ABSTRACT
Cumulative verified experimental studies have demonstrated that microRNAs (miRNAs) could be closely related with the development and progression of human complex diseases. Based on the assumption that functional similar miRNAs may have a strong correlation with phenotypically similar diseases and vice versa, researchers developed various effective computational models which combine heterogeneous biologic data sets including disease similarity network, miRNA similarity network, and known disease-miRNA association network to identify potential relationships between miRNAs and diseases in biomedical research. Considering the limitations in previous computational study, we introduced a novel computational method of Ranking-based KNN for miRNA-Disease Association prediction (RKNNMDA) to predict potential related miRNAs for diseases, and our method obtained an AUC of 0.8221 based on leave-one-out cross validation. In addition, RKNNMDA was applied to 3 kinds of important human cancers for further performance evaluation. The results showed that 96%, 80% and 94% of predicted top 50 potential related miRNAs for Colon Neoplasms, Esophageal Neoplasms, and Prostate Neoplasms have been confirmed by experimental literatures, respectively. Moreover, RKNNMDA could be used to predict potential miRNAs for diseases without any known miRNAs, and it is anticipated that RKNNMDA would be of great use for novel miRNA-disease association identification.
KEYWORDS: Disease, disease semantic similarity, KNN algorithm, miRNAs, miRNA-disease association, SVM Ranking model
Introduction
MicroRNAs (miRNAs) are one class of small non-coding RNAs (∼22 nt) that functions in post-transcriptional regulation of gene expression. It normally suppresses the expression of the target mRNAs (mRNAs) by binding to the 3′ untranslated regions (UTRs) of the target mRNAs through sequence-specific base pairing.1-4 However, some scientific studies have shown that miRNAs also act as positive regulators.5,6 In recent years, the research about the associations between miRNAs and diseases has aroused more attention than ever before especially considering that miRNAs have already been confirmed to play an essential role in many significant biologic processes including cell proliferation,7 development,8 differentiation,9 and apoptosis,10 metabolism,11,12 aging,11,12 signal transduction,13 viral infection9 and so on. For example, the serum levels of miR-103 expression in the breast cancer patients were significantly higher than healthy control group.14 Further study on these cancer patients showed that high miR-103 expression was significantly correlated with advanced clinical stage and lymph node metastasis. Also, researchers observed that miR-126 were expressed with significantly higher levels in the blood from patients with Crohn Disease compared with the healthy controls.15 Furthermore, the circulating levels of miR-15b were significantly reduced in patients with end-stage renal disease.16 Therefore, identifying disease-related miRNAs can enhance the study of biomarker detection for prognosis, diagnosis and treatment of human complex diseases.
In the past few years, a large number of computational models for predicting potential associations between miRNAs and diseases have been developed. For example, Jiang el al.17 developed a hypergeometric distribution-based computational model which combined miRNA similarity network, disease similarity network, and known miRNA-disease interactions and finally prioritized the human miRNAs retrieved from miRBase database for diseases of interest. Shi et al.18 further proposed a method which focused on the functional link between miRNA targets and disease genes by implementing random walk algorithm on protein-protein interaction (PPI) network. Meanwhile, Mørk et al.19 developed a method in which miRNAs were linked to diseases via proteins involved. This method ranked candidate miRNA-disease associations based on known and predicted miRNA-protein associations and protein-disease associations in the framework of a novel scoring scheme. However, these methods relied much on miRNA-target interactions which have a pretty high ratio of false-positive and false-negative results, thus they did not have satisfying prediction performance. Furthermore, Xuan et al.20 presented a new computational method called HDMP for predicting potential disease-related miRNAs, which was based on the weighted k most similar neighbors. After calculating the functional similarity scores between any 2 miRNAs, HDMP assigned higher weights to members in the same miRNA family or cluster since they were more likely associated with similar diseases. At last, all miRNAs were ranked by their relevance weight score assigned by HDMP with a certain disease, and the higher score a miRNA obtained, the more likely this miRNA is related to the disease. However, using local network similarity would potentially decrease their prediction accuracy. Additionally, the number of neighbors was a huge influence factor to the prediction results and there were no parameter differences among different diseases. Xu et al.21 introduced an approach which combined the expression files of miRNAs in tumor or non-tumor tissues. This method constructed the miRNA target-dysregulated network (MTDN), extracted network topological features, and distinguished positive or negative disease related miRNAs by Support Vector Machine (SVM). Nevertheless, it is difficult even impossible to obtain negative associations. Chen et al.22 proposed the first global network similarity based computational method of RWRMDA which implemented random walk on the miRNA-miRNA functional similarity network, but it cannot be applied to diseases without any known related miRNAs. Xuan et al.23 also developed a novel method based on random walk. Unlike RWRMDA, this method extended the walking on the miRNA-disease bilayer network which was composed of the miRNA network derived from the miRNA-associated diseases (Mnet), the disease network (Dnet), and the edges between 2 networks, thus it can be used for diseases without any related associations. There are also some other methods that can predict potential miRNAs for diseases without known associated miRNAs. RLSMDA24 is a semi-supervised learning method based on regularized least squares. One important improvement is that it did not require negative known samples. WBSMDA25 introduced Gaussian interaction profile kernel similarity, which was later combined with miRNA functional similarity, disease semantic similarity and known miRNA-disease associations, and finally ranked the potential associations by within and between score. RBMMMDA26 is a method that used the model of Restricted Boltzmann machine, and it could predict not only novel miRNA-disease associations but also the category of the new association. The categories are divided according to the different supporting evidences, including the evidences of genetics, epigenetics, circulating miRNAs, and miRNA-target interactions. Recently, the method of HGIMDA27 integrated miRNA functional similarity, disease semantic similarity, Gaussian interaction profile kernel similarity and known miRNA-disease associations into a heterogeneous graph. HGIMDA calculated the probability between disease d and miRNA m by summarizing all paths with the same length.
It is obvious that using traditional experimental methods for disease detection is an extremely time-consuming task, and the previously developed computational methods have some aforementioned limitations. Therefore, we developed a novel computational model of Ranking-based KNN for MiRNA-Disease Association prediction (RKNNMDA). We first combined miRNA functional similarity, disease semantic similarity, Gaussian interaction profile kernel similarity, and known miRNA-disease associations to search for k-nearest-neighbors both for miRNAs and diseases by using K-Nearest Neighbors (KNN) algorithm. The k-nearest-neighbors were obtained in a descending order from the similarity scores between other miRNAs (diseases) and the central miRNA (disease). Then we reranked these k-nearest-neighbors according to SVM Ranking model. Finally we did weighted voting and obtained the final ranking of all possible miRNA-disease associations. As a result, RKNNMDA showed superior performance to 5 classical miRNA-disease association prediction methods (HGIMDA,27 WBSMDA,25 RLSMDA,24 HDMP,13 and RWRMDA22) according to leave-one-out cross validation (LOOCV). This method has obtained an AUC of 0.8221, showing the reliability and precision of RKNNMDA. Besides, RKNNMDA can be applied to diseases without any known related miRNAs. Moreover, it has a satisfying result in case studies as well: 48, 40, and 47 out of top 50 predicted miRNA-disease associations for Colon Neoplasms, Esophageal Neoplasms, and Prostate Neoplasms have been validated by experimental reports respectively.
