ABSTRACT
The precise spatial and temporal regulation of gene expression orchestrates the many intricate processes during brain development. In the present study we examined the role of the brain-enriched microRNA-338 (miR-338) during mouse cortical development. Reduction of miR-338 levels in the developing mouse cortex, using a sequence-specific miR-sponge, resulted in a loss of neuronal polarity in the cortical plate and significantly reduced the number of neurons within this cortical layer. Conversely, miR-338 overexpression in developing mouse cortex increased the number of neurons, which exhibited a multipolar morphology. All together, our results raise the possibility for a direct role for this non-coding RNA, which was recently associated with schizophrenia, in the regulation of cortical neuronal polarity and layer placement.
KEYWORDS: Epigenetic gene regulation, in utero electroporation, neurodevelopment, neuronal migration, schizophrenia
Introduction
The cerebral cortex executes key brain functions, including the processing of sensory information, cognitive function and consciousness. Proliferation, migration, and differentiation of cortical progenitor cells during development generate functionally relevant neural networks for these critical tasks.1,2 The formation of these neural structures is tightly regulated and depends on the coordinated expression of a large number of genes.3 Moreover, accumulating evidence has implicated aberrant temporal or spatial gene expression patterns underlying the ethology of various neurodevelopmental disorders, such as autism spectrum disorder, schizophrenia and intellectual disability.3-6 MicroRNAs (miRs) have emerged as an important class of small non-coding RNAs (ncRNAs) that bind to complementary sequences within target transcripts followed by post-transcriptional regulation of mRNA stability and/or translation. miRs are expressed at high levels in the developing nervous system, and their expression is essential for the proper development of the brain, including the cortex.7-10 Indeed, miR-dependent regulation of gene networks in the brain has been shown to affect a wide set of cellular processes, which are important for cortical neurogenesis, neuronal survival and differentiation.11-15
Previous studies have identified the CNS-enriched miR-338 as a regulator of oligodendrocyte maturation and differentiation by repressing the transcription factors SOX6 and HES5.16 Furthermore, miR-338 controls axonal outgrowth in cortical and superior cervical ganglion (SCG) neurons through the regulation of axon guidance and nuclear-encoded mitochondrial genes, respectively.17,18 miR-338 is encoded within the intron of the AATK gene. Previously, we showed that miR-338 has the capacity to regulate the mRNA levels of its host-gene by binding to the AATK 3′ untranslated region (3′ UTR).19 Interestingly, AATK was identified as a regulator of axon growth, as well as dendrite formation and arborisation in cortical neurons.20,21 A recent study has linked this miR to neurodevelopmental deficits. The 22q11 deletion syndrome, which is associated with early-life behavioral abnormalities, and affecting individuals at high risk for the development of schizophrenia symptoms, is associated with age-related declines in miR-338–3p in the auditory thalamus. Levels of miR-338–3p were lower in the thalamus of individuals with schizophrenia compared with individuals of the same age and sex without the diagnosis. The decline was associated with an increase in Drd2 and reduced signaling in the circuit that links the thalamus and auditory cortex, a brain region implicated in auditory hallucination.22
Since miR-338 is abundantly expressed within the central nervous system, including the cortex,23 we aimed at delineating the in vivo function of miR-338 in early corticogenesis by modulating miR-338 expression using in utero electroporation (IUE)-mediated cortical gene transfer. Using this approach, the outcome of our investigation suggests that miR-338 overexpression results in an enhanced number of multipolar neurons within the upper layers of the cerebral cortex, whereas inhibition of miR-338 decreased the number of upper cortical layers neurons, which displayed a non-polar phenotype. Collectively, these findings support the notion that miR-338 functions as a novel regulator of early cortical neurodevelopment.
Results
Sequence-specific miR-sponge reduces endogenous miR-338 levels
To investigate the ability of miR-338 to modulate cortical development we used genetic tools to alter endogenous miR-338 expression levels. Previously, a miR-338 overexpression vector was used to enhance cellular miR-338 expression levels.19 To examine miR-338 overexpression following IUE, we introduced the precursor-miR-338 vector into the lateral ventricular wall of E13.5 mouse brains by IUE and analyzed the levels if this miR in the cortical tissue of electroporated brains at E17.5 using qRT-PCR. Introduction of miR-338 overexpression vector resulted in a 15.7% increase in the levels of mature miR-338 levels in developing mouse cortex, as compared with the levels of this miR in the mCherry control vector injected mouse cortex (Fig. 1A, mean 1.0 ± 0.0369 versus 1.158 ± 0.03743 relative expression; p = 0.016).
Figure 1.
