ABSTRACT
Noncoding RNAs (ncRNAs), particularly microRNAs (miRNAs) and long ncRNAs (lncRNAs), are important players in diseases and emerge as novel drug targets. Thus, unraveling the relationships between ncRNAs and other biomedical entities in cells are critical for better understanding ncRNA roles that may eventually help develop their use in medicine. To support ncRNA research and facilitate retrieval of relevant information regarding miRNAs and lncRNAs from the plethora of published ncRNA-related research, we developed DES-ncRNA (www.cbrc.kaust.edu.sa/des_ncrna). DES-ncRNA is a knowledgebase containing text- and data-mined information from public scientific literature and other public resources. Exploration of mined information is enabled through terms and pairs of terms from 19 topic-specific dictionaries including, for example, antibiotics, toxins, drugs, enzymes, mutations, pathways, human genes and proteins, drug indications and side effects, mutations, diseases, etc. DES-ncRNA contains approximately 878,000 associations of terms from these dictionaries of which 36,222 (5,373) are with regards to miRNAs (lncRNAs). We provide several ways to explore information regarding ncRNAs to users including controlled generation of association networks as well as hypotheses generation. We show an example how DES-ncRNA can aid research on Alzheimer disease and suggest potential therapeutic role for Fasudil. DES-ncRNA is a powerful tool that can be used on its own or as a complement to the existing resources, to support research in human ncRNA. To our knowledge, this is the only knowledgebase dedicated to human miRNAs and lncRNAs derived primarily through literature-mining enabling exploration of a broad spectrum of associated biomedical entities, not paralleled by any other resource.
KEYWORDS: Noncoding RNA, microRNA, long noncoding RNA, knowledgebase, literature-mining, text-mining, data-mining, information integration, bioinformatics, Alzheimer disease
Abbreviations
- AD
Alzheimer disease
- AJAX
Asynchronous JavaScript and XML
- ChEBI
Chemical Entities of Biological Interest
- CSS
Cascading Style Sheets
- DES
Dragon Exploration System
- DO
Disease Ontology
- EC
Enzyme Commission
- FARNA
Functional Annotation of RNA transcripts
- FDR
False Discovery Rate
- GO
Gene Ontology
- HTML
HyperText Markup Language
- IntEnz
Integrated relational Enzyme database
- JSON
JavaScript Object Notation
- KB
KnowledgeBase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KOBAS
KO-Based Annotation System
- lncRNA
long ncRNA
- miRNA
microRNA
- ncRNA
Noncoding RNA
- NCBI
National Center for Biotechnology Information
- PANTHER
Protein ANalysis THrough Evolutionary Relationships
- SIDER
Side Effect Resource
- sncRNA
small ncRNAs
- SNP
Single Nucleotide Polymorphisms
- T3DB
Toxin and Toxin Target Database
- TcoF-DB
Transcription co-Factors DataBase
- tmVar
Text-Mining for sequence Variants
- XML
eXtensible Markup Language
Introduction
Noncoding RNAs (ncRNAs), compared with mRNAs, are RNA molecules that do not encode protein information. Many ncRNAs have unknown function and are transcribed from various places within the genome.1,2 Through high-throughput sequencing technologies a considerable number of ncRNAs have been identified, many more than expected. New members and classes of ncRNAs are frequently detected and found to play crucial roles in a wide variety of important biologic processes.3-7 Among them, microRNAs (miRNAs), small ∼22 nt in length ncRNAs and long noncoding RNAs (lncRNAs) that are typically longer than 200 nt, are 2 popular and well-studied classes, featuring large quantity, ubiquitous distribution, significant functions in gene expression and regulation, and close associations with various human diseases.7-12
To facilitate in-depth investigations of human miRNAs and lncRNAs, several databases have been developed for collecting and managing their sequences and annotations. For miRNAs, representative examples include miRBase,13 miRDB,14 miRTarBase15 and DASHR.16 Specially, miRBase is a central repository for miRNA sequence information, containing miRNAs from more than 200 species.13 miRDB is a database for miRNA target predictions and functional annotations, currently including 947,941 associations of predicted gene targets regulated by 2,588 human miRNAs.14 Unlike miRDB, miRTarBase is a database collecting experimentally validated miRNA-target interactions, containing 12,738 human genes targeted by 2,619 miRNAs.15 DASHR is a dedicated database for human small ncRNAs (sncRNA) and contains information on ∼48,000 human sncRNA genes.16 Likewise, there are also several representative examples for lncRNAs. NONCODE features a large collection of ncRNAs and contains 141,353 human lncRNA transcripts.17 DIANA-LncBase is a database indexing miRNA targets on ncRNAs and integrates an in silico predicted collection of ∼51,000 miRNA-lncRNA interaction pairs for human and mouse.18 LncRNA2Target stores lncRNA-“target gene” associations under the assumption that a gene is targeted by the lncRNA if the gene is differentially expressed after lncRNA knockdown or overexpression.19 LncRNAWiki, as a core resource in the BIG Data Center,20 is a wiki-based database for community curation of human lncRNAs, which currently integrates a total of 105,824 lncRNAs and computationally identified 9,387 lncRNAs that potentially encode short proteins.21 RNAcentral provides a single-entry point to access all types of ncRNA sequences that integrates information from 22 participating resources.22 However, there are many other related ncRNA databases of interest.23-29 It has been documented that roles of miRNAs and lncRNAs are critical for better understanding molecular regulation in humans that affects a variety of biologic processes.12,30-32 However, to get deeper insight into the roles that miRNA and lncRNA may have in living cells, it is helpful to be able to explore associations of these ncRNAs to other biomedical entities. Interesting examples of such resources include ChemiRs,33 miRegulome,34 DisGeNET,35 miRCancer,36 LncRNADisease37 and FARNA.38 Each of these resources deal with few and/or specific types of these associations and would benefit if broader spectrum of associations is enabled. This issue could be leveraged to some extent by literature text-mining.
However, efficient exploration of information in the biomedical field is not an easy task, as the volume of published information is substantial and is continuously growing. The sheer volume of associations/links between relevant biomedical terms further exacerbated the problem. One approach to tackle this issue is to create topic-specific knowledgebases (KBs) that contains pre-computed term associations and is equipped with built-in information exploration capabilities to ease the task for researchers. Several such topic-specific KBs for life science have been developed,39-49 where text analysis has been used for titles and abstracts of PubMed records. Although titles and abstracts of published scientific articles contain the highest information density,50 full-text articles provide significantly more information overall.51 To date, the use of text-mining to identify and extract term associations has been used in the field of ncRNA research (see for example.35, 36, 52, 53 Although these attempts at using literature-mining for ncRNA research have been effective in identifying very specific associations, the scope of these resources could be expanded to support broader information exploration.
