cellular factors, e.g.,cyclin T1 (pink) and P-TEFb (blue) for transactivation.
In conclusion, consistent with intermolecular contact mediated intra-molecular stabilization of both binding partners, the HIV TAR RNA provides a chaperoning function to its interacting Tat protein (Fig. 6), and a crucial role of virally encoded RNA in establishing proteome link at the viral-host interface into successful viral replication in infected cells. We suggest that this aspect should be considered and reflected into current efforts on targeting HIV transcription for the intervention of HIV infection.32 The mechanism of RNA-mediated folding competence further enhances our understanding of chaperone-assisted protein folding inside the cells. The interference or modulation of RNA-mediated protein folding either by metabolic dysregulation or by infection into pathological consequences, remains to be further explored.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
JM Kim and HS Choi designed the research and conducted experiments. JM Kim wrote the paper. BL Seong discussed and analyzed the results. We thank Dr. Han Yong Lee at Purdue University for live cell experiments and valuable comments to the work, Soo Jeong Park and Dr. Joo Hee Jung at the KBSI (Seoul), Jae Won Kirk at Seoul National University, Young Mi Kang at Yonsei University College of Medicine for technical assistance and discussions.
Funding
This work was supported in part by the Brain Korea 21 (BK21) PLUS program (J. Kim), and the Grants from Korean Government (BL Seong): Ministry of Health and Welfare (HI13C0826), ICT and Future Planning, Ministry of Science (NRF-2014M3A9E4064580), and the Ministry of Agriculture, Food and Rural Affairs (MAFRA; 716002–7).
References
- 1.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature 2002; 418:222-8; PMID:12110898; https://doi.org/ 10.1038/418222a [DOI] [PubMed] [Google Scholar]
- 2.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982; 31:147-57; PMID:6297745; https://doi.org/ 10.1016/0092-8674(82)90414-7 [DOI] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983; 35:849-57; PMID:6197186; https://doi.org/ 10.1016/0092-8674(83)90117-4 [DOI] [PubMed] [Google Scholar]
- 4.Guerrier-Takada C, Altman S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science 1984; 223:285-6; PMID:6199841; https://doi.org/ 10.1126/science.6199841 [DOI] [PubMed] [Google Scholar]
- 5.Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 1981; 27:487-96; PMID:6101203; https://doi.org/ 10.1016/0092-8674(81)90390-1 [DOI] [PubMed] [Google Scholar]
- 6.Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol 2014; 15:749-60; PMID:25269475; https://doi.org/ 10.1038/nrm3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zlotorynski E. Non-coding RNA: Circular RNAs promote transcription. Nat Rev Mol Cell Biol 2015; 16:206; PMID:25714680; https://doi.org/ 10.1038/nrm3967 [DOI] [PubMed] [Google Scholar]
- 8.Fu XD. Non-coding RNA: a new frontier in regulatory biology. Natl Sci Rev 2014; 1:190-204; PMID:25821635; https://doi.org/ 10.1093/nsr/nwu008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calabretta S, Richard S. Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem Sci 2015; 40:662-72; PMID:26481498; https://doi.org/ 10.1016/j.tibs.2015.08.012 [DOI] [PubMed] [Google Scholar]
- 10.Hartl FU, Martin J, Neupert W. Protein folding in the cell - the role of molecular chaperones Hsp70 and Hsp60. Ann Rev Biophys Biomol Struct 1992; 21:293-322; PMID:1525471; https://doi.org/ 10.1146/annurev.bb.21.060192.001453 [DOI] [PubMed] [Google Scholar]
- 11.Borges JC, Ramos CH. Protein folding assisted by chaperones. Protein Pept Lett 2005; 12:257-61; PMID:15777275; https://doi.org/ 10.2174/0929866053587165 [DOI] [PubMed] [Google Scholar]
- 12.Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell 2010; 142:112-22; PMID:20603018; https://doi.org/ 10.1016/j.cell.2010.05.027 [DOI] [PubMed] [Google Scholar]