Results
Leave-one-out cross validation
LOOCV is a validation method using one known association as the test sample and the remaining known associations as the training samples. In this study, we have already obtained 5430 known miRNA-disease associations from HMDD database.28 In the gold standard data set, each disease is associated with 14.18 miRNA-disease associations on average, which means that there exists little difference between leave-one-out cross validation and 10-fold cross validation. Out of 383 diseases investigated in this paper, 105, 45, 23, 23, 16 diseases have 1, 2, 3, 4, 5 known related associations respectively. In consideration of the substantial proportion of diseases that have limited associations, it is not feasible for us to implement multi-fold cross validation. Therefore, here we use leave-one-out cross validation to evaluate the performance of RKNNMDA. Assume that a certain disease d is associated with n miRNAs (seed miRNAs), each time a seed miRNA is regarded as a test miRNA and the association between this miRNA and disease d is considered unrelated, the remaining n-1 miRNAs are considered as training miRNAs, and other miRNAs that are currently irrelevant to the disease d are considered as candidate miRNAs. Accordingly, we rank test miRNA among all the candidate miRNAs. If the ranking of the test miRNA is higher than the given threshold, it is thought to be a successful prediction of this miRNA-disease association.
Receiver-Operating Characteristics (ROC) curve is a 2 dimensional plotting graphic that shows the performance of a binary classifier system as its discrimination threshold varies. The vertical axis displays the true positive rate (TPR), also known as sensitivity, which equals to the ratio of the test miRNA-disease associations that exceed the threshold compared with all the associations; while the horizontal axis displays the false positive rate (FPR), which can be calculated as 1-specificity. Specificity refers to the percentage of the negative miRNA-disease associations that are ranked below the threshold. The corresponding values of TPR and FPR could be calculated by changing different thresholds. The area under the curve (AUC) refers to the probability that a randomly chosen positive association ranks higher than a randomly chosen negative one, which evaluates the performance of computational models. To be specific, AUC = 1 indicates that the prediction has a perfectly precise performance, and AUC = 0.5 demonstrates that the prediction has random performance. In this case, the AUC of 0.8221 demonstrated the reliable performance of RKNNMDA.
Compared with other methods
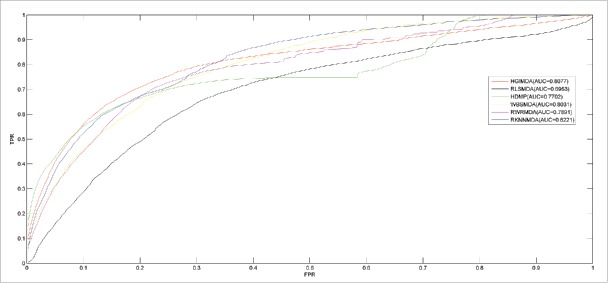
We further compare RKNNMDA with 5 other classical methods and then we are able to make a more objective evaluation for RKNNMDA:1 HGIMDA,27 which predicted potential associations between disease d and miRNA m by summarizing all paths with the length equal to 3;2 WBSMDA,25 which calculated Within-Scores and Between-scores for miRNA-disease pairs to calculate their association probabilities;3 RLSMDA,24 which is a semi-supervised method based on the framework of regularized least squares;4 HDMP,23 which took advantage of weighted k most similar neighbors and new similarity measures;5 RWRMDA,22 which implemented global network similarity-based random walk on the miRNA functional similarity network. Fig. 1 shows the comparison performances in the framework of LOOCV. HGIMDA, RLSMDA, HDMP, WBSMDA, RWRMDA and RKNNMDA achieved AUCs of 0.8077, 0.6953, 0.7702, 0.8031, 0.7891 and 0.8221, respectively. In conclusion, RKNNMDA has made advance in improving the prediction accuracy.
Figure 1.
The comparison results between RKNNMDA and other 5 classical computational methods (HGIMDA, RLSMDA, HDMP, WBDMDA, and RWRMDA). As a result, RKNNMDA achieved AUC of 0.8221 which significantly outperformed all the previous classical models.
Case studies
To further evaluate the performance of RKNNMDA, we here applied this method to predict 3 major human cancers: Colon Neoplasms, Esophageal Neoplasms, and Prostate Neoplasms. Training samples were downloaded from HMDD database,28 while potential predicted miRNA-disease associations were validated according to other 2 independent databases: miR2Disease29 and dbDEMC.30 The entire prediction list for miRNA-disease association contains 184155 miRNA-disease pairs ranked by RKNNMDA model, which could be used for further experimental validation (See Table S1).
Colon Neoplasms, also known as colon cancer or colorectal cancer, is the third most common type of cancer constituting about 10% of all cancer cases. Since colon neoplasms is a fairly hard-to-detect cancer at an early stage,31,32 there is an increasing need of novel sensitive biomarkers that could help improve the early detection of Colon Neoplasms. Studies have confirmed some miRNAs have been related to Colon Neoplasms, for instance, miR-145 is constantly downregulated in colorectal tumors.33 Moreover, miR-127 was reported to play a role as a possible tumor suppressor gene.34 By implementing RKNNMDA to prioritize candidate miRNAs for Colon Neoplasms, 20 out of the top 20, and 48 out of the top 50 predicted potential Colon Neoplasms related miRNAs were validated based on miR2Disease and dbDEMC (See Table 1). For example, in our prediction outcome, miR-143 expression level was extremely reduced in the colon cancer cells.35 What's more, miR-21, as an oncogene, may lead to the downregulation of transforming growth factor β receptor 2 (TGFβR2) and mediate the expression of Sprouty2 protein which was identified as a tumor suppressor in colon cancer cells.36,37
Table 1.
Here we implemented RKNNMDA to predict potential Colon Neoplasms-related miRNAs. As a result, 10 out of the top 10, 20 out of the top 20, and 48 out of the top 50 predicted Colon Neoplasms related miRNAs were confirmed based on dbDEMC and miR2Disease (1st column: top 1–25; 2nd column: top 26–50).