To enable a more comprehensive exploration of pairs of associated terms related to human miRNAs and lncRNAs, we developed DES-ncRNA KB. This KB makes use of dictionaries that are pre-compiled and contain terms and phrases belonging to different thematic categories (e.g. pathways, genes, diseases, miRNAs, lncRNAs, chemicals, drugs, drug indications and side effects, etc.). These dictionaries are used to index text. DES-ncRNA enables exploration and discovery of statistically enriched terms from these dictionaries as found in the analyzed text, but more importantly the statistically enriched associations among the enriched terms. As the primary source of textual data, DES-ncRNA identified dictionary terms in titles and abstracts (retrieved from PubMed54), as well as open-access full-length articles (retrieved from PubMed Central,54 and BioMed Central55). Moreover, due to the importance of miRNAs and lncRNAs in studies of human diseases, relevant dictionaries have been included, so that users can explore different aspects of potential ncRNA roles in living cells to have better insights into their roles in diseases. To illustrate how DES-ncRNA can assist researchers in the ncRNA domain, we present an example related to Alzheimer disease.
To our knowledge, DES-ncRNA is the first KB dedicated to human miRNAs and lncRNAs derived through literature-mining that focuses on term associations, which has comprehensive information exploration capabilities, comprehensive spectrum of associations and offers users a variety of options to localize information of interest. There is no similar KB available for support of ncRNA research.
Materials and methods
Server architecture and underlying systems
The server architecture is based on a 3-tier model: data, logic and presentation tiers, implemented as shown in Fig. 1. The traditional SQL-based data tier is expanded with an NoSQL document storage. Technologies used are PostgreSQL and MongoDB.
Figure 1.
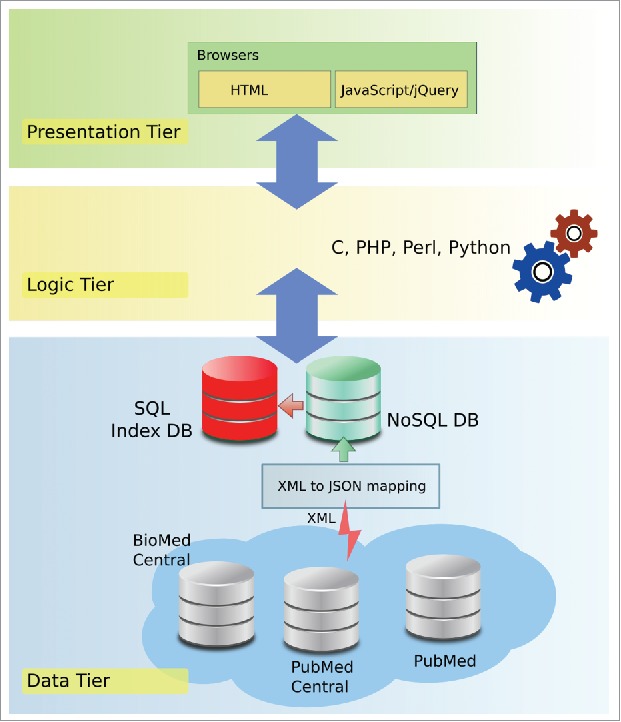
DES-ncRNA 3-tier server architecture.
NoSQL document storage is populated with publicly available biomedical literature from BioMed Central, PubMed and PubMed Central. Specifically written web-crawlers regularly gather data from these sources and provide cross-system data federalization. JSON (JavaScript Object Notation) format is used for data federalization since it has less syntactic overhead for similar amounts of information, is understood and processed by all tiers without much transformation and can be easily expanded if new data sources are included. NoSQL document storage was chosen since it is JSON-based, and as such has a flexible dynamic structure with no schema constraints. The resulting “Bio-Text” repository is fault tolerant, and horizontally scalable. However, a SQL database is still used for indexes related to the KB, since it has a more flexible and powerful query system for relational data.
The logic tier responds to user queries with data provided by the data tier. In addition, this tier handles data integration from SQL and NoSQL data sources (e.g., the literature view is constructed by fetching the relevant documents from the text repository (NoSQL) and annotating it using the index from the SQL store). Software modules in this tier respond to various AJAX (Asynchronous JavaScript and XML) calls from the presentation layer.
The presentation tier uses jQuery to provide separation between presentation related HTML/CSS code and server side logic. JQuery provides AJAX for asynchronous background calls to the logic tier, native JSON parsing, and dynamic rendering of the user's browser display.
Implementation and testing
DES-ncRNA is hosted on a CentOS-7 operating system using an Apache (2.4.6) web server. The literature repository is hosted on a MongoDB (2.6.11) database, and the KB index and related tables are stored in a PostgreSQL (9.2.15) database. DES-ncRNA was created using an Apache Lucene text index for fast querying of the text. Different components of the KB were developed using various programming languages/tools, namely: Java (openjdk 1.8.0_91), C/C++ (gcc 4.8.5), Perl (v5.16.3), PHP (5.4.16), JavaScript, and JQuery (3.0.0).
DES-ncRNA is functional across major web-browsers on Linux, Windows, and Mac OS platforms. It was specifically tested for Firefox, Chrome and Safari. DES-ncRNA was not tested for hand-held devices, and is not currently intended for such use.
Preparing the literature corpus
To create DES-ncRNA, we first queried our local literature repository, a MongoDB repository hosting PubMed, PubMed Central, and BioMed Central articles. The following DES-ncRNA query was used to create the ncRNA literature corpus: [((long AND (ncRNAs OR ncRNA OR “non-coding” OR noncoding OR “non coding”)) OR lncRNA OR lncRNAs OR “linc RNA” OR “linc RNAs” OR lincRNA OR lincRNAs OR microRNA OR miRNA OR microRNAs OR miRNAs OR “micro RNA” OR “micro RNAs”) AND (human OR humans OR “homo sapiens”)]. The query was made on 2016-12-25, and retrieved 31,074 articles.