- 13.Clare DK, Vasishtan D, Stagg S, Quispe J, Farr GW, Topf M, Horwich AL, Saibil HR. ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell 2012; 149:113-23; PMID:22445172; https://doi.org/ 10.1016/j.cell.2012.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis RJ, Hartl FU. Protein folding in the cell: competing models of chaperonin function. FASEB J 1996; 10:20-6; PMID:8566542 [DOI] [PubMed] [Google Scholar]
- 15.Anfinsen CB. Principles that govern the folding of protein chains. Science 1973; 181:223-30; PMID:4124164; https://doi.org/ 10.1126/science.181.4096.223 [DOI] [PubMed] [Google Scholar]
- 16.Dobson CM. Protein folding and misfolding. Nature 2003; 426:884-90; PMID:14685248; https://doi.org/ 10.1038/nature02261 [DOI] [PubMed] [Google Scholar]
- 17.Hartl FU. Molecular chaperones in the cytosol: from nascent Chain to folded protein. Science 2002; 295:1852-8; PMID:11884745; https://doi.org/ 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- 18.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang H-C, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al.. Proteome-wide analysis of chaperonin-dependent protein folding in escherichia coli. Cell 2005; 122:209-20; PMID:16051146; https://doi.org/ 10.1016/j.cell.2005.05.028 [DOI] [PubMed] [Google Scholar]
- 19.Vorderwülbecke S, Kramer G, Merz F, Kurz TA, Rauch T, Zachmann-Brand B, Bukau B, Deuerling E. Low temperature or GroEL/ES overproduction permits growth of Escherichia coli cells lacking trigger factor and DnaK. FEBS Lett 2004; 559:181-7; PMID:14960329; https://doi.org/ 10.1016/S0014-5793(04)00052-3 [DOI] [PubMed] [Google Scholar]
- 20.Ullers RS. SecB is a bona fide generalized chaperone in Escherichia coli. Proc Natl Acad Sci U S A 2004; 101:7583-8; PMID:15128935; https://doi.org/ 10.1073/pnas.0402398101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi SI, Ryu K, Seong BL. RNA-mediated chaperone type for de novo protein folding. RNA Biol 2009; 6:21-4; PMID:19106620; https://doi.org/ 10.4161/rna.6.1.7441 [DOI] [PubMed] [Google Scholar]
- 22.Rentzeperis D, Jonsson T, Sauer RT. Acceleration of the refolding of Arc repressor by nucleic acids and other polyanions. Nat Struct Biol 1999; 6:569-73; PMID:10360363; https://doi.org/ 10.1038/9353 [DOI] [PubMed] [Google Scholar]
- 23.Docter BE, Horowitz S, Gray MJ, Jakob U, Bardwell JC. Do nucleic acids moonlight as molecular chaperones? Nucleic Acids Res 2016; 44:4835-45; PMID:27105849; https://doi.org/ 10.1093/nar/gkw291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudlicki W. Ribosomes and ribosomal RNA as chaperones for folding of proteins. Fold Des 1997; 2:101-8; PMID:9135982; https://doi.org/ 10.1016/S1359-0278(97)00014-X [DOI] [PubMed] [Google Scholar]
- 25.Choi SI, Han KS, Kim CW, Ryu KS, Kim BH, Kim KH, Kim SI, Kang TH, Shin HC, Lim KH, et al.. Protein solubility and folding enhancement by interaction with RNA. Plos One 2008; 3:e2677; PMID:18628952; https://doi.org/ 10.1371/journal.pone.0002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi SI, Ryu K, Seong BL. RNA-mediated chaperone type for de novo protein folding. RNA Biol 2009; 6:21-4; PMID:19106620; https://doi.org/ 10.4161/rna.6.1.7441 [DOI] [PubMed] [Google Scholar]
- 27.Uversky VN, Gillespie JR, Fink AL. Why are ‘natively unfolded' proteins unstructured under physiologic conditions? Proteins 2000; 41:415-27; ; PMID:11025552; https://doi.org/ 10.1002/1097-0134(20001115)41:3%3c415::AID-PROT130%3e3.0.CO;2-7 [DOI] [PubMed] [Google Scholar]
- 28.Frankel AD, Smith CA. Induced folding in RNA-protein recognition: more than a simple molecular handshake. Cell 1998; 92:149-51; PMID:9458038; https://doi.org/ 10.1016/S0092-8674(00)80908-3 [DOI] [PubMed] [Google Scholar]
- 29.Tan R, Frankel AD. Costabilization of peptide and RNA structure in an HIV Rev peptide-RRE complex. Biochemistry 1994; 33:14579-85; PMID:7981219; https://doi.org/ 10.1021/bi00252a025 [DOI] [PubMed] [Google Scholar]
