Abstract
The genetic diversity in Plasmodium falciparum antigens is a major hurdle in developing an effective malaria vaccine. Protective efficacy of the vaccine is dependent on the polymorphic alleles of the vaccine candidate antigens. Therefore, we investigated the genetic diversity of the potential vaccine candidate antigens i.e. msp-1, msp-2, glurp, csp and pfs25 from field isolates of P.falciparum and determined the natural immune response against the synthetic peptide of these antigens. Genotyping was performed using Sanger method and size of alleles, multiplicity of infection, heterogeneity and recombination rate were analyzed. Asexual stage antigens were highly polymorphic with 55 and 50 unique alleles in msp-1 and msp-2 genes, respectively. The MOI for msp-1 and msp-2 were 1.67 and 1.28 respectively. A total 59 genotype was found in glurp gene with 8 types of amino acid repeats in the conserved part of RII repeat region. The number of NANP repeats from 40 to 44 was found among 55% samples in csp gene while pfs25 was found almost conserved with only two amino acid substitution site. The level of genetic diversity in the present study population was very similar to that from Asian countries. A higher IgG response was found in the B-cell epitopes of msp-1 and csp antigens and higher level of antibodies against csp B-cell epitope and glurp antigen were recorded with increasing age groups. Significantly, higher positive responses were observed in the csp antigen among the samples with ≥42 NANP repeats. The present finding showed extensive diversity in the asexual stage antigens.
Introduction
Globally about 3.2 billion people are at risk of malaria and 214 million new malaria cases and 438,000 deaths was reported in 2015[1]. South East Asia Region contributed 10% of the global malaria cases, of which India alone accounts for 70% of the cases [1]. Plasmodium falciparum contributes 67% of the total malaria cases in India with a greatly varied proportion from 0% to 93% in different states [2,3]. Malaria is a major health problem in rural/tribal areas of the Central Eastern and North Eastern States of India, which are having large groups of ethnic population [4]. Currently, an increasing number of countries including India are in the process of eliminating malaria. However in India, despite of scaling up of interventions such as use of insecticides treated net (ITN), indoor residual spray (IRS), improved diagnostic test and treatment using artemisnin combination therapy (ACT), malaria positivity is increasing 0.78% to 0.95% from 2013 to 2015 [2].
Therefore, highly effective malaria vaccine is definitely needed to achieve the target of malaria elimination. Although there is no effective vaccine as on date, however a number of potential stage specific vaccine candidate antigens of P. falciparum are under various stages of the development [5]. Understanding, the genetic diversity and population structure of the parasite is crucial as high genetic diversity especially at the surface- exposed antigens posses great challenge in developing effective malaria vaccine [6]. Almost all P. falciparum antigens currently under consideration for vaccine development, exhibit polymorphism in the field isolates from various part of the world [7–14]. Furthermore, antibodies induced by parasitic infection are in large part directed against the specific allele. Several studies have demonstrated the presence of B cell epitopes in the repeats of circumsporozoite protein (csp), merozoite surface protein 1 (msp1), merozoite surface protein 2 (msp2) and glutamate rich protien (glurp) of P. falciparum [15–16]. The immune status of the individuals and exposure to infection play essential role in the clinical response and development of natural immunity to malaria infection, respectively [17]. In endemic areas, immunity to malaria develops slowly and repeated parasite exposure gradually reinforces protective immunity, but disappear within a few months or non-exposure [18]. In addition, if immunity to infection is strain specific, then a state of generalized immunity would develop once exposure had occurred to a large enough sample of the many distinct parasite strains circulating in that region [19]. Therefore, the extensive polymorphisms of surface antigens contribute to the immune evasion and also appear to restrict the effectiveness of vaccine against the P.falciparum polymorphic proteins. Given the importance of antigenic diversity in influencing the outcome of any vaccine, we have conducted genetic polymorphism of vaccine candidate antigens (pfmsp1, pfmsp2, pfglurp, pfcsp and pfs25) and antibody responses in P. falciparum infected individual from Chhattisgarh state, Central India which is second malarious state contributing 12% of malaria cases in India.