Preparing the dictionaries
To ensure relevance and comprehensiveness, we used 19 dictionaries from the pre-existing DES v2.0 vocabularies (Table 1). Frequently, dictionary terms have several synonymous words/phrases. Where possible, we normalize them to the same internal identifier in DES-ncRNA. Such an approach allows for the universal identification of terms, for example using IDs from authoritative sources such as EntrezGene,56 ChEBI57 or UniProt ID,58 enables to complement text-mined information with information from such external sources as well as links to these sources.
Table 1.
List of dictionaries used in DES-ncRNA, with the number of terms that each dictionary contains and the number of statistically significantly enriched normalized terms identified in the analyzed documents.
| Dictionary | # of terms in dictionaries | # of statistically significant terms in documents | Source |
|---|---|---|---|
| Chemicals/Compounds | |||
| Antibiotics | 6,768 | 203 | pre-existing in DES |
| Chemical Entities of Biological Interest (ChEBI) | 164,419 | 3,943 | pre-existing in DES |
| Drugs (DrugBank + Chembl) | 40,131 | 1,563 | updated from Chembl |
| Enzymes (IntEnz) | 29,993 | 1,192 | pre-existing in DES |
| Metabolites (MetaboLights) | 59,569 | 1,139 | pre-existing in DES |
| Toxins (T3DB) | 47,140 | 728 | pre-existing in DES |
| Functional Annotation | |||
| Biological Process (GO) | 27,801 | 2,816 | pre-existing in DES |
| Cellular Component (GO) | 3,842 | 894 | pre-existing in DES |
| Disease Ontology (DO) | 23,553 | 1,701 | pre-existing in DES |
| Molecular Function (GO) | 10,796 | 717 | pre-existing in DES |
| Pathways (KEGG, Reactome, UniPathway, PANTHER) | 9,650 | 896 | pre-existing in DES |
| General | |||
| Drug Indications and Side Effects (SIDER) | 7,058 | 1,382 | Newly compiled |
| Human Anatomy | 7,167 | 1,476 | pre-existing in DES |
| Genes/Proteins/Transcripts | |||
| Human Genes & Proteins (EntrezGene) | 206,179 | 16,214 | pre-existing in DES |
| Human Long Non-Coding RNAs (FARNA) | 176,516 | 230 | Updated |
| Human MicroRNAs (miRBase) | 9,471 | 931 | Updated |
| Human Transcription Factors (TcoF-DB) | 12,280 | 1,273 | Updated |
| Human Transcription Co-Factors (TcoF-DB) | 3,850 | 345 | Updated |
| Mutations (tmVar) | 192,936 | 7,447 | pre-existing in DES |
References for the data sources indicated in Table 1 are as follows: ChEBI,57 DrugBank,66 Chembl,67 MetaboLights,68 IntEnz,69 T3DB,70 Industrially Important EnzymesEC,71,72 GO,73 KEGG,74 Reactome,75 PANTHER,76 UniPathways,77 EntrezGene,56 NCBI Taxonomy,78 KOBAS,79 FARNA,38 mirBase,13 TcoF-DB,80 tmVar,81 SIDER.59
Terms in these dictionaries are mined in the retrieved articles, highlighted and color-coded according to dictionary. This process is enabled by the back-end index that matches terms to their occurrences, up to the character level, within the mined articles. A term is defined as enriched when it is overrepresented in DES-ncRNA documents as compared with all PubMed, PubMed Central, and BioMed Central articles in our local repository. We used a false discovery rate (FDR) <0.05, which was calculated based on the Benjamini-Hochberg procedure to correct for multiplicity testing. Terms in all dictionaries are normalized, i.e. names, symbols and synonyms referring to the same concept are represented by a single entity when analyzed.
Results
Knowledgebase statistics
Table 1 shows the dictionaries used to mine the text documents, as well as the statistically significantly enriched unique terms found in these documents. While there are only 45,090 unique normalized statistically enriched terms identified from 1,039,119 terms contained in the used dictionaries, the number of statistically significantly enriched pairs of these terms are 877,977. Table 2 summarizes the associations between miRNAs and lncRNAs with terms from 19 dictionaries.
Table 2.
Statistically significantly enriched pairs of terms as identified in the analyzed set of documents (pairs with FDR < = 0.05), when one member of the pair is miRNA or lncRNA.
| Dictionary | # of statistically significantly enriched pairs of terms containing miRNAs | # of statistically significantly enriched pairs of terms containing lncRNAs |
|---|---|---|
| Chemicals/Compounds | ||
| Antibiotics | 32 | 10 |
| Chemical Entities of Biological Interest (ChEBI) | 728 | 152 |
| Drugs (DrugBank + Chembl) | 357 | 56 |
| Enzymes (IntEnz) | 267 | 60 |
| Metabolites (MetaboLights) | 234 | 51 |
| Toxins (T3DB) | 236 | 57 |
| Functional Annotation | ||
| Biological Process (GO) | 913 | 102 |
| Cellular Component (GO) | 111 | 84 |
| Disease Ontology (DO) | 1,412 | 274 |
| Molecular Function (GO) | 112 | 43 |
| Pathways (KEGG, Reactome, UniPathway, PANTHER) | 518 | 71 |
| General | ||
| Drug Indications and Side Effects (SIDER) | 1,048 | 202 |
| Human Anatomy | 1,099 | 241 |
| Genes/Proteins/Transcripts | ||
| Human Genes & Proteins (EntrezGene) | 7,303 | 3,055 |
| Human Long Non-Coding RNAs (FARNA) | 70 | 364 |
| Human MicroRNAs (miRBase) | 19,724 | 70 |
| Human Transcription Co-Factors (TcoF-DB) | 268 | 56 |
| Human Transcription Factors (TcoF-DB) | 1,123 | 345 |
| Mutations (tmVar) | 192,936 | 7,447 |
| Total | 36,222 | 5373 |
| Searchable records (includes redundant inverse pairs for the same dictionary associations, i.e., for miRNA-miRNA and lncRNA-lncRNA associations) | 36,222+19,724 = 55,966 | 5,373 + 364 = 5,737 |
Using the identified human genes ad proteins that statistically significantly enriched, we determined GO, Reactome pathway, KOBAS pathway and KOBAS disease terms to which these genes and proteins are associated. The results are summarized in Table 3. These mapped entities are not necessarily present in the text documents analyzed.
Table 3.