- 30.Goldgur Y, Mosyak L, Reshetnikova L, Ankilova V, Lavrik O, Khodyreva S, Safro M. The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNA(Phe). Structure 1997; 5:59-68; PMID:9016717; https://doi.org/ 10.1016/S0969-2126(97)00166-4 [DOI] [PubMed] [Google Scholar]
- 31.Shojania S, O'Neil JD. HIV-1 Tat is a natively unfolded protein: the solution conformation and dynamics of reduced HIV-1 Tat-(1-72) by NMR spectroscopy. J Biol Chem 2006; 281:8347-56; PMID:16423825; https://doi.org/ 10.1074/jbc.M510748200 [DOI] [PubMed] [Google Scholar]
- 32.Mousseau G, Mediouni S, Valente ST. Targeting HIV transcription: the quest for a functional cure. Curr Top Microbiol Immunol 2015; 389:121-45; PMID:25731772; https://doi.org/ 10.1007/82_2015_435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuciak M, Gabus C, Ivanyi-Nagy R, Semrad K, Storchak R, Chaloin O, Muller S, Mély Y, Darlix JL. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res 2008; 36:3389-400; PMID:18442994; https://doi.org/ 10.1093/nar/gkn177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res 2008; 36:712-25; PMID:18033802; https://doi.org/ 10.1093/nar/gkm1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Arginine-mediated RNA recognition: The arginine fork. Science 1991; 252:1167-71; PMID:1709522; https://doi.org/ 10.1126/science.252.5009.1167 [DOI] [PubMed] [Google Scholar]
- 36.Frankel AD. Peptide models of the Tat-TAR protein-RNA interaction. Protein Sci 1992; 1:1539-42; PMID:1304886; https://doi.org/ 10.1002/pro.5560011202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weeks KM, Crothers DM. RNA recognition by Tat-derived peptides: Interaction in the major groove? Cell 1991; 66:577-88; PMID:1907891; https://doi.org/ 10.1016/0092-8674(81)90020-9 [DOI] [PubMed] [Google Scholar]
- 38.Karn J. Tackling tat. J Mol Biol 1999; 293:235-54; PMID:10550206; https://doi.org/ 10.1006/jmbi.1999.3060 [DOI] [PubMed] [Google Scholar]
- 39.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 1989; 59:273-82; PMID:2478293; https://doi.org/ 10.1016/0092-8674(89)90289-4 [DOI] [PubMed] [Google Scholar]
- 40.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J 1990; 9:4145-53; PMID:2249668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weeks KM, Ampe C, Schultz SC, Steitz TA, Crothers DM. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science 1990; 249:1281-5; PMID:2205002; https://doi.org/ 10.1126/science.2205002 [DOI] [PubMed] [Google Scholar]
- 42.Marozzi A, Meneveri R, Giacca M, Gutierrez MI, Siccardi AG, Ginelli E. In vitro selection of HIV-1 TAR variants by the Tat protein. J Biotechnol 1998; 61:117-28; PMID:9654745; https://doi.org/ 10.1016/S0168-1656(98)00017-0 [DOI] [PubMed] [Google Scholar]
- 43.Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 2010; 465:747-51; PMID:20535204; https://doi.org/ 10.1038/nature09131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uversky VN, Na I, Landau KS, Schenk RO. Highly disordered proteins in prostate cancer. Curr Protein Pept Sci 2016; PMID:27804860; https://doi.org/ 10.2174/1389203717666161028145848 [DOI] [PubMed] [Google Scholar]
- 45.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol 1999; 17:691-5; PMID:10404163; https://doi.org/ 10.1038/10904 [DOI] [PubMed] [Google Scholar]
- 46.Sacchetti A, Alberti S. Protein tags enhance GFP folding in eukaryotic cells. Nat Biotechnol 1999; 17:1046; PMID:10545870; https://doi.org/ 10.1038/14990 [DOI] [PubMed] [Google Scholar]
- 47.Cabantous S, Rogers Y, Terwilliger TC, Waldo GS. New molecular reporters for rapid protein folding assays. PloS One 2008; 3:e2387; PMID:18545698; https://doi.org/ 10.1371/journal.pone.0002387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zahler AM, Damgaard CK, Kjems J, Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem 2004; 279:10077-84; PMID:14703516; https://doi.org/ 10.1074/jbc.M312743200 [DOI] [PubMed] [Google Scholar]