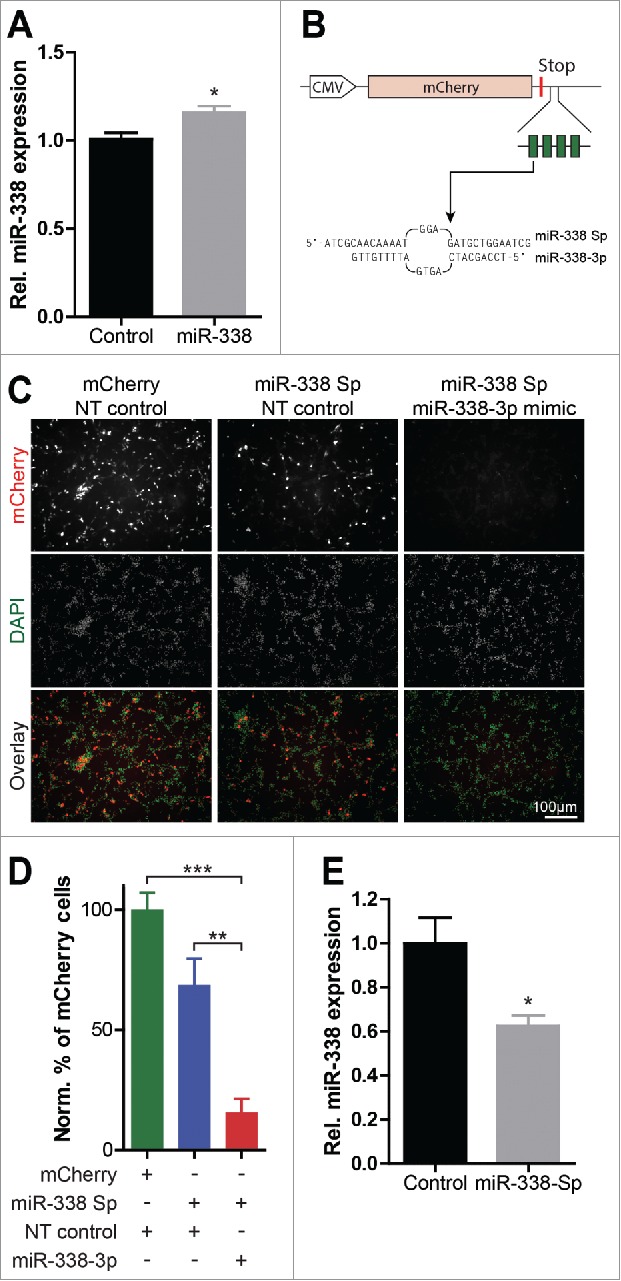
Characterization of the miR-338 overexpression vector and the miR-338-inhibiting sponge construct. (A) Analysis of miR-338 overexpression on mature miR-338 levels following IUE. Bars represent mean relative values to control with error bars ± s.e.m. Two-tailed unpaired student's t test, n = 6–7 samples; *P < 0.05. (B) The cytomegalovirus (CMV) promoter-driven miR-338-inhibiting sponge design. The 4 partially-complementary miR-338–3p binding sites are depicted as green blocks within the 3′ UTR of the mCherry gene. The sequence of both the miR-338 sponge (Sp) and mature miR-338–3p is shown, containing a central bulge to increase the efficiency of miR inhibition and a 4 nucleotide arbitrary linker sequence between each binding site. (C) Representative images of B35 neuroblastoma cells transfected with a control empty vector or the miR-338-sponge, co-transfected with a NT control or a miR-338 mimic to show the sensing capacity of this sponge for miR-338–3p activity. (D) Quantification of the percentage of cells displaying a fluorescent signal normalized to the total number of cells relative to the mCherry transfected control. Figure shows an expected decrease in the number of bright fluorescent cells under increasing levels of miR-338. Data represents mean percentage with error bars ± s.e.m. One-way ANOVA with Bonferroni multiple comparison test, average cell numbers collected from n = 3 experiments; ***P < 0.0001. (E) qPCR analysis of DIV 4 primary cortical neurons electroporated at DIV 1 with the miR-338 sponge or a GFP control vector. Bars represent mean relative values to control with error bars ± s.e.m. Two-tailed unpaired student's t test, n = 5 samples from independent experiments; *P < 0.05.
To inhibit miR-338 function, we used a mCherry-tagged miR-338 sponge vector, aimed at repressing mature miR-338–3p levels and function in developing cortex. The miR-338 sponge encoded a sequence comprising 4 partially complementary miR-338–3p binding sites and was embedded within the 3′ untranslated region (UTR) of the mCherry cDNA (Fig. 1B). To examine the functional selectivity of this sponge to sequester miR-338, we performed a miR-338 competition experiment in B35 rat neuroblastoma cell lines using synthetic miR-338 mimic and scramble control probes. B35 neuroblastoma cells were transfected with the control empty vector or the miR-338-sponge vector, co-transfected with either a non-targeting control or a miR-338 mimic, respectively, to verify the sensing capacity of the sponge for miR-338–3p activity (Fig. 1C). Cells co-transfected with the miR-338-sponge and miR-338 mimic showed reduced fluorescence signals as compared with the fluorescent signals observed in cells transfected with the sponge vector or the control vector. Relative quantification of the mCherry fluorescent signal as a mean of the total number of cells revealed a 50% decrease in the fluorescent signal in cells expressing the mCherry-tagged miR-338 sponge along with the miR-338 mimic, suggesting a high selectivity of the sponge to bind and repress miR-338 (Fig. 1D).
To examine the ability of the miR-338 sponge to repress miR-338–3p levels in neurons, dissociated primary cortical neurons were electroporated with the mCherry control or the miR-338 sponge vector. qPCR was used to examine the expression of miR-338 levels at 4 d in vitro (DIV). Electroporation of the miR-338 sponge resulted in a ∼40% decrease in mature miR-338 levels relative to the miR-338 levels in the mCherry eletroporated control cells (Fig. 1E, mean 1.0 ± 0.1160 vs. 0.6244 ± 0.04687 relative expression; p = 0.017). These results are in agreement with previous findings that miR-338 sponge efficiently reduces endogenous miR-338 levels in cortical neurons.18
In vivo modulation of miR-338 levels affects neuronal migration in the developing cortex
Since previous studies suggested that miR-338 might have the capacity to modulate axonal development in neurons, we aimed at determining the effects of miR-338 expression changes on cortical development. To examine the expression profile of miR-338 during brain development, total RNA was isolated from brain tissue obtained at several developmental time points, namely, embryonic (E) day E10.5, E13.5, E14.5, E15.5, E16.5, E18.5, and at postnatal day (P) 0, followed by qRT-PCR quantification of mature miR-338 levels at these time points. Levels of mature miR-338 were elevated 16-fold from E10.5 to E15.5, and remained at significantly high levels at later stages (Fig. 2A).
Figure 2.