Mapped entities from GO, Reactome and KOBAS resources.
| # of total inferred hits | # of statistically enriched inferred hits | |
|---|---|---|
| GO Terms | 12,755 | 2,893 |
| Reactome Pathways | 693 | 313 |
| KOBAS Pathways | 2,827 | 825 |
| KOBAS Diseases | 10,366 | 178 |
Utilization
DES-ncRNA provides users with tools that allow them to easily explore literature using statistically enriched single terms and pairs of terms, as well as potential hypotheses. These exploration tools are located in the main menu on the top of the DES-ncRNA homepage, and include “Enriched Terms,” “Enriched Term Pairs,” “Explore Hypotheses,” “GO Terms,” Reactome Pathways,” “KOBAS Pathways,” “KOBAS Diseases” and “Show Literature.”
An in-depth description of each exploration tool was previously detailed in Salhi et al.59 Briefly, the “Enriched Terms” option allows users to mine ncRNA-related literature using single biologic terms/keywords (such as XIST, HOTAIR, MIR21, plasma membrane, bone marrow etc.) organized into thematic dictionaries. The “Enriched Term Pairs” tool allows users in-depth literature-mining for pairs of biologic terms/keywords co-occurring in the same text (identified at the title and abstract level of all available documents, and at sentence level in the remaining full-text document if available) thereby inferring possible biologic connections. The third tool, “Explore Hypotheses,” can be used to check if the inferred biologic connections/associations in “Enriched Term Pairs” are known or are novel and can serve as a starting point for further investigation. Each of these exploration tools has a “Help” link that provides simple instruction on how to use the tool. Exploration via these tools allow users to view enriched terms in pre-compiled theme-based dictionaries that can be viewed and restricted by several types of ranking options. Additionally, when a user hovers the mouse pointer over a term, a hover box is activated with “Network,” “Term Co-occurrences” and “Term Link Sources” links that are generated for the term of interest.
On the pages for “Enriched Terms,” “Enriched Term Pairs,” and “Explore Hypotheses,” we added a link to FARNA, when the term in question is a miRNA or lncRNA, where users can find more information and a comprehensive annotation for the ncRNA. This is achieved through FARNA's RESTful API, where the ncRNA is used as the query term. However, it is worth noting that not all ncRNAs identified within DES-ncRNA have annotations in FARNA.
“KOBAS Pathways” integrates several pathway databases including “Reactome Pathways,” however, due to the large number of metabolic pathways for more than 2500 organisms, metabolic pathways in humans (in “Reactome Pathways”) are usually not enriched. In DES-ncRNA we provide “Reactome Pathways,” as a separate exploration tool to ensure that metabolic pathways in humans are fairly enriched as several ncRNAs have been linked to disease states. Accordingly, “GO Terms” and “KOBAS Diseases” are also presented as exploration tools. Using “GO Terms,” Reactome Pathways,” “KOBAS Pathways” and “KOBAS Diseases” tools allows users to easily identify GO terms, pathways and diseases frequently associated with ncRNAs. The basic functionalities of the DES-ncRNA are demonstrated in a short introductory video on the “Home” page, and a detailed “Software Manual” can be downloaded from the website.
Case study that illustrate the usefulness of DES-ncRNA as an effective research support system: Progression of Alzheimer disease and potential therapeutic use of Fasudil
Using DES-ncRNA we can extract information regarding a specific disease, such as Alzheimer disease (AD), and the suggested involvement of ncRNAs in the disease pathology for exploring new target strategies. Here, we will explore the associations of ncRNAs and their role in AD. Deciphering molecular signatures triggered by the amyloid cascade that plays important role in AD has been a daunting task due to the complexity of the regulatory networks involving transcription factors and other regulatory proteins, ncRNAs, and numerous interactions between these entities.60
To explore potential ncRNAs involved in this process, we start by clicking “Enriched Term Pairs” (Fig. 2, Step 1). This opens a page with all enriched associated terms for all dictionaries. However, since the miRNA miR-485-5p transcript is known to be deregulated in AD patients,61 we filter the first dictionary (term A) for miR-485-5p by typing mir485 in the white input field above the “Select the first dictionary” button. The top 2 scored associated terms build the pairs ‘mir485’-‘BACE1’ and ‘mir485’-‘BACE1-AS’ (Fig. 2, Step 2). Both terms ‘BACE1’ and ‘BACE1-AS’ are known to play key roles in AD progression. Since the focus of this exploration is ncRNAs, we chose to expand our search through ‘BACE1-AS’. In the “Network” window, we clicked on “Dictionaries” and checked off the “Disease Ontology,” “Human Genes and Proteins” and “Human MicroRNAs” dictionaries we right clicked on ‘BACE1-AS’, and selected “Expand from the term” (Fig. 2, Step 2). We then repeated this process by expanding from ‘MIR485’, the only miRNA retrieved with ‘BACE1-AS’, using the “Disease Ontology,” “Human Genes and Proteins” and “Human Long Non-Coding RNAs” dictionaries. All nodes retrieved from ‘MIR485’ were further expanded with the “Cellular Component,” “Disease Ontology” and “Human Genes and Proteins” dictionaries. The resulting network was simplified by removing nodes with a single link (Fig. 2, Step 3).
Figure 2.
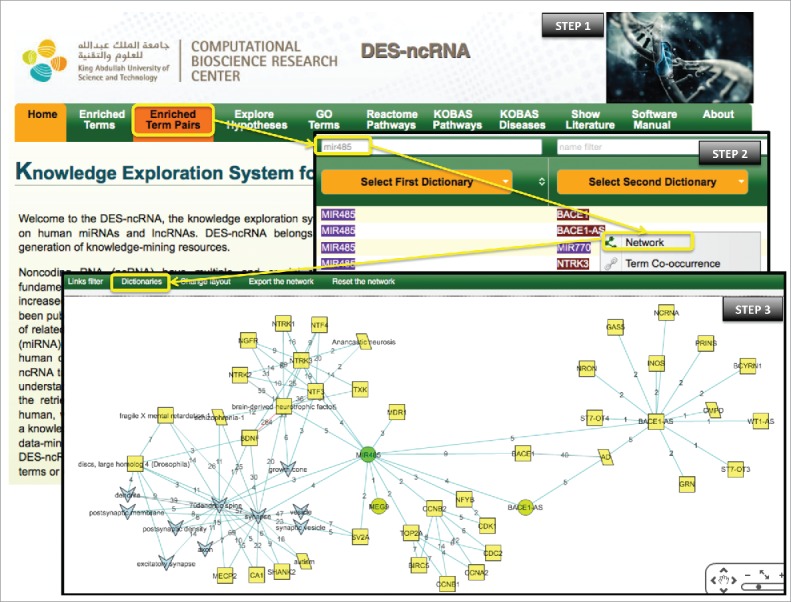
Step-by-step illustration of how DES-ncRNA can be used to identify ncRNA-related components involved in AD progression. The green circles represent the “Human MicroRNAs” dictionary; the gray upside-down triangles represent the “Cellular Component” dictionary; the yellow parallelograms represent ““Disease Ontology”” dictionary; the yellow squares represent the “Human Genes and Proteins” dictionary; and the lime circles represent “Human Long Non-Coding RNAs” dictionary. The edge color is distributed across a color spectrum from hot/red (high frequency co-occurrence/strong association) to cold/blue (small number of co-occurrences, weaker association). The numbers on the edges provide the number of publications that link the associated nodes.