Materials and methods
Study area, population and sample collection
This study was carried out at Janakpur Community Health Care (CHC), district Baikunthpur, Chhattisgarh, Central India (Fig 1). This is a secondary health care facility situated in the remote area of the district (23.7191° N, 81.7883° E and 550 M height above sea level) surround by dense forest (60%) and majority of the population is tribal (65%). P. falciparum is the predominant species followed by P. vivax and both Anopheles culicifacies and An. fluviatilis are responsible for transmission the disease in this area. A malaria clinic of National Institute for Research in Tribal Health (NIRTH) of Indian Council of Medical Research (ICMR) was established at Janakpur CHC (60 bedded hospital), Baikunthpur district, during August 2013 to March 2015. Symptomatic patients were screened for malaria parasites by microscopy using thick and thin blood smears stained with JSB stain [20]. Patients were given treatment as per National Vector Borne disease control programme [21]. The patient’s socio-demographics and clinical parameters such as age, gender, social groups, clinical history (Headache, Vomiting, Diarrhoea) history of malarial fever, and parasitaemia was recorded on the structured questionnaire. The intravenous blood samples, in sterile conditions, were collected from the patients positive for P. falciparum malaria after taking written informed consent. Plasma and red blood cells were separated and stored at -80°C for further use in immunological and molecular assays, respectively.
Fig 1. Map of India showing the study site.
Ethical approval
The study protocol for patient participation and collection of blood samples for laboratory testing was reviewed and approved by the institutional ethics committee of NIRTH, Jabalpur. All study participants’ provided written informed consent prior to their participation, accroding to ICMR guidelines. A copy of the consent form in the local language was also provided and explained to the patients or parents/Guardian of children. The participation of other institutes was approved by the ICMR, New Delhi, India.
Genomic DNA extraction and PCR based species identification
Genomic DNA was isolated by QIAamp DNA blood mini kit as per the manufacturer's instructions (Qiagen, CA, USA) and stored at -20°C for further use. Genus and species-specific nested PCR was performed using 18S rRNA gene to detect malaria parasite species (P. falciparum, P. vivax, P. ovale, and P. malariae), as described previously [22].
Genotyping of P. falciparum antigenic markers
The amplification of highly polymorphic antigens; pfmsp1 (block -2 region), pfmsp2 (central repeat region), pfglurp (polymorphic RII region), pfcsp (central repeat region) and pfs25 was performed in GeneAmp® PCR System 2700 (Applied Biosystems). The PCR reaction mixture and amplification conditions were summarized in S1 Table. Briefly, PCR amplification was carried out in a 25 μL reaction mixture containing 50–100 ng genomic DNA template and 1X of MgCl2 free buffer, 1.5 mM of MgCl2, 200 μM of dNTPs, 0.2 mM of each primer and 2.5U of Taq polymerase enzyme (Invitrogen Life Sciences, Carlsbad, CA, USA). The appropriate bands excised from the gel and purified using AccuPrep gel extraction kit, according to manufacturer’s protocol (Bioneer Corporation). The purified products were sequenced using both forward and reverse primers, using ABI Big Dye Terminator Ready Reaction Kit Version 3.1 (PE Applied Biosystems, CA, USA) on an ABI-3130xL genetic analyzer. The BioEdit Sequence Alignment Editor and Gene Doc Version 2.6.002 were used to analyze the sequencing electropherograms and generate sequence alignment, respectively. The sequence data generated in this study have been submitted into the NCBI GenBank database. Amino acid sequences of P. falciparum msp1 (block-2), msp2 (central repeat region), csp (central repeat region) from present study and partial/complete amino acid sequences from NCBI database were retrieved using BATCH Entrez and considered for further analysis. Blast clust tool was used to identify the variants from globally available sequence. The details about these isolates have been provided in S2 Table. To study the genetic relationship among the global sequences of pfmsp1, pfmsp2 and pfcsp, Minimal Spanning Tree (MST) was constructed using MLST Clustering algorithm implemented in BioNumerics version 7.6.1 (Applied-Maths, Inc. Austin, TX). The haplotype with the highest numbers of single locus variants (SLVs) was considered as a root haplotype and all other haplotypes as relatives.
Multiplicity of infection (MOI)
The multiplicity of infection (MOI) was defined as the mean number of P. falciparum genotypes per infected individual. The MOI was calculated as the proportion of the total number of P. falciparum genotypes for the same gene and the number of PCR positive isolates.
Tests of neutrality, recombination and Statistical analysis
Genetic parameters of P. falciparum gene specific allelic types were determined by different methods implemented MEGA 7 Software [23]. The average number of substitutions (π) per site between any two sequence and heterozygosity per site (θ) was determined as described earlier (Nei and Tamura) [24,25]. Intra and Inter specific transitions and transversion was estimated using the pair-wise differences without any correction. The average number nonsynonymous (dN) and of synonymous substitutions (dS) per site, was estimated using the method of Nei and Gojobori with the Jukes and Cantor [26,27]. Tajima test was performed for detecting the positive natural selection in maintaining genetic polymorphism based on Statistic D and PHI test was performed to detect the overall recombination at each locus the with SPLITSTREE (ver. 4.13) [28]. Window size (W) was set to 100 bases (Φw = 100), and the statistical significance of Φw values were assessed using a permutation test with 1,000 iterations {H0 (no recombination, Φw ≠0) rejected at α = 0.05 in favor of H1 (recombination)} [29].