| miRNA | score | Evidence | miRNA | score | Evidence |
|---|---|---|---|---|---|
| hsa-mir-143 | 12 | dbdemc; miR2Disease | hsa-mir-31 | 4 | dbdemc; miR2Disease |
| hsa-mir-21 | 10 | dbdemc; miR2Disease | hsa-mir-449a | 4 | unconfirmed |
| hsa-mir-155 | 10 | dbdemc; miR2Disease | hsa-mir-92a | 4 | dbdemc |
| hsa-mir-20a | 10 | dbdemc; miR2Disease | hsa-let-7e | 4 | dbdemc; miR2Disease |
| hsa-mir-223 | 6 | dbdemc; miR2Disease | hsa-mir-101 | 4 | unconfirmed |
| hsa-mir-125b | 6 | dbdemc; miR2Disease | hsa-mir-18a | 4 | dbdemc; miR2Disease |
| hsa-mir-132 | 6 | miR2Disease | hsa-mir-19b | 4 | dbdemc; miR2Disease |
| hsa-mir-125a | 6 | dbdemc; miR2Disease | hsa-mir-30a | 4 | dbdemc; miR2Disease |
| hsa-mir-29a | 6 | dbdemc; miR2Disease | hsa-mir-30c | 4 | dbdemc; miR2Disease |
| hsa-mir-29b | 6 | dbdemc; miR2Disease | hsa-mir-34a | 4 | dbdemc; miR2Disease |
| hsa-let-7a | 6 | dbdemc; miR2Disease | hsa-let-7b | 4 | dbdemc; miR2Disease |
| hsa-mir-141 | 4 | dbdemc; miR2Disease | hsa-let-7c | 4 | dbdemc; miR2Disease |
| hsa-mir-15a | 4 | dbdemc; miR2Disease | hsa-let-7d | 4 | dbdemc; miR2Disease |
| hsa-mir-16 | 4 | dbdemc | hsa-mir-106b | 4 | dbdemc; miR2Disease |
| hsa-mir-1 | 4 | dbdemc; miR2Disease | hsa-mir-137 | 4 | dbdemc; miR2Disease |
| hsa-mir-133b | 4 | dbdemc; miR2Disease | hsa-mir-23b | 4 | miR2Disease |
| hsa-mir-146a | 4 | dbdemc | hsa-mir-424 | 4 | dbdemc |
| hsa-mir-10b | 4 | dbdemc; miR2Disease | hsa-mir-107 | 4 | dbdemc; miR2Disease |
| hsa-mir-152 | 4 | dbdemc | hsa-mir-128 | 4 | dbdemc; miR2Disease |
| hsa-mir-191 | 4 | dbdemc; miR2Disease | hsa-mir-9 | 4 | dbdemc; miR2Disease |
| hsa-mir-192 | 4 | dbdemc; miR2Disease | hsa-mir-140 | 4 | miR2Disease |
| hsa-mir-200b | 4 | dbdemc; miR2Disease | hsa-mir-19a | 4 | dbdemc; miR2Disease |
| hsa-mir-200c | 4 | dbdemc; miR2Disease | hsa-mir-498 | 4 | dbdemc |
| hsa-mir-205 | 4 | dbdemc | hsa-let-7f | 4 | dbdemc; miR2Disease |
| hsa-mir-221 | 4 | dbdemc; miR2Disease | hsa-let-7 g | 4 | dbdemc; miR2Disease |
Esophageal Neoplasms, or esophageal cancer, can be divided into 2 main categories: esophageal squamous-cell carcinoma (ESCC), which is more common in the developing world, and esophageal adenocarcinoma (EAC), which is more common in the developed world.38 In 2012, Esophageal Neoplasms ranked the eighth most common cancer globally with 456,000 new cases during that year. By the time when imperceptible symptoms such as difficulty in swallowing and weight loss first appear, the cancer has already further exacerbated.39 Besides, treatment of surgical removal is a difficult operation with a relatively high risk of mortality or post-surgical difficulties.40 Therefore, reliable prediction model for potential Esophageal Neoplasms related miRNAs is seriously needed. Up to now, experiments have already confirmed several related miRNAs, for instance, miR-148a improved response to chemotherapy in sensitive and resistant squamous-cell carcinoma cells.41 Also, overexpression of miR-200c induced chemoresistance in esophageal cancers through the Akt signaling pathway.42 Taking Esophageal Neoplasms as a case to implement RKNNMDA, we successfully verified all the top 20 predicted miRNA-disease associations by scientific literatures. Moreover, among the top 50 pairs predicted by RKNNMDA, the accuracy reached 80% (See Table 2). For example, miR-194 was elevated in cancerous tissue from patients with Esophageal Neoplasms compared with noncancerous tissue from normal people.43 Also, Esophageal Neoplasms patients with high levels of miR-135 in the post treatment biopsy specimens had significantly shorter median disease-free survival (DFS) than did those with low levels.44
Table 2.
Here we implemented RKNNMDA to predict potential Esophageal Neoplasms-related miRNAs. As a result, 10 out of the top 10, 20 out of the top 20, and 40 out of the top 50 predicted Esophageal Neoplasms related miRNAs were confirmed based on dbDEMC and miR2Disease (1st column: top 1-25; 2nd column: top 26-50).
| miRNA | Score | Evidence | miRNA | Score | Evidence |
|---|---|---|---|---|---|
| hsa-mir-660 | 10 | dbdemc | hsa-mir-638 | 10 | unconfirmed |
| hsa-mir-16 | 10 | dbdemc | hsa-mir-96 | 10 | dbdemc |
| hsa-mir-1 | 10 | dbdemc | hsa-mir-302e | 10 | dbdemc |
| hsa-mir-135a | 10 | dbdemc | hsa-mir-370 | 10 | dbdemc |
| hsa-mir-17 | 10 | dbdemc | hsa-mir-602 | 10 | dbdemc |
| hsa-mir-191 | 10 | dbdemc | hsa-mir-612 | 10 | dbdemc |
| hsa-mir-194 | 10 | dbdemc; miR2Disease | hsa-mir-615 | 8 | dbdemc |
| hsa-mir-200b | 10 | dbdemc | hsa-mir-637 | 8 | unconfirmed |
| hsa-mir-429 | 10 | dbdemc | hsa-mir-657 | 8 | unconfirmed |
| hsa-mir-93 | 10 | dbdemc | hsa-mir-185 | 8 | dbdemc |
| hsa-mir-125b | 10 | dbdemc | hsa-mir-518c | 8 | dbdemc |
| hsa-mir-148b | 10 | dbdemc | hsa-mir-622 | 8 | dbdemc |
| hsa-mir-18a | 10 | dbdemc | hsa-mir-596 | 8 | dbdemc |
| hsa-mir-30a | 10 | dbdemc | hsa-mir-583 | 8 | dbdemc |
| hsa-mir-324 | 10 | dbdemc | hsa-mir-557 | 8 | dbdemc |
| hsa-mir-139 | 10 | dbdemc | hsa-mir-600 | 8 | unconfirmed |
| hsa-mir-335 | 10 | dbdemc | hsa-mir-601 | 8 | unconfirmed |
| hsa-mir-376c | 10 | dbdemc | hsa-mir-611 | 8 | unconfirmed |
| hsa-mir-30d | 10 | dbdemc | hsa-mir-654 | 8 | unconfirmed |
| hsa-mir-30e | 10 | dbdemc | hsa-mir-662 | 8 | unconfirmed |
| hsa-mir-23a | 10 | dbdemc | hsa-mir-769 | 8 | dbdemc |
| hsa-mir-127 | 10 | dbdemc | hsa-mir-125a | 8 | dbdemc |
| hsa-mir-142 | 10 | dbdemc | hsa-mir-198 | 8 | dbdemc |
| hsa-mir-608 | 10 | unconfirmed | hsa-mir-29a | 8 | dbdemc |
| hsa-mir-629 | 10 | unconfirmed | hsa-mir-29b | 8 | dbdemc |
Prostate Neoplasms, also known as Prostate Cancer, is a kind of cancer that develops in a gland in the male reproductive system. Though most Prostate Neoplasms are slow-growing, the potential risks lie in that once it begins to develop, it may spread to other parts of human body particularly bones and lymph nodes. Besides, Prostate Neoplasms initially causes no symptoms, which easily leads to delayed treatment.38 Thus, scientists developed computational models that identified the associations between Prostate Neoplasms and miRNA together. Some miRNAs have been validated to be related to Prostate Neoplasms. For instance, bioinformatics analysis confirmed that miR-99a/let-7c/miR-125b-2 were enriched in androgen-induced gene sets which stimulate and repress gene expression to promote the initiation and development of Prostate Cancer.45 Moreover, miR-183 expression was significantly higher in Prostate Cancer cells and tissues, while its knockdown decreased cell growth and mobility of Prostate Cancer cells.46 By applying RKNNMDA to the case study of Prostate Neoplasms, 9 out of top 10, 19 out of top 20, and 47 out of top 50 predicted potential related miRNAs were confirmed (See Table 3). For example, the aberrant expression of miR-143 has been detected in Prostate Neoplasms.47 Also, ectopic expression of miR-126* regulated protein translation and led to invasiveness of prostate cancer LNCaP cells.48
Table 3.