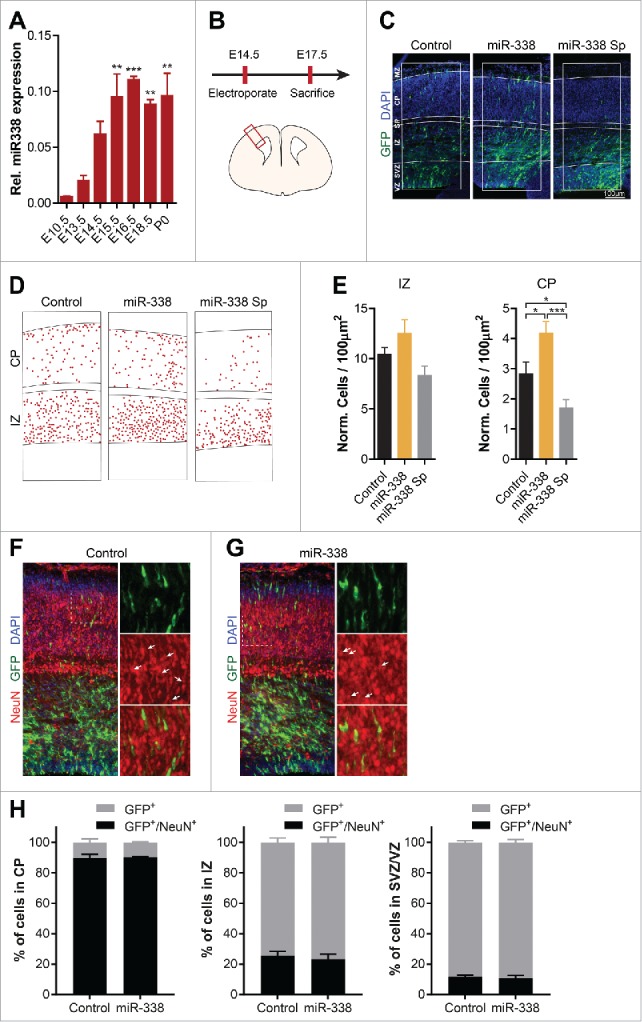
miR-338 affects neuronal migration within the developing cortex. (A) Expression level of mature miR-338 in the whole mouse brain from embryonic day 10.5 (E10.5) up to postnatal day 0 (P0). Data represents the relative fold change to E10.5 ± s.e.m. One-way ANOVA with Bonferroni multiple comparison test, n = 3 samples collected from independently isolated brains; ** P< 0.01, ***P < 0.001. (B) Outline of the performed IUE experiment and a graphic illustration of a cortical slice with the red box denoting the analyzed cortical area. (C) Representative micrographs of control GFP, miR-338 overexpression and miR-338 sponge in electroporated cortical slices with 300 µm wide masks highlighting the cortical layers ventricular zone (VZ); subventricular zone (SVZ); intermediate zone (IZ); subplate (SP); cortical plate (CP); marginal zone (MZ). The IUE targeted cells are depicted in green and DAPI nuclear staining is shown in blue. (D) Representative analysis of miR-338 overexpression and inhibition compared with the control with each dot defining a single neuron. (E) Quantification of IUE neurons within 100 µm2 IZ and CP corrected for the total number of cells. Bars represent mean relative numbers to control with error bars ± s.e.m. Two-tailed unpaired student's t test, n = 4–5 brains per condition from independent experiments; *P < 0.05 and ***P < 0.001. (F-G) Representative images of cortical slices electroporated with (F) a control GFP vector or (G) the miR-338 overexpresssion vector with DAPI in blue, GFP in red and NeuN depicted in red. The dotted white box illustrates the zoomed in areas on the right. The white arrows highlight electroporated cells which are positive for NeuN. (H) Quantification of the percentage of GFP-positive control or miR-338 electroporated cells within the CP, IZ and VZ/SVZ, which are positive for NeuN. The error bars represent ± s.e.m, n = 3 brains per condition from independent experiments.
Subsequently, the role of miR-338 in cortical development was examined by targeting ventricular proliferating cells in the somatosensory cortex with either a precursor miR-338 vector or a miR-338 sponge vector using IUE. As controls, corresponding empty precursor and sponge vectors were used. Mouse embryos were electroporated with vectors encoding control, miR-338-precursor or miR-338-sponge vectors at E14.5, and killed at E17.5 followed by coronal cryo-sectioning and GFP immuno-labeling (Fig. 2B, C). The number of fluorescently labeled cells in the intermediate zone (IZ) and in the cortical plate (CP) was quantified to assess the role of miR-338 in modulating the radial neuronal migration (Fig. 2D). This investigation revealed a modest increase in the number of neurons migrating toward the IZ when overexpressing miR-338. In mouse embryos with an attenuated miR-338 function, fewer neurons moved toward the IZ (Fig. 2E). Quantification of the number of GFP-positive cells that localized in the CP suggested a significant increase in the number of neurons overexpressing miR-338 (mean 2.840 ± 0.3756 vs. 4.188 ± 0.3788; p = 0.0449), whereas sponge-mediated miR-338 repression resulted in a significant decrease in the number of migrating cortical neurons toward the CP (mean 2.840 ± 0.3756 vs. 1.704 ± 0.2634; p = 0.0380) (Fig. 2E). This outcome raises the possibility that miR-338 expression can either modulate the number of neurons or affect the neuronal migration rate within the upper layers of the cerebral cortex.
To specifically examine if the changes in the number of cells seen in the CP are due to changes in the number of neurons, we used an antibody against NeuN, a neuronal biomarker, and monitored the co-localization of GFP-positive cells with NeuN (Fig. 2F, G). Quantitation of the cells revealed that over 80% of GFP-positive cells are immunostained for NeuN, suggesting that the majority of the electroporated cells are of neuronal origin. However, miR-338 overexpression failed to alter the percentage of cells that were positively labeled for the incorporation of NeuN in the CP. Moreover, overexpression of this miR did not result in changes in the number of neurons in the IZ and SVZ/VZ, as examined by NeuN immune-staining (Fig. 2H). Collectively, this analysis suggests that alteration of miR-338 level have no gross impact on the number of neurons.