Enzyme-linked immunosorbent assay
The total IgG response against the synthetic peptides of pfmsp1 B & T cell epitopes, pfmsp2 B cell epitope, pfcsp B & T cell epitopes and pfglurp were quantified by direct ELISA. Details of peptide sequence were given in the supplementary table (S3 Table). Briefly, 96-well microtitre plates (Nunc-Immuno, Thermo Scientific) were coated overnight at 4°C with 5 μg/ml of each antigenic peptides. After washing, the plates were blocked with 1% skimmed milk for 1 hour at 37°C. Subsequently the plasma samples were diluted (1:100) in PBS-5% skimmed milk, 0.1% Tweeen-20 and incubated for 1 hour at 37°C. The plates were washed four times in PBS 0.1% Tween-20 and incubated for 1 hour at 37°C following the addition of horseradish peroxidase-conjugated goat-antihuman-IgG (1:4000 in PBS-5% skimmed milk, 0.1% Tweeen-20). The assay was developed by adding the tetra-methylbenzidine enzyme substrate and incubated for 30 minutes at 37°C. The reaction was stopped with 2M H2SO4. The OD was measured at 450 nm using an ELISA plate reader.
To determine the specificity of the assay, sera samples of 16 healthy individuals from non-endemic area (not exposed to malaria) were used as negative control. Prevalence of total IgG immune response for the different antigens were considered positive if their OD values were higher than the mean plus two standard deviations of negative control after subtraction. The percentage prevalence was calculated as follows: (total no. of positive samples / total no. of sample tested) x 100. Odd ratio (OR) was calculated for association between the different age groups.
Results
Demographic profiles of the study population
A total of 6718 patients were screened, of which 5.2% (n = 352) were found positive for malaria parasite {P.falciparam (n = 271), P.vivax (n = 79) and two mixed infection of P.falciparam and P.vivax}. Out of 271, polymerase chain reaction (PCR) positive 180 mono-infected P. falciparam patients who fulfilled the enrolment criteria were included in the study. The age range across the sample was 01 to 75 years with a mean age of 20.34±15.19 year. Fifty two percent (n = 93) of the patients were female and the rest were male, only 7% (n = 13) of the patients had history of previous malaria infection. Additionally, majority of the participants were from the tribal community (75%, n = 134). Majority (94%) of the patients had fever at the time of enrollment followed by headache (90%), vomiting (75%) and diarrhoea (5%). The mean parasite density was 5827.05 ± 17295(95%CI, 3283.29–8370.80).
Allelic diversity of pfmsp1 and pfmsp2
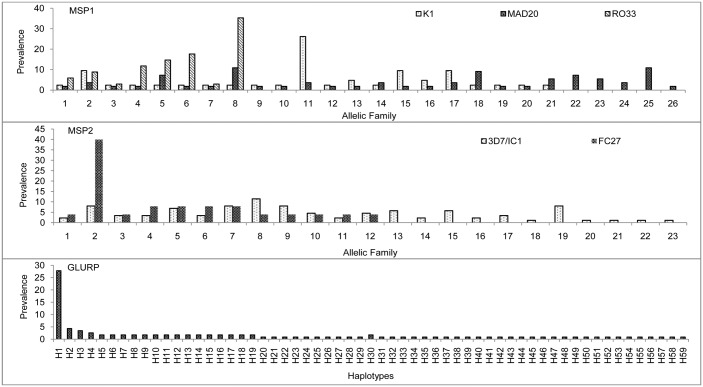
A total 131(73%) samples were successfully sequenced and analyzed for block-2 region of pfmsp1 gene. The overall allelic prevalence was higher in MAD20 (42%) followed by K1 (32%) and RO33 (26%) (Table 1). Fifty five unique alleles were observed, of which majority were found in MAD20 (26 alleles) followed by K1 (21 alleles) and RO33 (8 alleles) (Fig 2). The average number of amino acid was significantly higher in K1 and MAD 20 when it compared to 3D7 and HN2 reference strains, respectively (Table 1). The different alleles of RO33 were based on only substitutions while in the K1 and MAD20 family both insertion/deletion as well as substitutions were observed. The dN/dS ratio showed the positive selection for K1 and MAD20 alleles while RO33 was found to be neutral (Table 2). Thirty seven percent of the samples were polyclonal infection and the MOI for msp1 was 1.67. The NCBI GenBank database accession number msp1 allelic family is KY425824—KY425878.