Here, we implemented RKNNMDA to predict potential Prostate Neoplasms-related miRNAs. As a result, 9 out of the top 10, 19 out of the top 20, and 47 out of the top 50 predicted Esophageal Neoplasms related miRNAs were confirmed based on dbDEMC and miR2Disease (1st column: top 1–25; 2nd column: top 26–50).
| miRNA | Score | Evidence | miRNA | Score | Evidence |
|---|---|---|---|---|---|
| hsa-mir-143 | 10 | dbdemc; miR2Disease | hsa-mir-194 | 2 | dbdemc; miR2Disease |
| hsa-mir-126 | 10 | dbdemc; miR2Disease | hsa-mir-195 | 2 | dbdemc; miR2Disease |
| hsa-mir-203 | 10 | unconfirmed | hsa-mir-200a | 2 | dbdemc |
| hsa-mir-223 | 10 | dbdemc; miR2Disease | hsa-mir-200b | 2 | dbdemc |
| hsa-mir-96 | 10 | dbdemc; miR2Disease | hsa-mir-200c | 2 | dbdemc |
| hsa-mir-198 | 4 | dbdemc; miR2Disease | hsa-mir-204 | 2 | dbdemc |
| hsa-mir-29a | 4 | dbdemc; miR2Disease | hsa-mir-205 | 2 | dbdemc; miR2Disease |
| hsa-mir-29b | 4 | dbdemc; miR2Disease | hsa-mir-20a | 2 | dbdemc; miR2Disease |
| hsa-let-7a | 4 | dbdemc; miR2Disease | hsa-mir-221 | 2 | dbdemc; miR2Disease |
| hsa-mir-141 | 4 | miR2Disease | hsa-mir-25 | 2 | dbdemc; miR2Disease |
| hsa-mir-15a | 2 | dbdemc; miR2Disease | hsa-mir-31 | 2 | dbdemc; miR2Disease |
| hsa-mir-16 | 2 | dbdemc; miR2Disease | hsa-mir-34b | 2 | dbdemc; miR2Disease |
| hsa-mir-21 | 2 | dbdemc; miR2Disease | hsa-mir-449a | 2 | miR2Disease |
| hsa-mir-1 | 2 | dbdemc | hsa-mir-92a | 2 | dbdemc; miR2Disease |
| hsa-mir-133a | 2 | dbdemc | hsa-mir-93 | 2 | unconfirmed |
| hsa-mir-133b | 2 | dbdemc | hsa-mir-99b | 2 | dbdemc; miR2Disease |
| hsa-mir-146a | 2 | dbdemc; miR2Disease | hsa-mir-101 | 2 | dbdemc; miR2Disease |
| hsa-mir-106a | 2 | dbdemc; miR2Disease | hsa-mir-146b | 2 | dbdemc; miR2Disease |
| hsa-mir-151a | 2 | dbdemc; miR2Disease | hsa-mir-148a | 2 | miR2Disease |
| hsa-mir-152 | 2 | dbdemc | hsa-mir-196b | 2 | dbdemc |
| hsa-mir-17 | 2 | miR2Disease | hsa-mir-27a | 2 | dbdemc; miR2Disease |
| hsa-mir-181b | 2 | dbdemc; miR2Disease | hsa-mir-27b | 2 | dbdemc; miR2Disease |
| hsa-mir-182 | 2 | dbdemc; miR2Disease | hsa-mir-30c | 2 | dbdemc; miR2Disease |
| hsa-mir-191 | 2 | dbdemc; miR2Disease | hsa-mir-34a | 2 | dbdemc; miR2Disease |
| hsa-mir-193b | 2 | dbdemc | hsa-mir-378a | 2 | unconfirmed |
Discussion
Nowadays, plenty of researches have confirmed that miRNAs are closely related with the development and progression of various human complex diseases, thus more attention has been paid to identify potential miRNA-disease associations to better understand the pathogenesis of disease at the miRNA level. Potential miRNA-disease associations could be predicted and ranked by computational models, and the most possible associations could be given priority for further experimental validation, which greatly speeds up the experimental validation processes. On the basis of the hypothesis that functional similar miRNAs are expected to be involved in similar diseases and vice versa, we developed a novel computational method of RKNNMDA which integrated miRNA functional similarity, disease semantic similarity, Gaussian interaction profile kernel similarity, and known miRNA-disease associations. First, we took advantage of KNN algorithm to obtain each miRNA's or each disease's k-nearest-neighbors which were sorted by the similarity scores between other miRNAs (diseases) and central miRNA (disease). Second, we calculated the Hamming loss of every miRNA pair, as well as the Hamming loss of every disease pair. Third, we constructed the SVM Ranking model on the basis of Hamming loss to rerank previously obtained k-nearest-neighbors of each miRNA and k-nearest-neighbors of each disease. Therefore, we obtained a group of miRNAs which contains each miRNA and its reranked k-nearest-neighbors, and a group of diseases which contains each disease and its reranked k-nearest-neighbors. Finally, we could infer possible diseases derived from miRNA's k-nearest-neighbors for each miRNA, as well as infer possible miRNAs derived from disease's k-nearest-neighbors for each disease. Thus we obtained 2 miRNA-disease association matrices. Finally, we assigned corresponding weight score to the 2 miRNA-disease association matrices respectively according to weighted voting and added the weight scores up. The weight score sum represents the possibility of associations which we used as the basis to obtain the final ranking list. RKNNMDA obtained a reliable AUC = 0.8221 in the LOOCV, indicating its reliable performance. Furthermore, 96% (Colon Neoplasms), 80% (Esophageal Neoplasms), and 94% (Prostate Neoplasms) of top 50 predicted miRNA-disease associations have been confirmed in case studies of 3 important human cancers. It is believed that RKNNMDA will be a useful tool with potential value in human disease prognosis, treatment, and prevention.