The final network is clearly divided into 2 sub-networks; one is centered on ‘MIR485’ while the other is centered on ‘BACE1-AS’ (Fig. 1), which is expected as BACE1-AS prevents miRNA- induced repression of BACE1 by masking the binding site for miR-485-5p. Both, BACE1-AS and miR-485-5p are deregulated in RNA samples from AD patients,61 however their opposing roles suggest that they each part of separate smaller networks focused on a common goal. With in the ‘MIR485’ subnetwork, smaller networks centered on ‘NTRK3’ and ‘discs, large homolog 4 (Drosophila)’ are visible. The ‘discs, large homolog 4 (Drosophila)’ (also known as PSD-95 or SAP-90) is a molecule known to regulate synaptic plasticity, thus its impairment is crucial to memory symptomatology in AD.62 Also, PSD-95 was demonstrated to play a key role in aging and other psychiatric disorders, thus making it an interesting molecule for chemical management.62 Moreover, a correlation was found between miR-485 and increased expression at pre synapses and prevented clustering of PSD-95, a post-synaptic protein.63 Mouse models have revealed that the levels of PSD-95 are reduced by Aβ deposition in brain vulnerable arias, in excitatory synapses in the hippocampus.64 Similar relevant connections can be made for ‘NTRK3’ (not discussed).
To possibly address a therapeutic strategy given by our query, we found that Fasudil may act on more than 2 targets, in our case, the BACE and PSD-95 protein.65 Using the APP/PS1 Tg mice model Yu et al.65 demonstrated that Fasudil decreases levels of BACE and increases levels of PSD-95 suggesting that using the multitarget approach is an appropriate strategy to pursue. Fasudil also inhibited several inflammatory factors, suggesting its role in neuroprotection and immunomodulation.65
These results suggest a hypothesis that repression of neurogenesis “at the right time” may help to delay the onset of AD. This also suggests the importance of finding early biomarkers for increased neurogenesis in non-symptomatic stages of dementia.
Discussions and concluding remarks
DES-ncRNA is able to extract specific information on links between terms form 19 topic-specific dictionaries from these documents, as well as to link the extracted information to information in external resources. Such inferred information may not necessarily be present in the analyzed text. Overall, with the provided exploration interface and several tools integrated, DES-ncRNA can support exploration of many research questions that ncRNA researchers may have. It is worth mentioning that no similar system exists for support of miRNA and lncRNA research in humans.
With the increasing complexity of rapidly growing information corpus related to molecular processes in living cells and the control mechanisms they are subjected to, it becomes more and more difficult to analyze that information. This is even more emphasized in relation to diseases and the understanding of their molecular functioning, as this would help in developing efficient diagnosis and treatment. The miRNA and lncRNA research fields are relatively new. However, these molecules have been found to be critical components in regulating fundamental cellular processes and many diseases. With the advent of advanced experimental high-throughput technologies, the volume of information derived through them is already enormous and is quickly increasing. It appears infeasible even for a large research group to follow all developments even in a selected topic. Thus, there is a need for tools that can assist researchers in summarizing the key information on selected topics and enabling them to explore potential associations of terms as suggested by the mined text. For this reason we believe that systems such as DES-ncRNA will be of help to the ncRNA research community. DES-ncRNA allows for linking numerous types of entities (as defined in the dictionaries used), generating association networks of these entities, and allows for hypotheses exploration that no other system currently provides.
Nonetheless, it should be acknowledged that every existing resource aimed at supporting research in particular fields of science has numerous limitations. DES-ncRNA has the same types of shortcomings as other text-mining-based resources. For example, it is confined to information presented in the available documents. It should be noted that many of the full-text documents are not allowed for text-mining, which also limits what information can be extracted. In addition, all text-mining systems are far from being able to extract all useful information from the available texts. This is a field that will require significant improvements.
For future developments of DES-ncRNA system, we intend to expand it by additional types of ncRNAs and the construction of ncRNA regulatory networks based on large-scale data sets and high-volume scientific literature. DES-ncRNA will be updated regularly.
Availability and requirements
DES-ncRNA is free for academic and nonprofit users and can be accessed through the portal (www.cbrc.kaust.edu.sa/des_ncrna). Users can access the KB using any of the mainstream web browsers, including Firefox, Safari and Chrome.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The computational analysis for this study was performed on Dragon and Snapdragon compute clusters of the Computational Bioscience Research Center (CBRC) at King Abdullah University of Science and Technology (KAUST). The King Abdullah University of Science and Technology (KAUST) Base Research Funds [BAS/1/1606-01-01] to VBB supported research reported in this publication.
Funding
This work was also supported by grants from Strategic Priority Research Program of the Chinese Academy of Sciences [XDB13040500 to ZZ], International Partnership Program of the Chinese Academy of Sciences [153F11KYSB20160008], National Programs for High Technology Research and Development [2015AA020108 to ZZ] and The 100 Talent Program of the Chinese Academy of Sciences (to ZZ). Ministry of Education, Science and Technological Development of the Republic of Serbia, Project No 173034 support VPB.
Author contributions
V.B.B. and M.E. conceived the study; V.B.B and Z.Z. designed the study; A.S. conducted the main technical development; A.R. and B.M. worked on some aspects of technical implementation; T.A. and S.S. updated dictionaries; V.P.B. and M.E. developed the example; V.B.B., A.S., M.E., T.A., V.P.B., A.R., L.M. S.S. and Z.Z. wrote the paper.