Table 1. Distribution of allelic family and size variations of vaccine candidate genes in Indian P. falciparum isolates.
| Genes (n) |
Allelic family type | No. of samples | Percentage | No. of variants | Allele Size | Standard strain (amino acid) | MOI |
|---|---|---|---|---|---|---|---|
|
pfmsp1 (131) |
K1 | 42 | 32.06 | 21 | 148.48 ± 8.99* (132–171) |
3D7 (133) | 1.67 |
| MAD20 | 55 | 41.98 | 26 | 143.18 ± 8.61* (126–156) |
HN2 (127) | ||
| RO33 | 34 | 25.95 | 8 | 132.00 ± 0.00 (132–132) |
RO33 (132) | ||
| Mixed | 48 | ||||||
|
pfmsp2 (112) |
3D7/IC1 | 87 | 77.68 | 38 | 210.46 ± 11.69* (193–248) |
3D7 (245) | 1.28 |
| FC27 | 25 | 22.32 | 12 | 191.04 ± 10.86* (184–225) |
IGH-CR14 (185) | ||
| Mixed | 31 | ||||||
|
pfglurp (115) |
- | - | - | 59 | 257.04 ± 28.31* (156–317) |
3D7 (303) | |
|
pfcsp (97) |
- | - | - | 22 | 289.30 ± 15.24* (266–321) |
3D7 (277) | |
|
pfs25 (155) |
Wild type | 84 | 54.29 | - | 155 | 3D7 (155) | |
| A131G | 45 | 29.03 | - | 155 | 3D7 (155) | ||
| V143A | 26 | 16.77 | - | 155 | 3D7 (155) |
* P<0.05
Fig 2. Frequency distribution of different allelic variations of P. falciparum msp1, msp2 and glurp genes in Indian isolates.
Table 2. Nucleotide diversity test of neutrality and rate of recombination test of vaccine candidate genes in Indian P. falciparum isolates.
| Gene | No. of Allelic types | S | θ | π | Transitions | Transversions | dN | dS | dN/dS ratio | Tajima's D | PHI test P value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| pfcsp | 22 | 55 | 0.02473 | 0.02143 | 6.329 | 6.7402 | 0.0087 | 0.0788 | 0.110 | -0.529924 | 0.000 |
| pfglurp | 59 | 21 | 0.00854 | 0.00524 | 0.6166 | 2.15546 | 0.0038 | 0.0072 | 0.528 | -1.209877 | 0.3584 |
| pfmsp1- K1 | 21 | 24 | 0.01728 | 0.00899 | 1.3857 | 2.0857 | 0.0103 | 0.0055 | 1.873 | -1.830012 | 0.0380 |
| pfmsp1- MAD20 | 26 | 38 | 0.02853 | 0.03030 | 5.329 | 5.246 | 0.0330 | 0.0295 | 1.119 | 0.233564 | 0.000 |
| pfmsp1- RO33 | 8 | 12 | 0.01169 | 0.01380 | 3.607 | 1.857 | 0.0137 | 0.0149 | 0.919 | 0.909062 | 0.0150 |
| pfmsp2-3D7/IC1 | 38 | 53 | 0.02502 | 0.033894 | 9.3058 | 7.776 | 0.0415 | 0.0039 | 10.641 | 1.275263 | 0.000 |
| pfmsp2-FC27 | 12 | 59 | 0.03527 | 0.03457 | 7.015 | 12.136 | 0.0376 | 0.0318 | 1.182 | -0.091029 | 0.0248 |
| pfs25 | 3 | 2 | 0.00287 | 0.00287 | 0.6666 | 0.6666 | 0.0036 | 0.0000 | ∞ | n/c |
S: Segregating sites; θ: Hetrozygosity per site; π: Average Nucleotide diversity; dN: Average number of non synonymous mutation; dS: Average number of synonymous mutation
Sequencing of the central repeat region of pfmsp2 gene was successfully analyzed in 112 (62%) samples (Table 1). The predominant alleles was 3D7/ICI (78%) with 38 allelic variant while FC27 was formed among 22% samples with only 12 allelic variants (Fig 2). The amino acid length was significantly lower in both the allelic family when compared to the reference strain (Table 1). The dN/dS ratio indicated the gene under positive selection (Table 2). The transversion for FC27 alleles was much higher as compared to the transitions. Twenty eight percent of the samples were polyclonal infection and the MOI for pfmsp2 was 1.28. The NCBI GenBank database accession number for msp2 allelic family is KY425879—KY42928.