One important advantage of RKNNMDA is that it integrated several trustable biologic data sets and thus we obtained a much larger data pool compared with previous methods. Secondly, our method is a bilateral process which means that we adopted the same procedure of KNN, SVM Ranking, and weighted voting to both miRNA and disease to reduce prediction bias. A unilateral process might lead to false high weights for some miRNA-disease associations due to the incompleteness of initial data set from HMDD database. However, through our method, we were able to obtain 2 sets of weighted voting scores for miRNA-disease associations. Sequentially we added these 2 weighted voting scores together, and treated the sum as the final ranking basis. Furthermore, RKNNMDA could be applied to diseases without any known related miRNAs and miRNAs without any known related diseases which greatly expanded the application scope. Of course, RKNNMDA has some limitations and need improvements in the future. Firstly, obtaining sufficient experimental verified miRNA-disease associations still are in need, thus combing larger and heterogeneous biologic data sets will enhance the model effectiveness and accuracy.49-53 Secondly, RKNNMDA may cause bias to miRNAs with more known associated diseases. Furthermore, it needs further study to better integrate similarity networks, KNN algorithm, and SVM Ranking model to calculate association scores, such as introducing a practical algorithm to decide weighting parameters during calculation.54 In additional, some state-of-the-art computational models of other research fields could be introduced to the prediction of miRNA-disease association.55-57 Finally, we would further improve the current version of RKNNMDA to realize the miRNA-disease association types,54 disease-related miRNA-target interactions, and disease-related miRNA-environmental factors.58,59
A cancer hallmark network framework provides solutions to solve the mentioned limitations of RKNNMDA, which can effectively evaluate cancer risks based on miRNA profiles. As for personalized medicine, there remain 3 crucial problems in the term of miRNAs that could be considered in the future:1 how to obtain the tumor recurrence and metastases probability of patients;2 how to predict potential consequences after applying a specific drug to patients;3 how to identify molecular signatures to evaluate and predict chemotherapeutic results after cancer treatment.60,61 In addition, scientists discovered that tumors often contain subclones, which has already been confirmed by tumor sequencing. Through tumor genome sequencing, scientists are able to quantify and computationally dissect clones, and then clone-based analysis is available. By using miRNA-disease prediction method like RKNNMDA and combining the clone-based similarity network (eg. using the phenotypic data of different cancer subpopulations to construct cancer subclone similarity network) with miRNA similarity network, we are able to predict potential miRNAs-subclone associations.62,63
Materials and methods
MiRNA-disease associations
In the past few years, some experiment-supported evidences have already confirmed a certain number of miRNA-disease associations. Here, we downloaded 5430 verified miRNA-disease associations from the Human microRNA Disease Database (HMDD),28 which includes 495 miRNAs and 383 diseases. Furthermore, we constructed an adjacency matrix A, in which the entity A(i,j) represented whether miRNA m(i) was related to disease d(j). If there was association between m(i) and d(j), then the entity A(i,j) was equal to 1, otherwise 0.
MiRNA functional similarity
Based on the hypothesis that miRNAs with similar functions tend to be associated with diseases with similar phenotypes, we constructed miRNA functional similarity matrix FS.64 MiRNA functional similarity data set was downloaded from http://www.cuilab.cn/files/images/cuilab/misim.zip in January 2010. In miRNA functional similarity matrix FS, the entity FS(i,j) represented the functional similarity score between miRNA m(i) and m(j).
Disease semantic similarity
The relationship of different diseases can be described through a structure of Directed Acyclic Graph (DAG). A disease d can be described as DAG(d) = (d, T(d), E(d)), where T(d) is the node set including all ancestors of d and d itself, and E(d) is the edge set including the direct edges linking parent nodes to child nodes.49,50,64-66 We defined the contribution value of a disease t to disease d as follows:
| (1) |
Here Δ is the semantic contribution factor. For disease d, the contribution of itself is 1, while the contribution of another disease t decreases as the distance between d and t increases. Hence, the semantic value of disease d can be calculated according to the contribution of ancestor diseases and disease d itself, i.e.
| (2) |
According to the assumption that similar diseases tend to share more parts of their DAGs, we define the semantic similarity between disease a and b as follows:
| (3) |
Gaussian interaction profile kernel similarity for miRNAs
Based on the assumption that functionally similar miRNAs tend to be associated with similar diseases, we constructed Gaussian interaction profile kernel similarity for further similarity calculation.67 First, we introduced a binary vector IP(m(i)) to denote the interaction profile of miRNA m(i). IP(m(i)) represented the presence or absence of associations between each disease and miRNA m(i) in the known miRNA-disease association network, namely, the row i of the adjacency matrix A. Then we can calculate the Gaussian interaction profile kernel similarity between miRNA m(i) and m(j) as follows:
| (4) |
Here, the parameter γm was used to control the kernel bandwidth, and was calculated by normalizing a new bandwidth parameter γm’ by the average number of associations with diseases for all miRNAs. Therefore, the bandwidth parameter γm was defined as follows:
| (5) |
In principle, the new bandwidth parameter γm′ could be set with further cross-validation. In this paper we simply set γm′ = 1 according to some previous researches.67,68
Gaussian interaction profile kernel similarity for diseases
Similar to Gaussian interaction profile kernel similarity calculation for miRNAs, we used binary vector IP(d(i)) to denote the interaction profiles of disease d(i). Therefore, disease Gaussian interaction profile kernel similarity matrix could be calculated as follows:
| (6) |
| (7) |
Here, KD refers to the Gaussian interaction similarity for diseases, and the bandwidth parameter γd is obtained through the normalization that a new bandwidth parameter γd’ is divided by the average number of associations with miRNAs for all the diseases. Here, the new bandwidth γd’ was similarly set equal to 1.67,68
Integrated similarity for miRNAs and diseases
Given that miRNA functional similarity and Gaussian interaction similarity for miRNAs do not cover the whole miRNA-miRNA similarity network, we then integrated these 2 similarity matrices to enhance the prediction performance. That is to say, if miRNA m(i) and m(j) have functional similarity, we use the miRNA functional similarity value. Otherwise, we use the Gaussian kernel similarity value. Hence, the integrated similarity matrix for miRNAs is constructed as follows:
| (8) |
Similarly, the integrated similarity matrix for diseases could be constructed as follows:
| (9) |
RKNNMDA
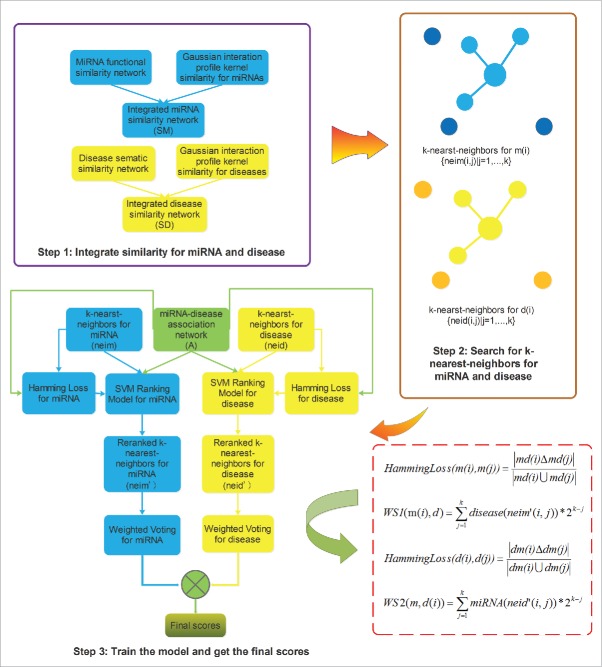
We developed the computational model of RKNNMDA to predict potential miRNA-disease associations by integrating miRNA-disease association adjacency matrix A, miRNA functional similarity matrix FS, disease semantic similarity matrix SS, Gaussian interaction profile kernel similarity matrices for miRNAs (KM) and diseases (KD) (the code of this method could be downloaded from http://www.escience.cn/system/file?fileId=87100). Fig. 2 demonstrates the entire process in the form of a flowchart. To begin with, based on the KNN algorithm, we could find k-nearest-neighbors neim(i) of a selected miRNA m(i). However, the initial similarity-based ranking of k-nearest-neighbors was not reliable for further prediction because KNN is a type of instance-based learning, or lazy learning, whose drawback lies in that the class which contains larger training examples tends to dominate the prediction of the new example and leads to the unbalanced outcome for the new example. Therefore, we introduced the SVM Ranking model to rerank these previously sorted neighbors.69-72 SVM Ranking model is an variant of the SVM algorithm, and is used to solve certain ranking problems via learning, i.e., it extracts special features from training data sets and ranks the given examples according to previously learned features. Hamming loss, which calculates the inconsistent proportion of 2 examples, acts as an essential training data set which the SVM Ranking model learns from. Given the selected miRNA m(i), we need to obtain associated disease label sets for each neighbor m(j), marked as md(j), and the associated disease label set for m(i) itself, marked as md(i), from adjacency matrix A, respectively. Then the Hamming loss could be defined as follows:
| (10) |
Figure 2.