miR-338 affects neuronal morphologies and polarity in vivo
During their radial migration phase, cortical neurons undergo extensive shape remodelling: while neurons within the IZ typically have a multipolar morphology, neurons located within the CP change their morphology to a bipolar state.24-26 It was previously shown that neuronal migration defects can affect the multipolar-bipolar transition of cortical neurons which is important for correct placement of these cells within the cortex.27 Due to the finding that miR-338 has the capacity to influence the migration of neurons toward the CP, we examined whether modulation of miR-338 expression alters the morphology of neurons within the CP. Using the same IUE protocol described above, morphological analysis of targeted cells was performed. This study revealed that miR-338 overexpression led to an increase in the total cell area (mean 98.08 ± 2.514 µm2 vs. 138.3 ± 4.239 µm2; p < 0.0001), whereas miR-338 repression decreased the cell area (mean 98.08 ± 2.514 vs. 64.20 ± 1.630; p < 0.0001) (Fig. 3A, B). Further analysis of these neurons suggested that miR-338-mediated changes resulted in altered neuronal polarity. miR-338 overexpression caused significantly more multipolar neurons (mean 0.07227 ± 0.01387 vs. 0.2301 ± 0.04054; p = 0.0021), with an increased number of neurons having 4–5 neurite endpoints (mean 0.05678 ± 0.01596 vs. 0.1518 ± 0.01341; p = 0.0006). miR-338 inhibition produced more rounded, nonpolar neurons (mean miR-338 Sp, 0.1119 ± 0.03104 vs. control, 0.3108 ± 0.02257; p = 0.0006) exhibiting fewer process endpoints (mean 0 endpoints 0.1094 ± 0.03701 vs. 0.2984 ± 0.01957; p = 0.0013) (Fig. 3C, D). These observations suggest that miR-338 mediated morphological differences in the cortical neurons were not due to an increase in apoptosis, since neurons in which miR-338 was either overexpressed or repressed were not immune-positive for cleaved Caspase-3 (data not shown). Collectively, the outcome of these studies raise the possibility that miR-338 is capable of modulating both neuronal morphologies and increases the number of neurons within the CP, suggesting a post-transcriptional regulatory role in the developing cortex.
Figure 3.
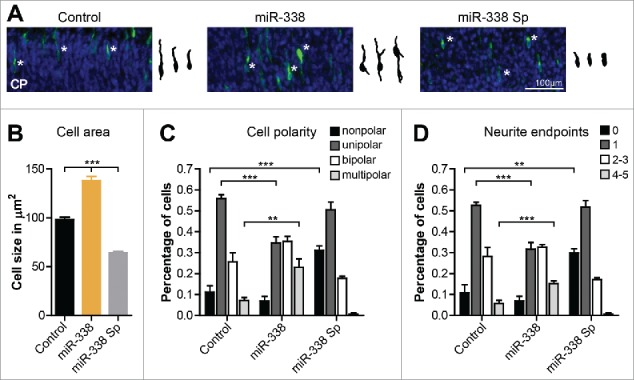
Modulation of miR-338 levels leads to alteration of neuronal morphologies within the cortical plate. (A) Representative images of neurons electroporated at E14.5 with either the control, miR-338 overexpression or the miR-338-sponge vector and analyzed at E17.5. The manipulated neurons are shown in green and the DAPI nuclear staining is shown in blue. Corresponding traced neurons within the CP are highlighted with a white asterisk within each image. (B) The total cellular area in µm2. Mean values with error bars ± s.e.m. One-way ANOVA with Bonferroni multiple comparison test; ***P < 0.001. (C) Neuronal polarity classified in the percentage of nonpolar, unipolar, bipolar and multipolar cells. (D) The percentage of neurons having no, 1, 2–3 or more number of neurite endpoints. Graphs represent mean values with error bars ± s.e.m. One-way ANOVA with Bonferroni multiple comparison test, average of 150 cells measured collected from n = 3 brains per condition from independent experiments; *P < 0.05, **P < 0.01 and ***P < 0.001.
Discussion
miR-338 is an evolutionarily conserved RNA and its expression is strongly increased at early neurodevelopmental stages. Overexpression of miR-338 in targeted cortical cells during early mouse brain development resulted in an increased number of neurons within the CP, whereas inhibition of miR-338 led to a reduced number of neurons within the CP. Since neurons divide and radially migrate from the deeper cortical layers (VZ and SVZ), the changes in the number of neurons within the upper cortical layers may indicate that miR-338 acts to modulate either the neuronal cell division or their radial migration toward the CP. Morphological analysis of the targeted neurons within the CP revealed morphological changes, as cortical neurons overexpressing miR-338 manifested a multipolar phenotype within the CP and an increase in the number of neurite protrusions. Conversely, inhibition of miR-338 resulted in an increase in the number of non-polar neurons highlighted by a lack of neurite processes. Moreover, these morphological alterations were not due to changes in the apoptotic state of the cells, since the levels of cleaved Caspase 3 were unaltered.
From embryonic day 10 onwards, a time period critical for genesis and further development of the cortical neuronal cell population,28,29 miR-338s expression levels increase steadily, suggesting its role in neuronal migration and further development of cortical neurons during this neurodevelopmental period. Indeed, miR-sponge mediated inhibition of endogenous miR-338 at these time points raises the possibility that it affects the number of neurons within the upper cortical layers. This observation raises the possibility that miR-338 acts upon downstream gene transcripts involved in the radial migration of neurons from deeper cortical layers. Alternatively, miR-338 could modulate the cell-cycle of the neural stem cells (NSCs) within the VZ and SVZ. Enhancing the number of neurons exiting the cell cycle may lead to an increase in the number of neurons prematurely moving up toward the upper cortical layers.