References
- 1.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol 2010; 8:e1000371; PMID:20502517; https://doi.org/ 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takemata N, Ohta K. Role of non-coding RNA transcription around gene regulatory elements in transcription factor recruitment. RNA Biol 2016; 14:1-5; PMID:27763805; https://doi.org/ 10.1080/15476286.2016.1248020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013; 10:924-33; PMID:23696037; https://doi.org/ 10.4161/rna.24604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011; 21:354-61; PMID:21550244; https://doi.org/ 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 5.Findeiß S, Schmidtke C, Stadler PF, Bonas U. A novel family of plasmid-transferred anti-sense ncRNAs. RNA Biol 2010; 7:120-4; PMID:20220307; https://doi.org/ 10.4161/rna.7.2.11184 [DOI] [PubMed] [Google Scholar]
- 6.Pircher A, Gebetsberger J, Polacek N. Ribosome-associated ncRNAs: An emerging class of translation regulators. RNA Biol 2014; 11:1335-9; PMID:25692232; https://doi.org/ 10.1080/15476286.2014.996459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit. Rev. Biochem. Mol. Biol 2014; 49:16-32; PMID:24164576; https://doi.org/ 10.3109/10409238.2013.844092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J. Pathol 2010; 220:126-39; PMID:19882673; https://doi.org/ 10.1002/path.2638 [DOI] [PubMed] [Google Scholar]
- 9.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016; 539:452-5; PMID:27783602; https://doi.org/ 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain S, Thakkar N, Chhatai J, Bhadra MP, Bhadra U. Long non-coding RNA: Functional agent for disease traits. RNA Biol 2016; 26:1-14; PMID:27229269; https://doi.org/ 10.1080/15476286.2016.1172756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huynh NP, Anderson B, Guilak F, McAlinden A. Emerging roles for long non-coding RNAs in skeletal biology and disease. Connect Tissue Res 2016; 58:116-141; PMID:27254479; https://doi.org/ 10.1080/03008207.2016.1194406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Roosbroeck K, Pollet J, Calin GA. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev Mol Diagn 2013; 13:183-204; PMID:23477558; https://doi.org/ 10.1586/erm.12.134 [DOI] [PubMed] [Google Scholar]
- 13.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014; 42:D68-D73; PMID:24275495; https://doi.org/ 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2014; 43:gku1104; PMID:25378301; https://doi.org/ 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou C-H, Chang N-W, Shrestha S, Hsu S-D, Lin Y-L, Lee W-H, Yang CD, Hong HC, Wei TY, Tu SJ, et al.. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016; 44:D239-D47; PMID:26590260; https://doi.org/ 10.1093/nar/gkv1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung YY, Kuksa PP, Amlie-Wolf A, Valladares O, Ungar LH, Kannan S, Gregory BD, Wang LS. DASHR: Database of small human noncoding RNAs. Nucleic Acids Res 2015; 44:gkv1188; PMID:26553799; https://doi.org/ 10.1093/nar/gkv1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res 2014; 42:D98-D103; PMID:24285305; https://doi.org/ 10.1093/nar/gkt1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T, et al.. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res 2016; 44:D231-D8; PMID:26612864; https://doi.org/ 10.1093/nar/gkv1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Q, Wang J, Wu X, Ma R, Zhang T, Jin S, et al.. LncRNA2Target: A database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res 2015; 43:D193-D6; PMID:25399422; https://doi.org/ 10.1093/nar/gku1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.BIG DCM The BIG Data Center: From deposition to integration to translation. Nucleic Acids Res 2016; 45:D18-D24; PMID:27899658; https://doi.org/ 10.1093/nar/gkw1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Li A, Zou D, Xu X, Xia L, Yu J, Bajic VB, Zhang Z. LncRNAWiki: Harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res 2014; 43:gku1167; PMID:25399417; https://doi.org/ 10.1093/nar/gku1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium R. RNAcentral: A comprehensive database of non-coding RNA sequences. Nucleic Acids Res 2016; 45:gkw1008; PMID:27794554; https://doi.org/ 10.1093/nar/gkw1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volders P-J, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P. LNCipedia: A database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res 2013; 41:D246-D51; PMID:23042674; https://doi.org/ 10.1093/nar/gks915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res 2011; 39:D146-D51; PMID:21112873; https://doi.org/ 10.1093/nar/gkq1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 2012; 41:gks1005; PMID:23125362; https://doi.org/ 10.1093/nar/gks1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denisenko E, Ho D, Tamgue O, Ozturk M, Suzuki H, Brombacher F, et al.. IRNdb: The database of immunologically relevant non-coding RNAs. bioRxiv 2016:037911; https://doi.org/ 10.1093/database/baw138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J-H, Li J-H, Shao P, Zhou H, Chen Y-Q, Qu L-H. starBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 2011; 39:D202-D9; PMID:21037263; https://doi.org/ 10.1093/nar/gkq1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou K-R, Liu S, Sun W-J, Zheng L-L, Zhou H, Yang J-H, Qu LH. ChIPBase v2. 0: Decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res 2016; 45:gkw965; PMID:27924033; https://doi.org/ 10.1093/nar/gkw965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L-L, Li J-H, Wu J, Sun W-J, Liu S, Wang Z-L, Zhou H, Yang JH, Qu LH. deepBase v2. 0: Identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res 2015; 44:gkv1273; PMID:26590255; https://doi.org/ 10.1093/nar/gkv1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia W, Chen W, Kang J. The functions of microRNAs and long non-coding RNAs in embryonic and induced pluripotent stem cells. Genomics Proteomics Bioinformatics 2013; 11:275-83; PMID:24096129; https://doi.org/ 10.1016/j.gpb.