Flowchart of potential miRNA-disease association prediction based on the computational model of RKNNMDA.1 Obtaining integrated similarity matrices by combining miRNA functional similarity, disease semantic similarity, and Gaussian interaction profile kernel similarity;2 Applying KNN algorithm and searching for k-nearest-neighbors for miRNA and disease;3 Calculating Hamming loss, reranking k-nearest-neighbors by SVM Ranking model, and implementing prediction based on weighted voting.
Here, the denominator refers to the number of elements of union set of md(i) and md(j), while the numerator refers to the number of elements in the symmetric difference set between md(i) and md(j). After calculating the Hamming loss between m(i) and all its neighbors, we inputted these outcomes as training data set to the SVM Ranking software which we downloaded from http://www.cs.cornell.edu/people/tj/svm_light/svm_rank.html. Then, the SVM Ranking model outputted reranked reliable k-nearest-neighbors neim’(i) for miRNA m(i). Next, by examining adjacency matrix A, we identified diseases having known associations with the neighboring miRNAs of m(i), i.e., miRNAs in neim’(i), thus obtained corresponding association probability between m(i) and these diseases. The next step is to give the corresponding weight score to each miRNA-disease association that we obtained from the last step for the final possibility sorting by means of weighted voting. The weight score (WS) between m(i) and disease d was defined as the following formula:
| (11) |
Here, neim’(i,j) refers to the jth neighboring miRNA of m(i) and disease(neim’(i,j)) refers to disease d's feature score with regard to miRNA m(i) and its neighbor m(j). Because we lacked real feature data for diseases, we used miRNA functional similarity score between m(i) and m(j) as the feature score of d since m(j) was related to d and thus m(j) represented one of d's features, so this similarity score between m(i) and m(j) to some extant represented the relationship between m(i) and d. As the weight score value increases, the miRNA m(i) is more likely to be associated with disease d.
To further enhance the accuracy of the computational model, we also applied similar methodology to achieve k-nearest-neighbors of disease d(i), reranked these neighbors by the SVM Ranking model based on Hamming loss input and implemented weighted voting to each predicted miRNA-disease association. Hamming loss for disease d(i) and d(j), and weight score by means of weighted voting between d(i) and miRNA m could be defined as follows:
| (12) |
| (13) |
Here, dm(i) and dm(j) refers to the related miRNA label sets of disease d(i) and d(j), respectively. The denominator of HammingLoss(d(i),d(j)) refers to the number of elements of union set of dm(i) and dm(i), while the numerator refers to the number of elements in the symmetric difference set between dm(i) and dm(i). Besides, neid’(i,j) represents jth neighboring disease of d(i), and miRNA(neid’(i,j)) represents miRNA m's feature score with regard to disease d(i) and its neighbor d(j). Similarly, as the lack of real feature data for miRNAs, we used disease semantic similarity score between d(i) and d(j) as the feature score of m since d(j) related to m and thus d(j) represented one of m's features, so this similarity score between d(i) and d(j) to some extant represented the relationship between d(i) and m.
Finally, we added WS1 and WS2 together and ranked the potential miRNA-disease associations.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
XC was supported by the National Natural Science Foundation of China under Grant No. 11631014. GYY was supported by National Natural Science Foundation of China under Grant No. 11371355 and 11631014.
References
- 1.Ambros V. Tiny regulators with great potential. Cell 2001; 107(7):823-6; PMID:11779458; https://doi.org/ 10.1016/S0092-8674(01)00616-X [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature 2004; 431(7006):350-5; PMID:15372042; https://doi.org/ 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004; 116(2):281-97; PMID:14744438; https://doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 4.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004; 431(7006):343-9; PMID:15372041; https://doi.org/ 10.1038/nature02873 [DOI] [PubMed] [Google Scholar]
- 5.Vasudevan S, Tong YC, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007; 318(5858):1931-4; PMID:18048652; https://doi.org/ 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 6.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005; 309(5740):1577-81; PMID:16141076; https://doi.org/ 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- 7.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 2005; 33(4):1290-7; PMID:15741182; https://doi.org/ 10.1093/nar/gki200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karp X, Ambros V. Encountering MicroRNAs in cell fate signaling. Science 2005; 310(5752):1288-9; PMID:16311325; https://doi.org/ 10.1126/science.1121566 [DOI] [PubMed] [Google Scholar]
- 9.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 2005; 15(5):563-8; PMID:16099643; https://doi.org/ 10.1016/j.gde.2005.08.005 [DOI] [PubMed] [Google Scholar]
- 10.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet 2004; 20(12):617-24; PMID:15522457; https://doi.org/ 10.1016/j.tig.2004.09.010 [DOI] [PubMed] [Google Scholar]
- 11.Alshalalfa M, Alhajj R. Using context-specific effect of miRNAs to identify functional associations between miRNAs and gene signatures. Bmc Bioinformatics 2013; 14(Suppl 12):839-50; ; https://doi.org/ 10.1186/1471-2105-14-S12-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009; 136(2):215-33; PMID:19167326; https://doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol 2006; 2(1):46; PMID:16969338; https://doi.org/ 10.1038/msb4100089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Xingping WU, Yan L. Serum miR-103 as a potential diagnostic biomarker for breast cancer. Nan Fang Yi Ke Da Xue Xue Bao 2012; 32(5):631-4; PMID:22588912; https://doi.org/ 10.3969/j.issn.1673-4254.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 15.Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis 2012; 6(9):900-4; PMID:22386737; https://doi.org/ 10.1016/j.crohns.2012.02.006 [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Peng W, Ouyang X, Dai Y. Reduced circulating miR-15b is correlated with phosphate metabolism in patients with end-stage renal disease on maintenance hemodialysis. Ren Fail 2012; 34(6):685-90; PMID:22512691; https://doi.org/ 10.3109/0886022X.2012.676491 [DOI] [PubMed] [Google Scholar]