- 49.De Marco A, Dans PD, Knezevich A, Maiuri P, Pantano S, Marcello A. Subcellular localization of the interaction between the human immunodeficiency virus transactivator Tat and the nucleosome assembly protein 1. Amino Acids 2010; 38:1583-93; PMID:19888548; https://doi.org/ 10.1007/s00726-009-0378-9 [DOI] [PubMed] [Google Scholar]
- 50.Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol 2000; 20:1063-71; PMID:10629063; https://doi.org/ 10.1128/MCB.20.3.1063-1071.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hallay H, Locker N, Ayadi L, Ropers D, Guittet E, Branlant C. Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 Tat exon 2 splicing site. J Biol Chem 2006; 281:37159-74; PMID:16990281; https://doi.org/ 10.1074/jbc.M603864200 [DOI] [PubMed] [Google Scholar]
- 52.Jablonski JA, Amelio AL, Giacca M, Caputi M. The transcriptional transactivator Tat selectively regulates viral splicing. Nucleic Acids Res 2010; 38:1249-60; PMID:19966273; https://doi.org/ 10.1093/nar/gkp1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pendergrast PS, Wang C, Hernandez N, Huang S. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-P-TEFb-containing nuclear speckles. Mol Biol Cell 2002; 13:915-29; PMID:11907272; https://doi.org/ 10.1091/mbc.01-08-0383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herrmann CH, Mancini MA. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J Cell Sci 2001; 114:1491-503; PMID:11282025 [DOI] [PubMed] [Google Scholar]
- 55.Cullen BR, Hauber J, Campbell K, Sodroski JG, Haseltine WA, Rosen CA. Subcellular localization of the human immunodeficiency virus trans-acting art gene product. J Virol 1988; 62:2498-501; PMID:2836628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 1990; 343:437-41; PMID:2137203; https://doi.org/ 10.1038/343437a0 [DOI] [PubMed] [Google Scholar]
- 57.Siomi H, Shida H, Maki M, Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol 1990; 64:1803-7; PMID:2108259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lallena MJ, Correas I. Transcription-dependent redistribution of nuclear protein 4.1 to SC35-enriched nuclear domains. J Cell Sci 1997; 110(Pt 2):239-47; PMID:9044054 [DOI] [PubMed] [Google Scholar]
- 59.Jacquenet S, Decimo D, Muriaux D, Darlix JL. Dual effect of the SR proteins ASF/SF2, SC35 and 9G8 on HIV-1 RNA splicing and virion production. Retrovirology 2005; 2:33; PMID:15907217; https://doi.org/ 10.1186/1742-4690-2-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 2008; 15:819-26; PMID:18641664; https://doi.org/ 10.1038/nsmb.1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu ST, Blaser G, Jackson SE. The folding, stability and conformational dynamics of beta-barrel fluorescent proteins. Chem Soc Rev 2009; 38:2951-65; PMID:19771338; https://doi.org/ 10.1039/b908170b [DOI] [PubMed] [Google Scholar]
- 62.Calnan BJ, Biancalana S, Hudson D, Frankel AD. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev 1991; 5:201-10; PMID:1899841; https://doi.org/ 10.1101/gad.5.2.201 [DOI] [PubMed] [Google Scholar]
- 63.Choi SI, Kwon S, Son A, Jeong H, Kim KH, Seong BL. Protein folding in vivo revisited. Curr Protein Pept Sci 2013; 14:721-33; PMID:24384034; https://doi.org/ 10.2174/138920371408131227170544 [DOI] [PubMed] [Google Scholar]
- 64.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 1989; 59:273-82; PMID:2478293; https://doi.org/ 10.1016/0092-8674(89)90289-4 [DOI] [PubMed] [Google Scholar]
- 65.Berkhout B, Jeang KT. Detailed mutational analysis of TAR RNA: critical spacing between the bulge and loop recognition domains. Nucleic Acids Res 1991; 19:6169-76; PMID:1956776; https://doi.org/ 10.1093/nar/19.22.6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J 1990; 9:4145-53; PMID:2249668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harper JW, Logsdon NJ. Refolded HIV-1 tat protein protects both bulge and loop nucleotides in TAR RNA from ribonucleolytic cleavage. Biochemistry 1991; 30:8060-6; PMID:1868081; https://doi.org/ 10.1021/bi00246a026 [DOI] [PubMed] [Google Scholar]