The outcome of the studies also suggested that an increased number of neurons overexpressing miR-338 maintained their multipolar morphology, while inhibition of miR-338 led to non-polar neuronal appearance with fewer neurite extensions. During migration from the IZ to the CP, neurons transit from a multipolar state to a bipolar state. This multipolar-to-bipolar transition is important for the formation of the various cortical layers and ultimately affects the proper functioning of the cortex.30,31 Changes in morphology could be due to an altered differentiation state of the neurons, leading to extended maintenance of their multipolar state under increased miR-338 expression. The altered differentiation state could be a direct consequence of the aberrant migration pattern of the neurons in which miR-338 levels were manipulated. Indeed, previous studies have revealed that miRs are capable of controlling the migration and polarity of cortical neurons through direct regulation of critical downstream genes (Reviewed in26,32-35). For example, miR-22 and miR-124 co-jointly control the expression of components of the CoREST/REST transcriptional repressor complex, regulating the migration and polarity of cortical neurons.26 miR-9 post-transcriptionally regulates the expression of the nuclear receptor TLX, resulting in altered cell proliferation of the NSCs residing within the VZ, and increasing the number of neurons within the CP.14 Furthermore, miR-134 controls the proliferation, migration and differentiation of cortical neurons depending on the neurodevelopmental state of these cells through regulation of Dcx and/or Chrdl-1.36 Interestingly, the above-mentioned miRs directly or indirectly control similar neurodevelopmentally critical genes. miR-338 target analysis in primary cortical neurons raised the possibility that miR-338 could similarly modulate the expression of many genes fitting in pathways critical for programming neuronal development.18
Previously we have used anti-miR-338 with a relatively short half-life, and provided evidence that this miR attenuates neuronal outgrowth by locally regulating axonal mitochondrial function.17 Our studies suggested that the repressing effect of miR-338 on axonal outgrowth diminished over time as axonal outgrowth grew near the length of unaltered control neurons within 5–6 d (unpublished observation). Increased miR-338 levels during neuronal differentiation could outweigh the limited number of anti-miR molecules and weaken their impact. Since the present study revealed that the expression of miR-338 incrementally increased during different time courses of cortical development (Fig. 2A), we decided to use a miR-338-sponge to achieve a long-lasting repression of this miR throughout development. This approach raises the possibility that the constitutive overexpression or sponging of miR-338 by means a cDNA vector would decisively modulate miR levels over a longer period and alter the final positioning and morphology of these manipulated neurons at later stages.
Previous studies have identified several developmental roles for miR-338 within various nervous systems. Increased miR-338 expression within mouse tissue promoted oligodendrocyte maturation and differentiation, while inhibition of miR-338 in Zebrafish spinal cord reduced the number of cells positive for oligodendrocyte makers.16 Moreover, miR-338 modulates the expression of the nuclear-encoded mitochondrial genes CoxIV and Atp5g1 within the growing axons of SCG neurons, resulting in reduced mitochondrial ATP production, reduced axon outgrowth and diminished presynaptic function.17,37,38 In cortical neurons, this miR was identified as a potential regulator of axon development and dendrite growth, through expression regulation of several critical genes governing different steps of cortical outgrowth20,21: monitoring axon outgrowth in cortical cells revealed that miR-338 overexpression decreased, whereas inhibition of miR-338 increased axonal length. To identify gene targets mediating the observed phenotype, miR-338 was inhibited in dissociated primary cortical neurons, and whole-transcriptome analysis was performed. Pathway analysis revealed that miR-338 modulates a subset of transcripts involved in the axonal guidance machinery by means of direct and indirect gene targeting.18 Interestingly, the mouse models of 22q11.2 deletion syndrome, which constitutes one of the strongest genetic risks for schizophrenia and exhibits an age-dependent reduction of miR-338 levels, also suffer from mitochondrial dysfunction, which raises the possibility that mitochondrial dysfunction in this mouse model may be one of the many consequences of an aberrant miR-338 levels.39,40
In Conclusion, the outcome of this investigation suggests a direct role for miR-338 during early cortical development. Our finding raises the possibility that spatio-temporal expression of miR-338 is critical for proper cortical development. Future studies will aim at identifying novel gene networks, which act down stream of this miR during the critical periods of neuronal migration and placement, resulting in the identification of novel mechanisms critical for these developmental processes.
Materials and methods
Animals
For the IUE experiments, timed-pregnant C57BL/6 JolaHSD mice (Harlan Laboratories B.V., Boxmeer, The Netherlands) were used. Embryonic day 18 (E18) embryos from timed-pregnant Wistar rats (Harlan laboratories B.V., Boxmeer, The Netherlands) were used for the isolation of primary cortical neurons. Animals were housed 2–3 per cage with ad libitum food and water access with a 12 hr light cycle at controlled ambient temperature (21 ± 1°C). All animal use, care and experiments were performed according to protocols approved by the Committee for Animal Experiments of the Radboud university medical center, Nijmegen, The Netherlands.
Cell culture
Primary cortical neurons were isolated from E18 rat embryonic brains. The cortical region was isolated and placed in ice-cold Hanks’ Balanced Salt Solution (HBSS) containing 2 mmol/L GlutaMAX (Life Technology, Grand Island, NY) supplemented with Pen/Strep antibiotics (Life Technology, Grand Island, NY). Cortical tissue was washed 2 times using supplemented HBSS before adding 0.025% trypsin in HBSS solution followed by incubation at 37°C for 15 minutes. After incubation, the tissue was washed 3 times with the supplemented HBSS before adding Neurobasal (NB) medium (Life Technology, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) (Life Technology, Grand Island, NY) and 2 mmol/L GlutaMAX. Cells were dissociated with a glass Pasteur pipette, this treatment was repeated with a fire-polished glass Pasteur pipette to obtain fully dissociated cells. Separated neurons were seeded in cell culture plates, which were coated overnight with 0.1 g/L, mol wt 70,000–150,000 Poly-D-Lysine (PDL) (Sigma–Aldrich, St. Louis, MO). For the first 5 hours, the neurons were cultured in medium containing NB medium with 10% FBS and 2 mmol/L glutaMAX. Afterwards, the medium was replaced with culturing medium containing NB with the serum free neural supplement B27 (Life Technology, Grand Island, NY) and 2 mmol/L glutaMAX. Neuronal cell preparations were maintained at 37°C and with 5% CO2.