- 17.Jiang Q, Hao Y, Wang G, Juan L, Zhang T, Teng M, Liu Y, Wang Y. Prioritization of disease microRNAs through a human phenome-microRNAome network. BMC Syst Biol 2010; 4(Suppl 1):S2; PMID:20522252; https://doi.org/ 10.1186/1752-0509-4-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Xu J, Zhang G, Xu L, Li C, Li W, Zhao Z, Jiang W, Guo Z, Li X. Walking the interactome to identify human miRNA-disease associations through the functional link between miRNA targets and disease genes. BMC Syst Biol 2013; 7:101; PMID:24103777; https://doi.org/ 10.1186/1752-0509-7-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mørk S, Pletscherfrankild S, Caro AP, Gorodkin J, Jensen LJ. Protein-driven inference of miRNA-disease associations. Bioinformatics 2014; 30(3):392-7; PMID:24273243; https://doi.org/ 10.1093/bioinformatics/btt677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xuan P, Han K, Guo M, Guo Y, Li J, Ding J, Liu Y, Dai Q, Li J, Teng Z, et al.. Prediction of microRNAs associated with human diseases based on weighted k most similar neighbors. PloS One 2013; 8(8):e70204; PMID:23950912; https://doi.org/ 10.1371/journal.pone.0070204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Li CX, Lv JY, Li YS, Xiao Y, Shao TT, Huo X, Li X, Zou Y, Han QL, et al.. Prioritizing candidate disease miRNAs by topological features in the miRNA target-dysregulated network: Case study of prostate cancer. Mol Cancer Ther 2011; 10(10):1857-66; PMID:21768329; https://doi.org/ 10.1158/1535-7163.MCT-11-0055 [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Liu MX, Yan GY. RWRMDA: Predicting novel human microRNA-disease associations. Mol Biosyst 2012; 8(10):2792-8; PMID:22875290; https://doi.org/ 10.1039/c2mb25180a [DOI] [PubMed] [Google Scholar]
- 23.Xuan P, Han K, Guo Y, Li J, Li X, Zhong Y, Zhang Z, Ding J. Prediction of potential disease-associated microRNAs based on random walk. Bioinformatics 2015; 31(11):1805-15; PMID:25618864; https://doi.org/ 10.1093/bioinformatics/btv039 [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Yan GY. Semi-supervised learning for potential human microRNA-disease associations inference. Sci Rep 2014; 4:5501; PMID:24975600; https://doi.org/ 10.1038/srep05501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Yan CC, Zhang X, You ZH, Deng L, Liu Y, Zhang Y, Dai Q. WBSMDA: Within and between score for MiRNA-disease association prediction. Sci Rep 2016; 6:21106; PMID:26880032; https://doi.org/ 10.1038/srep21106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Clarence YC, Zhang X, Li Z, Deng L, Zhang Y, Dai Q. RBMMMDA: Predicting multiple types of disease-microRNA associations. Sci Rep 2015; 5:13877; PMID:26347258; https://doi.org/ 10.1038/srep13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Clarence Yan C, Zhang X, You ZH, Huang YA, Yan GY. HGIMDA: Heterogeneous graph inference for miRNA-disease association prediction. Oncotarget 2016; 7(40):65257-69; PMID:27533456; https://doi.org/ 10.18632/oncotarget.11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, Cui Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res 2014; 42(Database issue):D1070-4; PMID:24194601; https://doi.org/ 10.1093/nar/gkt1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 2009; 37(Database issue):D98-104; PMID:18927107; https://doi.org/ 10.1093/nar/gkn714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Ren F, Liu C, He S, Sun G, Gao Q, Yao L, Zhang Y, Miao R, Cao Y, et al.. dbDEMC: A database of differentially expressed miRNAs in human cancers. BMC Genomics 2010; 11(Suppl 4):S5; PMID:21143814; https://doi.org/ 10.1186/1471-2164-11-S4-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61(2):69-90; PMID:21296855; https://doi.org/ 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 32.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, et al.. Circulating exosomal microRNAs as biomarkers of colon cancer. PloS One 2014; 9(4):e92921; PMID:24705249; https://doi.org/ 10.1371/journal.pone.0092921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med 2006; 79(3–4):131-40; [PMC free article] [PubMed] [Google Scholar]
- 34.Cahill S, Smyth P, Denning K, Flavin R, Li J, Potratz A, Guenther SM, Henfrey R, O'Leary JJ, Sheils O. Effect of BRAF V600E mutation on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer 2007; 6:21; PMID:17355635; https://doi.org/ 10.1186/1476-4598-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 2007; 72(5–6):397-402; PMID:18196926; https://doi.org/ 10.1159/000113489 [DOI] [PubMed] [Google Scholar]
- 36.Feng YH, Wu CL, Shiau AL, Lee JC, Chang JG, Lu PJ, Tung CL, Feng LY, Huang WT, Tsao CJ. MicroRNA-21-mediated regulation of Sprouty2 protein expression enhances the cytotoxic effect of 5-fluorouracil and metformin in colon cancer cells. Int J Mol Med 2012; 29(5):920-6; PMID:22322462; https://doi.org/ 10.3892/ijmm.2012.910 [DOI] [PubMed] [Google Scholar]
- 37.Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH, Majumdar AP. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis 2012; 33(1):68-76; PMID:22072622; https://doi.org/ 10.1093/carcin/bgr246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart BW, Kleihues P. World cancer report: IARC press; 2003. [Google Scholar]
- 39.Polednak AP. Trends in survival for both histologic types of esophageal cancer in U.S. surveillance, epidemiology and end results areas. Int J Cancer 2003; 105(1):98-100; PMID:12672037; https://doi.org/ 10.1002/ijc.11029 [DOI] [PubMed] [Google Scholar]
- 40.Berry MF. Esophageal cancer: Staging system and guidelines for staging and treatment. J Thorac Dis 2014; 6(Suppl 3):S289-S97; PMID:24876933; https://doi.org/ 10.3978/j.issn.2072-1439.2014.03.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hummel R, Watson DI, Smith C, Kist J, Michael MZ, Haier J, Hussey DJ. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J Gastrointest Surg 2011; 15(3):429-38; PMID:21246413; https://doi.org/ 10.1007/s11605-011-1418-9 [DOI] [PubMed] [Google Scholar]
- 42.Hamano R, Miyata H, Yamasaki M, Kurokawa Y, Hara J, Moon JH, Nakajima K, Takiguchi S, Fujiwara Y, Mori M, et al.. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res 2011; 17(9):3029-38; PMID:21248297; https://doi.org/ 10.1158/1078-0432.CCR-10-2532 [DOI] [PubMed] [Google Scholar]
- 43.Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et al.. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: Associations with survival. Clin Cancer Res 2009; 15(19):6192-200; PMID:19789312; https://doi.org/ 10.1158/1078-0432.CCR-09-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ko MA, Guan Z, Virtanen C, Guindi M, Waddell TK, Keshavjee S, Darling GE. MicroRNA expression profiling of esophageal cancer before and after induction chemoradiotherapy. Ann Thorac Surg 2012; 94(4):1094-103; PMID:22939244; https://doi.org/ 10.1016/j.athoracsur.2012.04.145 [DOI] [PubMed] [Google Scholar]