- 68.Roy S, Delling U, Chen CH, Rosen CA, Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev 1990; 4:1365-73; PMID:1720714; https://doi.org/ 10.1101/gad.4.8.1365 [DOI] [PubMed] [Google Scholar]
- 69.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature 2011; 475:324-32; PMID:21776078; https://doi.org/ 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- 70.Brandvold KR, Morimoto RI. The Chemical Biology of Molecular Chaperones–Implications for Modulation of Proteostasis. J Mol Biol 2015; 427:2931-47; PMID:26003923; https://doi.org/ 10.1016/j.jmb.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Biro JC. Nucleic acid chaperons: a theory of an RNA-assisted protein folding. Theor Biol Med Model 2005; 2:35; https://doi.org/ 10.1186/1742-4682-2-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry 2005; 44:4957-70; PMID:15794634; https://doi.org/ 10.1021/bi047314+ [DOI] [PubMed] [Google Scholar]
- 73.Son A, Choi SI, Han G, Seong BL. M1 RNA is important for the in-cell solubility of its cognate C5 protein: Implications for RNA-mediated protein folding. RNA Biol 2015; 12:1198-208; PMID:26517763; https://doi.org/ 10.1080/15476286.2015.1096487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamamoto R, Katahira M, Nishikawa S, Baba T, Taira K, Kumar PK. A novel RNA motif that binds efficiently and specifically to the Ttat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells 2000; 5:371-88; PMID:10886365; https://doi.org/ 10.1046/j.1365-2443.2000.00330.x [DOI] [PubMed] [Google Scholar]
- 75.Karn J. Tackling tat. J Mol Biol 1999; 293:235-54; PMID:10550206; https://doi.org/ 10.1006/jmbi.1999.3060 [DOI] [PubMed] [Google Scholar]
- 76.Calabro V, Daugherty MD, Frankel AD. A single intermolecular contact mediates intramolecular stabilization of both RNA and protein. Proc Natl Acad Sci U S A 2005; 102:6849-54; PMID:15857951; https://doi.org/ 10.1073/pnas.0409282102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fernandes J, Jayaraman B, Frankel A. The HIV-1 Rev response element: an RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex. RNA Biol 2012; 9:6-11; PMID:22258145; https://doi.org/ 10.4161/rna.9.1.18178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD. RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex. eLife 2014; 3:e04120; PMID:25486594; https://doi.org/ 10.7554/eLife.04120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi SI, Han KS, Kim CW, Ryu KS, Kim BH, Kim KH, Kim SI, Kang TH, Shin HC, Lim KH, et al.. Protein solubility and folding enhancement by interaction with RNA. PloS One 2008; 3:e2677; PMID:18628952; https://doi.org/ 10.1371/journal.pone.0002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 1998; 12:3512-27; PMID:9832504; https://doi.org/ 10.1101/gad.12.22.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J 1998; 17:3681-91; PMID:9649438; https://doi.org/ 10.1093/emboj/17.13.3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anand K, Schulte A, Vogel-Bachmayr K, Scheffzek K, Geyer M. Structural insights into the cyclin T1-Tat-TAR RNA transcription activation complex from EIAV. Nat Struct Mol Biol 2008; 15:1287-92; PMID:19029897; https://doi.org/ 10.1038/nsmb.1513 [DOI] [PubMed] [Google Scholar]
- 83.Anand K, Schulte A, Fujinaga K, Scheffzek K, Geyer M. Cyclin box structure of the P-TEFb subunit cyclin T1 derived from a fusion complex with EIAV tat. J Mol Biol 2007; 370:826-36; PMID:17540406; https://doi.org/ 10.1016/j.jmb.2007.04.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Russell RS, Liang C, Wainberg MA. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology 2004; 1:23; PMID:15345057; https://doi.org/ 10.1186/1742-4690-1-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.