DNA constructs and transfection
The mammalian miR-338 overexpression vector and its corresponding control vector were described previously.19 The miR-338-sponge plasmid vector was generated by designing DNA oligonucleotides (Sigma–Aldrich, St. Louis, MO) described previously.41,42 miR Sponge sequences were designed to contain 4 complementary artificial binding sites for miR-338–3p (Table 1). Presynthesized DNA oligonucleotides were ligated 3′ to the stop codon of the cDNA mCherry. For cloning miR sponges into the pmR-mCherry vector (Clontech, Mountain View, CA), the restriction sites HindIII and BamHI were used. All DNA constructs were sequenced to confirm their correct sequence orientation and identity. Electroporation of primary neurons was performed using the Amaxa Nucleofector II combined with the Amaxa rat neuron Nucleofector kit (Lonza, Basel, Switzerland) according to the manufacturers protocol.
Table 1.
pmCherry miR-338 sponge oligonucleotides.
| miR-338-sp-Fw | 5′AGCTTCAACAAAATGGAGATGCTGGAATCGCAACAAAATGGAGATGCTGGAATCGCAACAAAATGGAGATGCTGGAATCGCAACAAAATGGAGATGCTGGAG 3′ |
| miR-338-sp-Rv | 5′GATCCTCCAGCATCTCCATTTTGTTGCGATTCCAGCATCTCCATTTTGTTGCGATTCCAGCATCTCCATTTTGTTGCGATTCCAGCATCTCCATTTTGTTGA 3′ |
RNA isolation, reverse transcription and real-time PCR
RNA was isolated using the miRNAeasy kit (Qiagen, Germantown, MD), according to the manufacturer's manual, which allows for the preservation of both the small RNA and mRNA fraction. RNA purity was determined using the Nanodrop ND1000 (Thermo Scientific, Waltham, MA) UV-spectrophotometer. The 260/280nm ratios were measured and samples with ratios of 2.0 ± 0.05 were considered pure. This was followed by RNA integrity assessment with a 1% agarose gel imaged with the Gel doc XR system (Bio-Rad, Hercules, CA). Samples showing clearly visible S28 and S18 rRNA bands were considered to be intact. cDNA synthesis of small RNAs was performed using the miScript reverse transcription kit (Qiagen, Germantown, MD) according to the supplied protocol. Following cDNA synthesis, samples were diluted 1:10 with water. Quantitative real-time PCR (qRT-PCR) detection of mature miRs was performed using the miScript SYBR green PCR kit (Qiagen, Germantown, MD) according to the manufacturer's instructions. Data was normalized to U6 housekeeping gene and the relative expression differences were calculated using the delta CT method.43 The following PCR primer were used: mature miR-338–3p forward, 5′ TCCAGCATCAGTGATTTTGTTG 3′; reverse, universal reverse primer supplied with the miScript PCR kit. SnoU6 forward, 3′ GCTTCGGCAGCACATATA 5′; reverse, 5′ CGCTTCACGAATTTGCGT 3′.
IUE and tissue section preparation
IUE survival surgeries were performed as described previously.44,45 Timed pregnant (E14.5) mice were anaesthetized with 100 mg/kg Ketamine and 10 mg/kg Xylazine. Following laparotomy, a solution containing the plasmid DNA (2 μg/μl of vector DNA of interest co-injected with pEBFP-N1 (Clontech, Mountain View, CA) in Tris-buffered [pH 7.4] 0.02% Fast Green was injected through the uterine wall into the lateral ventricle of each embryo. Squarewave low voltage current pulses (5 unipolar pulses of 0.0033V, 050ms with a 950ms interval) were delivered across the embryonic head using Tweezertrodes (BTX, Harvard Apparatus, Holliston, MA). Embryos were kept hydrated, placed back into the abdomen of the mother and gestation was allowed to proceed for 3 d. Brains were removed from the embryo and fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), pH 7.4, for 1 hour. After fixation, the brains were placed in 30% sucrose overnight, and frozen in Shandon M-1 Embedding Medium (Thermo Scientific, Waltham, MA) on dry ice, and stored at −80°C. Coronal 16 µm thick cryostat sections were prepared, mounted on Superfrost plus slides (Thermo Scientific, Waltham, MA), air-dried and stored desiccated at −20°C. Only embryos showing effective transfection, assessed by reporter gene expression in the somatosensory (S1) cortex were analyzed.
Immunolabeling and imaging
Immunohistochemistry on IUE brain slices was performed as described previously by.44 Slices were rehydrated in PBS and incubated in blocking buffer (2.5% normal donkey serum, 2.5% normal goat serum, 2.5% normal horse serum, 1% bovine serum albumin (BSA), 1% glycine, 0.1% lysine, 0.4% Triton X-100) for 30 minutes at room temperature (RT). Sections were incubated in primary antibody diluted in blocking buffer overnight (o/n) at 4°C in humidified chambers. Then, sections were washed 3 times, each 10 min in PBS at RT. Subsequently, sections were incubated with species-specific Alexa-conjugated secondary antibodies diluted in blocking buffer for 30 minutes to 1 hour at RT. After 3 washes in PBS for a total of 30 min at RT, sections were counterstained with DAPI (Sigma–Aldrich, St. Louis, MO at 1:2000) in PBS, washed extensively in PBS and embedded in 90% glycerol in PBS. Primary antibodies used included chicken anti-GFP (Abcam, Cambridge, UK at 1:500), rabbit anti-GFP (Molecular Probes, Life Technologies, Grand Island, NY at 1:1000), rabbit anti-cleaved Caspase 3 (Cell Signaling, Danvers, MA at 1:200), mouse anti-NeuN (Merck Millipore, Darmstadt, Germany at 1:100). Secondary antibodies included Alexa488 goat anti-chicken, Alexa488 goat anti-rabbit and Alexa568 goat anti-rabbit (Molecular Probes, Life Technologies, Grand Island, NY at 1:500). Fluorescent images of brain slices were obtained using a Leica DMRA fluorescence microscope fitted with a DFC340 FX CCD camera (Leica, Wetzlar, Germany) or a Leica TCS SP2 AOBS Confocal Laser Scanning Microscope (CLSM) (Leica, Wetzlar, Germany).