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M-T, Lin H-S, Shen C, Ma Y-N, Wang F, Zhao H-L, Yu J, Zhang JW. PU. 1-regulated long noncoding RNA lnc-MC controls human monocyte/macrophage differentiation through interaction with microRNA 199a-5p. Mol. Cell Biol 2015; 35:3212-24; PMID:26149389; https://doi.org/ 10.1128/MCB.00429-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010; 38:5366-83; PMID:20423907; https://doi.org/ 10.1093/nar/gkq285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su EC-Y, Chen Y-S, Tien Y-C, Liu J, Ho B-C, Yu S-L, Singh S. ChemiRs: A web application for microRNAs and chemicals. BMC Bioinformatics 2016; 17:1; PMID:26817711; https://doi.org/ 10.1186/s12859-015-0844-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barh D, Kamapantula B, Jain N, Nalluri J, Bhattacharya A, Juneja L, Barve N, Tiwari S, Miyoshi A, Azevedo V, et al.. miRegulome: A knowledge-base of miRNA regulomics and analysis. Sci Rep 2015; 5:12832; PMID:26243198; https://doi.org/ 10.1038/srep12832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F, Furlong LI. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015; 2015:bav028; PMID:25877637; https://doi.org/ 10.1093/database/bav028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie B, Ding Q, Han H, Wu D. miRCancer: A microRNA-cancer association database constructed by text mining on literature. Bioinformatics 2013; 29:btt014; PMID:23325619; https://doi.org/ 10.1093/bioinformatics/btt014 [DOI] [PubMed] [Google Scholar]
- 37.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res 2013; 41:D983-D6; PMID:23175614; https://doi.org/ 10.1093/nar/gks1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alam T, Uludag M, Essack M, Salhi A, Ashoor H, Hanks JB, Kapfer C, Mineta K, Gojobori T, Bajic VB. FARNA: Knowledgebase of inferred functions of non-coding RNA transcripts. Nucleic Acids Res 2016; 45:gkw973; PMID:27924038; https://doi.org/ 10.1093/nar/gkw973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawe AS, Radovanovic A, Kaur M, Sagar S, Seshadri SV, Schaefer U, Kamau AA, Christoffels A, Bajic VB. DESTAF: A database of text-mined associations for reproductive toxins potentially affecting human fertility. Reprod Toxicol 2012; 33:99-105; PMID:22198179; https://doi.org/ 10.1016/j.reprotox.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 40.Essack M, Radovanovic A, Bajic VB. Information exploration system for sickle cell disease and repurposing of hydroxyfasudil. PLoS One 2013; 8:e65190; PMID:23762313; https://doi.org/ 10.1371/journal.pone.0065190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Essack M, Radovanovic A, Schaefer U, Schmeier S, Seshadri SV, Christoffels A, Kaur M, Bajic VB. DDEC: Dragon database of genes implicated in esophageal cancer. BMC Cancer 2009; 9:219; PMID:19580656; https://doi.org/ 10.1186/1471-2407-9-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaur M, Radovanovic A, Essack M, Schaefer U, Maqungo M, Kibler T, Schmeier S, Christoffels A, Narasimhan K, Choolani M, et al.. Database for exploration of functional context of genes implicated in ovarian cancer. Nucleic Acids Res 2009; 37:D820-3; PMID:18790805; https://doi.org/ 10.1093/nar/gkn593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwofie SK, Radovanovic A, Sundararajan VS, Maqungo M, Christoffels A, Bajic VB. Dragon exploratory system on hepatitis C virus (DESHCV). Infect Genet Evol 2011; 11:734-9; PMID:21194573; https://doi.org/ 10.1016/j.meegid.2010.12.006 [DOI] [PubMed] [Google Scholar]
- 44.Kwofie SK, Schaefer U, Sundararajan VS, Bajic VB, Christoffels A. HCVpro: Hepatitis C virus protein interaction database. Infect Genet Evol 2011; 11:1971-7; PMID:21930248; https://doi.org/ 10.1016/j.meegid.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 45.Maqungo M, Kaur M, Kwofie SK, Radovanovic A, Schaefer U, Schmeier S, Oppon E, Christoffels A, Bajic VB. DDPC: Dragon database of genes associated with prostate cancer. Nucleic Acids Res 2011; 39:D980-5; PMID:20880996; https://doi.org/ 10.1093/nar/gkq849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sagar S, Kaur M, Dawe A, Seshadri SV, Christoffels A, Schaefer U, Radovanovic A, Bajic VB. DDESC: Dragon database for exploration of sodium channels in human. BMC Genomics 2008; 9:622; PMID:19099596; https://doi.org/ 10.1186/1471-2164-9-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sagar S, Kaur M, Radovanovic A, Bajic VB. Dragon exploration system on marine sponge compounds interactions. J Cheminform 2013; 5:11; PMID:23415072; https://doi.org/ 10.1186/1758-2946-5-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salhi A, Essack M, Radovanovic A, Marchand B, Bougouffa S, Antunes A, Simoes MF, Lafi FF, Motwalli OA, Bokhari A, et al.. DESM: Portal for microbial knowledge exploration systems. Nucleic Acids Res 2016; 44:D624-33; PMID:26546514; https://doi.org/ 10.1093/nar/gkv1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bajic VB, Veronika M, Veladandi PS, Meka A, Heng MW, Rajaraman K, Pan H, Swarup S. Dragon plant biology explorer. A text-mining tool for integrating associations between genetic and biochemical entities with genome annotation and biochemical terms lists. Plant Physiol 2005; 138:1914-25; PMID:16172098; https://doi.org/ 10.1104/pp.105.060863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuemie MJ, Weeber M, Schijvenaars BJ, van Mulligen EM, van der Eijk CC, Jelier R, Mons B, Kors JA. Distribution of information in biomedical abstracts and full-text publications. Bioinformatics 2004; 20:2597-604; PMID:15130936; https://doi.org/ 10.1093/bioinformatics/bth291 [DOI] [PubMed] [Google Scholar]
- 51.Shah PK, Perez-Iratxeta C, Bork P, Andrade MA. Information extraction from full text scientific articles: Where are the keywords?. BMC Bioinformatics 2003; 4:1; PMID:12513700; https://doi.org/ 10.1186/1471-2105-4-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li A, Zang Q, Sun D, Wang M. A text feature-based approach for literature mining of lncRNA-protein interactions. Neurocomputing 2016; 206:73-80; https://doi.org/ 10.1016/j.neucom.2015.11.110 [DOI] [Google Scholar]
- 53.Chen X, Yan CC, Luo C, Ji W, Zhang Y, Dai Q. Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Sci Rep 2015; 5:11338; PMID:26061969; https://doi.org/ 10.1038/srep11338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, et al.. Database resources of the national center for biotechnology. Nucleic Acids Res 2003; 31:28-33; PMID:12519941; https://doi.org/ 10.1093/nar/gkg033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler D. BioMed Central boosted by editorial board. Nature 2000; 405:384; PMID:10839505; https://doi.org/ 10.1038/35013218 [DOI] [PubMed] [Google Scholar]