- 45.Sun D, Layer R, Mueller AC, Cichewicz MA, Negishi M, Paschal BM, Dutta A. Regulation of several androgen-induced genes through the repression of the miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells. Oncogene 2014; 33(11):1448-57; PMID:23503464; https://doi.org/ 10.1038/onc.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueno K, Hirata H, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. microRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer 2013; 108(8):1659-67; PMID:23538390; https://doi.org/ 10.1038/bjc.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res 2007; 67(13):6130-5; PMID:17616669; https://doi.org/ 10.1158/0008-5472.CAN-07-0533 [DOI] [PubMed] [Google Scholar]
- 48.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med 2008; 86(3):313-22; PMID:18193184; https://doi.org/ 10.1007/s00109-007-0296-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Yan CC, Luo C, Ji W, Zhang Y, Dai Q. Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Sci Rep 2015; 5:11338; PMID:26061969; https://doi.org/ 10.1038/srep11338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Yan CC, Zhang X, You Z-H. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief Bioinform 2016:bbw060; PMID:27345524; https://doi.org/ 10.1093/bib/bbw060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, Ren B, Chen M, Wang Q, Yan G, Zhang L. NLLSS: Predicting synergistic drug combinations based on semi-supervised learning. PLOS Comput Biol 2016; 12(7):e1004975; PMID:27415801; https://doi.org/ 10.1371/journal.pcbi.1004975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y-A, You Z-H, Chen X, Chan K, Luo X. Sequence-based prediction of protein-protein interactions using weighted sparse representation model combined with global encoding. BMC Bioinformatics 2016; 17:184; PMID:27112932; https://doi.org/ 10.1186/s12859-016-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong L, You Z-H, Ming Z, Li J, Chen X, Huang Y-A. Detection of interactions between proteins through rotation forest and local phase quantization descriptors. Int J Mol Sci 2016; 17(1):21; ; https://doi.org/ 10.3390/ijms17010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, You Z, Yan G, Gong D. IRWRLDA: Improved random walk with restart for LncRNA-disease association prediction. Oncotarget 2016; 7(36):57919-31; PMID:27517318; https://doi.org/ 10.18632/oncotarget.11141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Z-A, Chen X, Zhu Z, Liu H, Yan G-Y, You Z-H, Wen Z. PBHMDA: Path-based human microbe-disease association prediction. Front Microbiol 2017; 8:233; PMID:28275370; https://doi.org/ 10.3389/fmicb.2017.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, Huang Y-A, You Z-H, Yan G-Y, Wang X-S. A novel approach based on KATZ measure to predict associations of human microbiota with non-infectious diseases. Bioinformatics 2017; 33(5):733-9; PMID:28025197; https://doi.org/ 10.1093/bioinformatics/btw715 [DOI] [PubMed] [Google Scholar]
- 57.Chen X. Drug-target interaction prediction: Databases, web servers and computational models. Brief Bioinform 2016; 17(4):696-712; PMID:26283676; https://doi.org/ 10.1093/bib/bbv066 [DOI] [PubMed] [Google Scholar]
- 58.Chen X. miREFRWR: A novel disease-related microRNA-environmental factor interactions prediction method. Mol Biosyst 2016; 12(2):624-33; PMID:26689259; https://doi.org/ 10.1039/C5MB00697J [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Liu MX, Cui QH, Yan GY. Prediction of disease-related interactions between MicroRNAs and environmental factors based on a semi-supervised classifier. PloS One 2012; 7(8):e43425; PMID:22937049; https://doi.org/ 10.1371/journal.pone.0043425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao S, Tibiche C, Zou J, Zaman N, Trifiro M, O'Connormccourt M, Wang E, et al.. Identification and construction of combinatory cancer hallmark-based gene signature sets to predict recurrence and chemotherapy benefit in stage ii colorectal cancer. Jama Oncol 2015; 2(1):1-9; ; https://doi.org/ 10.1001/jamaoncol.2015.3413 [DOI] [PubMed] [Google Scholar]
- 61.Wang E, Zaman N, Mcgee S, Milanese JS, Masoudinejad A, O'Connormccourt M. Predictive genomics: A cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol 2015; 30:4-12; PMID:24747696; https://doi.org/ 10.1016/j.semcancer.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 62.Wang E, Zou J, Zaman N, Beitel LK, Trifiro M, Paliouras M. Cancer systems biology in the genome sequencing era: Part 1, dissecting and modeling of tumor clones and their networks. Semin Cancer Biol 2014; 23(4):279-85; ; https://doi.org/ 10.1016/j.semcancer.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 63.Wang E, Zou J, Zaman N, Beitel LK, Trifiro M, Paliouras M. Cancer systems biology in the genome sequencing era: Part 2, evolutionary dynamics of tumor clonal networks and drug resistance. Semin Cancer Biol 2013; 23(4):286-92; PMID:23792107; https://doi.org/ 10.1016/j.semcancer.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 64.Wang D, Wang J, Lu M, Song F, Cui Q. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics 2010; 26(13):1644-50; PMID:20439255; https://doi.org/ 10.1093/bioinformatics/btq241 [DOI] [PubMed] [Google Scholar]
- 65.Chen X, Huang Y, Wang X, You Z, Chan K. FMLNCSIM: Fuzzy measure-based lncRNA functional similarity calculation model. Oncotarget 2016; 7(29):45948-58; PMID:27322210; https://doi.org/ 10.18632/oncotarget.10008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang Y, Chen X, You Z, Huang D, Chan K. ILNCSIM: Improved lncRNA functional similarity calculation model. Oncotarget 2016; 7(18):25902-14; PMID:27028993; https://doi.org/ 10.18632/oncotarget.8296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Laarhoven T, Nabuurs SB, Marchiori E. Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics 2011; 27(21):3036-43; PMID:21893517; https://doi.org/ 10.1093/bioinformatics/btr500 [DOI] [PubMed] [Google Scholar]
- 68.Chen X, Yan G-Y. Novel human lncRNA–disease association inference based on lncRNA expression profiles. Bioinformatics 2013; 29(20):2617-24; PMID:24002109; https://doi.org/ 10.1093/bioinformatics/btt426 [DOI] [PubMed] [Google Scholar]
- 69.Joachims T. Making large-scale SVM learning practical. Tech Rep 1998; 8(3):499-526; https://doi.org/ 10.17877/DE290R-14262 [DOI] [Google Scholar]
- 70.Joachims T. editor A support vector method for multivariate performance measures. Proceedings of the 22nd International Conference on Machine learning; 2005: ACM. [Google Scholar]
- 71.Joachims T. Training linear SVMs in linear time. Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining; New York: ACM; 2006. p. 217-26. [Google Scholar]
- 72.Joachims T, Finley T, Yu CNJ. Cutting-plane training of structural SVMs. Mach Learn 2009; 77(1):27-59; https://doi.org/ 10.1007/s10994-009-5108-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.