Neuronal morphology and migration analysis
Fluorescent images were randomized and analyzed in a blinded fashion. Imaging and analysis of the primary somatosensory (S1) cortex IUE brain sections was performed at the same neuroanatomical level for all conditions. The images were loaded into Adobe Illustrator in which a mask was made to highlight the various cortical layers identified on the basis of cell density (Nuclear staining by DAPI). The quantified number of electroporated cells was normalized to the total number of cells within the intermediate zone (IZ) and cortical plate (CP). Neuron outlines were traced using Adobe Illustrator software. The digitized neuronal tracings were analyzed using the NeuronJ plugin for ImageJ.46
Statistical analysis
Quantitative data are presented as the mean ± s.e.m. Non-paired 2-tailed Student's t-test was used to determine significant differences between 2 groups. One-way ANOVA with Bonferroni multiple comparison testing was used to analyze significant differences between multiple groups. P ≤ 0.05 was considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank N van Bakel for his technical assistance. We thank the RIMLS microscopy platform (http://ncmls.nl/technology-platform/microscope-imaging-center/) for support and maintenance of the equipment.
Funding
The research of the authors is supported by grants from the “FP7-Marie Curie International Reintegration Grant” [to AA grant number 276868]. This work was supported by the Division of Intramural Research Programs of the National Institute of Mental Health (MH002768).
References
- 1.Franco SJ, Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron 2013; 77:19-34; PMID:23312513; https://doi.org/ 10.1016/j.neuron.2012.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell 2011; 146:18-36; PMID:21729779; https://doi.org/ 10.1016/j.cell.2011.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schubert D, Martens GJ, Kolk SM. Molecular underpinnings of prefrontal cortex development in rodents provide insights into the etiology of neurodevelopmental disorders. Mol Psychiatry 2015; 20:795-809; PMID:25450230; https://doi.org/ 10.1038/mp.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet 2011; 45:81-104; PMID:21910631; https://doi.org/ 10.1146/annurev-genet-110410-132512 [DOI] [PubMed] [Google Scholar]
- 5.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, et al.. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet 2008; 40:751-60; PMID:18469815; https://doi.org/ 10.1038/ng.138 [DOI] [PubMed] [Google Scholar]
- 6.Willemsen MH, Vallès A, Kirkels LA, Mastebroek M, Olde Loohuis N, Kos A, Wissink-Lindhout WM, de Brouwer AP, Nillesen WM, Pfundt R, et al.. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet 2011; 48:810-8; PMID:22003227; https://doi.org/ 10.1136/jmedgenet-2011-100294 [DOI] [PubMed] [Google Scholar]
- 7.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003; 9:1274-81; PMID:13130141; https://doi.org/ 10.1261/rna.5980303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao MJ, Chen G, Zhao PP, Lu MH, Jian J, Liu MF, Yuan XB. Transcriptome analysis of microRNAs in developing cerebral cortex of rat. BMC Genomics 2012; 13:232; PMID:22691069; https://doi.org/ 10.1186/1471-2164-13-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 2008; 135:3911-21; PMID:18997113; https://doi.org/ 10.1242/dev.025080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn 2009; 238:2800-12; PMID:19806666; https://doi.org/ 10.1002/dvdy.22109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci 2012; 69:89-102; PMID:21833581; https://doi.org/ 10.1007/s00018-011-0788-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. MicroRNAs tune cerebral cortical neurogenesis. Cell Death Differ 2012; 19:1573-81; PMID:22858543; https://doi.org/ 10.1038/cdd.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 2009; 12:399-408; PMID:19287386; https://doi.org/ 10.1038/nn.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 2009; 16:365-71; PMID:19330006; https://doi.org/ 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 2004; 5:R13; PMID:15003116; https://doi.org/ 10.1186/gb-2004-5-3-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, et al.. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 2010; 65:612-26; PMID:20223198; https://doi.org/ 10.1016/j.neuron.2010.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci 2008; 28:12581-90; PMID:19020050; https://doi.org/ 10.1523/JNEUROSCI.3338-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kos A, Klein-Gunnewiek T, Meinhardt J, Loohuis NF, van Bokhoven H, Kaplan BB, Martens GJ, Kolk SM, Aschrafi A. MicroRNA-338 attenuates cortical neuronal outgrowth by modulating the expression of axon guidance genes. Mol Neurobiol 2016; PMID:27180071; https://doi.org/ 10.1007/s12035-016-9925-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kos A, Olde Loohuis NF, Wieczorek ML, Glennon JC, Martens GJ, Kolk SM, Aschrafi A. A potential regulatory role for intronic microRNA-338-3p for its host gene encoding apoptosis-associated tyrosine kinase. PLoS One 2012; 7:e31022; PMID:22363537; https://doi.org/ 10.1371/journal.pone.0031022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takano T, Urushibara T, Yoshioka N, Saito T, Fukuda M, Tomomura M, Hisanaga S. LMTK1 regulates dendritic formation by regulating movement of Rab11A-positive endosomes. Mol Biol Cell 2014; 25:1755-68; PMID:24672056; https://doi.org/ 10.1091/mbc.E14-01-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takano T, Tomomura M, Yoshioka N, Tsutsumi K, Terasawa Y, Saito T, Kawano H, Kamiguchi H, Fukuda M, Hisanaga S. LMTK1/AATYK1 is a novel regulator of axonal outgrowth that acts via Rab11 in a Cdk5-dependent manner. J Neurosci 2012; 32:6587-99; PMID:22573681; https://doi.org/ 10.1523/JNEUROSCI.5317-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chun S, Du F, Westmoreland JJ, Han SB, Wang YD, Eddins D, Bayazitov IT, Devaraju P, Yu J, Mellado Lagarde MM, et al.. Thalamic miR-338-3p mediates auditory thalamocortical disruption and its late onset in models of 22q11.2 microdeletion. Nat Med 2017; 23:39-48; PMID:27892953; https://doi.org/ 10.1038/nm.4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 2012; 73:35-48; PMID:22243745; https://doi.org/ 10.1016/j.neuron.