- 56.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res 2005; 33:D54-D8; PMID:15608257; https://doi.org/ 10.1093/nar/gki031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hastings J, de Matos P, Dekker A, Ennis M, Harsha B, Kale N, Muthukrishnan V, Owen G, Turner S, Williams M, et al.. The ChEBI reference database and ontology for biologically relevant chemistry: Enhancements for 2013. Nucleic Acids Res 2013; 41:D456-D63; PMID:23180789; https://doi.org/ 10.1093/nar/gks1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Consortium U. Activities at the universal protein resource (UniProt). Nucleic Acids Res 2014; 42:D191-D8; PMID:24253303; https://doi.org/ 10.1093/nar/gkt1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuhn M, Letunic I, Jensen LJ, Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res 2015; 44:gkv1075; PMID:26481350; https://doi.org/ 10.1093/nar/gkv1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Follert P, Cremer H, Béclin C. MicroRNAs in brain development and function: A matter of flexibility and stability. Front Mol Neurosci 2014; 7:26; PMID:24570654; https://doi.org/ 10.3389/fnmol.2014.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol 2010; 11:1; PMID:20507594; https://doi.org/ 10.1186/gb-2010-11-5-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Savioz A, Leuba G, Vallet PG. A framework to understand the variations of PSD-95 expression in brain aging and in Alzheimer's disease. Ageing Res Rev 2014; 18:86-94; PMID:25264360; https://doi.org/ 10.1016/j.arr.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 63.Cohen JW, Louneva N, Han LY, Hodes GE, Wilson RS, Bennett DA, Lucki I, Arnold SE. Chronic corticosterone exposure alters postsynaptic protein levels of PSD‐95, NR1, and synaptopodin in the mouse brain. Synapse 2011; 65:763-70; PMID:21190219; https://doi.org/ 10.1002/syn.20900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao CY, Mirra SS, Sait HB, Sacktor TC, Sigurdsson EM. Postsynaptic degeneration as revealed by PSD-95 reduction occurs after advanced Aβ and tau pathology in transgenic mouse models of Alzheimer's disease. Acta Neuropathol 2011; 122:285-92; PMID:21630115; https://doi.org/ 10.1007/s00401-011-0843-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu J-Z. Multitarget therapeutic effect of Fasudil in APP/PS1 transgenic mice. CNS Neurol Disord Drug Targets 2016; 15; PMID:27401064; https://doi.org/ 10.2174/1871527315666160711104719 [DOI] [PubMed] [Google Scholar]
- 66.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al.. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res 2014; 42:D1091-D7; PMID:24203711; https://doi.org/ 10.1093/nar/gkt1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrián-Uhalte E, et al.. The ChEMBL database in 2017. Nucleic Acids Res 2016; 45:gkw1074; PMID:27899562; https://doi.org/ 10.1093/nar/gkw1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kale NS, Haug K, Conesa P, Jayseelan K, Moreno P, Rocca-Serra P, Nainala VC, Spicer RA, Williams M, Li X, et al.. MetaboLights: An open-access database repository for metabolomics data. Curr Protoc Bioinformatics 2016; 53:14.3.1-.3.8; PMID:27010336; https://doi.org/ 10.1002/0471250953.bi1413s53 [DOI] [PubMed] [Google Scholar]
- 69.Fleischmann A, Darsow M, Degtyarenko K, Fleischmann W, Boyce S, Axelsen KB, Bairoch A, Schomburg D, Tipton KF, Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res 2004; 32:D434-7; PMID:14681451; https://doi.org/ 10.1093/nar/gkh119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wishart D, Arndt D, Pon A, Sajed T, Guo AC, Djoumbou Y, Knox C, Wilson M, Liang Y, Grant J, et al.. T3DB: the toxic exposome database. Nucleic Acids Res 2015; 43:D928-34; PMID:25378312; https://doi.org/ 10.1093/nar/gku1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alam I, Antunes A, Kamau AA, Ba Alawi W, Kalkatawi M, Stingl U, Bajic VB. INDIGO - INtegrated data warehouse of microbial genomes with examples from the red sea extremophiles. PLoS One 2013; 8:e82210; PMID:24324765; https://doi.org/ 10.1371/journal.pone.0082210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res 2000; 28:304-5; PMID:10592255; https://doi.org/ 10.1093/nar/28.1.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Consortium GO. Gene ontology consortium: Going forward. Nucleic Acids Res 2015; 43:D1049-D56; PMID:25428369; https://doi.org/ 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanehisa M. Molecular network analysis of diseases and drugs in KEGG In: Mamitsuka H, DeLisi C, Kanehisa M, eds. Data Mining for Systems Biology: Methods and Protocols. Totowa, NJ: Humana Press, 2013:263-75. [DOI] [PubMed] [Google Scholar]
- 75.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al.. The Reactome pathway knowledgebase. Nucleic Acids Res 2014; 42:D472-D7; PMID:24243840; https://doi.org/ 10.1093/nar/gkt1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al.. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 2005; 33:D284-D8; PMID:15608197; https://doi.org/ 10.1093/nar/gki078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morgat A, Coissac E, Coudert E, Axelsen KB, Keller G, Bairoch A, Bridge A, Bougueleret L, Xenarios I, Viari A. UniPathway: a resource for the exploration and annotation of metabolic pathways. Nucleic Acids Res 2011; 40:D761-D9; PMID:22102589; https://doi.org/ 10.1093/nar/gkr1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Federhen S. The NCBI taxonomy database. Nucleic Acids Res 2012; 40:D136-43; PMID:22139910; https://doi.org/ 10.1093/nar/gkr1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011; 39:W316-W22; PMID:21715386; https://doi.org/ 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schaefer U, Schmeier S, Bajic VB. TcoF-DB: dragon database for human transcription co-factors and transcription factor interacting proteins. Nucleic Acids Res 2011; 39:D106-D10; PMID:20965969; https://doi.org/ 10.1093/nar/gkq945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei C-H, Harris BR, Kao H-Y, Lu Z. tmVar: a text mining approach for extracting sequence variants in biomedical literature. Bioinformatics 2013; 29:btt156; PMID:23564842; https://doi.org/ 10.1093/bioinformatics/btt156 [DOI] [PMC free article] [PubMed] [Google Scholar]