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 2004; 7:136-44; PMID:14703572; https://doi.org/ 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- 25.Ye T, Ip JP, Fu AK, Ip NY. Cdk5-mediated phosphorylation of RapGEF2 controls neuronal migration in the developing cerebral cortex. Nat Commun 2014; 5:4826; PMID:25189171; https://doi.org/ 10.1038/ncomms5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volvert ML, Prévot PP, Close P, Laguesse S, Pirotte S, Hemphill J, Rogister F, Kruzy N, Sacheli R, Moonen G, et al.. MicroRNA targeting of CoREST controls polarization of migrating cortical neurons. Cell Rep 2014; 7:1168-83; PMID:24794437; https://doi.org/ 10.1016/j.celrep.2014.03.075 [DOI] [PubMed] [Google Scholar]
- 27.Ohtaka-Maruyama C, Hirai S, Miwa A, Heng JI, Shitara H, Ishii R, Taya C, Kawano H, Kasai M, Nakajima K, et al.. RP58 regulates the multipolar-bipolar transition of newborn neurons in the developing cerebral cortex. Cell Rep 2013; 3:458-71; PMID:23395638; https://doi.org/ 10.1016/j.celrep.2013.01.012 [DOI] [PubMed] [Google Scholar]
- 28.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol 2005; 6:777-88; PMID:16314867; https://doi.org/ 10.1038/nrm1739 [DOI] [PubMed] [Google Scholar]
- 29.Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci 2006; 7:883-90; PMID:17033683; https://doi.org/ 10.1038/nrn2008 [DOI] [PubMed] [Google Scholar]
- 30.Rakic P, Ayoub AE, Breunig JJ, Dominguez MH. Decision by division: making cortical maps. Trends Neurosci 2009; 32:291-301; PMID:19380167; https://doi.org/ 10.1016/j.tins.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.La Fata G, Gärtner A, Domínguez-Iturza N, Dresselaers T, Dawitz J, Poorthuis RB, Averna M, Himmelreich U, Meredith RM, Achsel T, et al.. FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat Neurosci 2014; 17:1693-700; PMID:25402856; https://doi.org/ 10.1038/nn.3870 [DOI] [PubMed] [Google Scholar]
- 32.Kos A, Aschrafi A, Nadif Kasri N. The Multifarious Hippocampal Functions of MicroRNA-137. Neuroscientist 2016; 22:440-6; PMID:26396150; https://doi.org/ 10.1177/1073858415608356 [DOI] [PubMed] [Google Scholar]
- 33.Barca-Mayo O, De Pietri Tonelli D. Convergent microRNA actions coordinate neocortical development. Cell Mol Life Sci 2014; 71:2975-95; PMID:24519472; https://doi.org/ 10.1007/s00018-014-1576-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Candiani S, Moronti L, De Pietri Tonelli D, Garbarino G, Pestarino M. A study of neural-related microRNAs in the developing amphioxus. Evodevo 2011; 2:15; PMID:21722366; https://doi.org/ 10.1186/2041-9139-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Pietri TD, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 2008; 135:3911-21; PMID:18997113; https://doi.org/ 10.1242/dev.025080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex 2011; 21:1857-69; PMID:21228099; https://doi.org/ 10.1093/cercor/bhq262 [DOI] [PubMed] [Google Scholar]
- 37.Aschrafi A, Kar AN, Natera-Naranjo O, MacGibeny MA, Gioio AE, Kaplan BB. MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol Life Sci 2012; 69:4017-27; PMID:22773120; https://doi.org/ 10.1007/s00018-012-1064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, Macgibeny MA, Gioio AE, Kaplan BB. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci 2012; 49:263-70; PMID:22209705; https://doi.org/ 10.1016/j.mcn.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 39.Devaraju P, Zakharenko SS. Mitochondria in complex psychiatric disorders: Lessons from mouse models of 22q11.2 deletion syndrome: Hemizygous deletion of several mitochondrial genes in the 22q11.2 genomic region can lead to symptoms associated with neuropsychiatric disease. Bioessays 2017; 39(2); PMID:28044359; https://doi.org/ 10.1002/bies.201600177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devaraju P, Yu J, Eddins D, Mellado-Lagarde MM, Earls LR, Westmoreland JJ, Quarato G, Green DR, Zakharenko SS. Haploinsufficiency of the 22q11.2 microdeletion gene Mrpl40 disrupts short-term synaptic plasticity and working memory through dysregulation of mitochondrial calcium. Mol Psychiatry 2016; p1-4; PMID:27184122; https://doi.org/ 10.1038/mp.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, et al.. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci 2011; 14:1240-2; PMID:21892155; https://doi.org/ 10.1038/nn.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, Ratnu VS, Boskovic Z, Kobor MS, Sun YE, et al.. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci 2011; 14:1115-7; PMID:21841775; https://doi.org/ 10.1038/nn.2891 [DOI] [PubMed] [Google Scholar]
- 43.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3:1101-8; PMID:18546601; https://doi.org/ 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 44.Kolk SM, Gunput RA, Tran TS, van den Heuvel DM, Prasad AA, Hellemons AJ, Adolfs Y, Ginty DD, Kolodkin AL, Burbach JP, et al.. Semaphorin 3F is a bifunctional guidance cue for dopaminergic axons and controls their fasciculation, channeling, rostral growth, and intracortical targeting. J Neurosci 2009; 29:12542-57; PMID:19812329; https://doi.org/ 10.1523/JNEUROSCI.2521-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolk SM, de Mooij-Malsen AJ, Martens GJ. Spatiotemporal molecular approach of in utero electroporation to functionally decipher endophenotypes in neurodevelopmental disorders. Front Mol Neurosci 2011; 4:37; PMID:22065947; https://doi.org/ 10.3389/fnmol.2011.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 2004; 58:167-76; PMID:15057970; https://doi.org/ 10.1002/cyto.a.20022 [DOI] [PubMed] [Google Scholar]