Sequence diversity of pfglurp
One hundred fifteen samples were successfully genotyped for pfglurp RII repeat region. A total 59 genotypes (Fig 2) were detected with significantly lower average amino acid length when compared to the reference strain 3D7 (Table 1). Tajima test shows that pfglurp gene is under negative selection (Table 2). There are 4 type of amino acid repeat sequence unit was present among the all isolates in the well conserved part of RII repeat region with varies in the repeat numbers (Table 3). Apart from these 4 types, another 4 types repeats were also found with minor amino acid substitution in the limited number of isolates (Table 3). However, an extra D or E amino acid was found in between the repeats (S1 Fig). The NCBI GenBank database accession number for glurp allelic family is KY425929—KY425987.
Table 3. Type of repeats sequence with limited polymorphism of P. falciparum glurp gene in the Indian isolates.
| Type | amino acid repeat sequence | Repeat length (amino acid) |
prevalence | average frequency of the repeats ± SD (Min—Max) |
|---|---|---|---|---|
| 1 | DKSAHIQHEIVEVEEILPE | 19 | 100 | 1.01 ± 0.09 (1.00–2.00) |
| 2 | DKNEKVEHEIVEVEEILPE | 19 | 100 | 1.79 ± 0.97 (1.00–4.00) |
| 3 | DKNEKGQHEIVEVEEILPE | 19 | 100 | 4.50 ± 0.91 (2.00–7.00) |
| 4 | DKNEKVQHEIVEVEEILPE | 19 | 100 | 3.14 ± 1.02 (1.00–6.00) |
| 5 | DKNEKVNMKIVEVEEILPE | 19 | 2.61 | 1.00 ± 0.00 (1.00–1.00) |
| 6 | DKNEKGQHEIVEGEEILPE | 19 | 1.74 | 1.00 ± 0.00 (1.00–1.00) |
| 7 | DKNEKVQHEIVEVEEVLPE | 19 | 0.89 | 1.00 |
| 8 | DKSEKGQHEIVEVEEILPE | 19 | 0.89 | 1.00 |
| 9 | DENEKGQHEIVEVEEILPE | 19 | 0.89 | 1.00 |
Diversity in the central repeat region of pfcsp
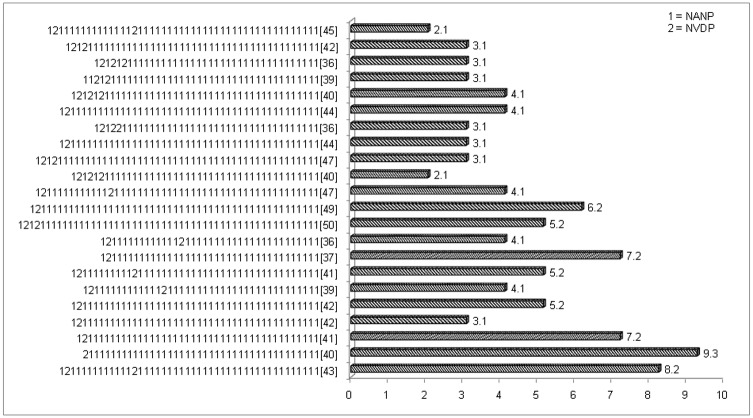
Twenty two unique haplotypes were observed among 97 samples in the central repeat region (Table 1). The number of tetra-peptide repeats that includes NANP and other minor variants, in this region varied from 36 to 50. Among these haplotypes with most common NANP repeat number was 40 (15%) with 55% samples had tetra-peptide repeats between 40 to 44 numbers. Some repeats contribute three haplotype with same number and some repeats were shared two haplotype however, they had variation in their sequence (Fig 3). GenBank accession number for these haplotypes are KY425988—KY426009
Fig 3. Prevalence of P. falciparum csp haplotype based on NANP and NVDP repeats and their arrangement.
Sequence diversity in pfs25
As expected, the pfs25 was highly conserved among the 155 successfully sequenced samples and resulted only two amino acid substitution sites (Table 1). Overall, 54% (n = 84) samples were perfectly matched with reference strain 3D7 while only 29% (n = 45) were found substitution at A131G followed by 17% (n = 26) at V143A position. GenBank accession number for these haplotypes are KY426010—KY426012.
Global analysis of Pfmsp1, Pfmsp2 and Pfcsp sequence
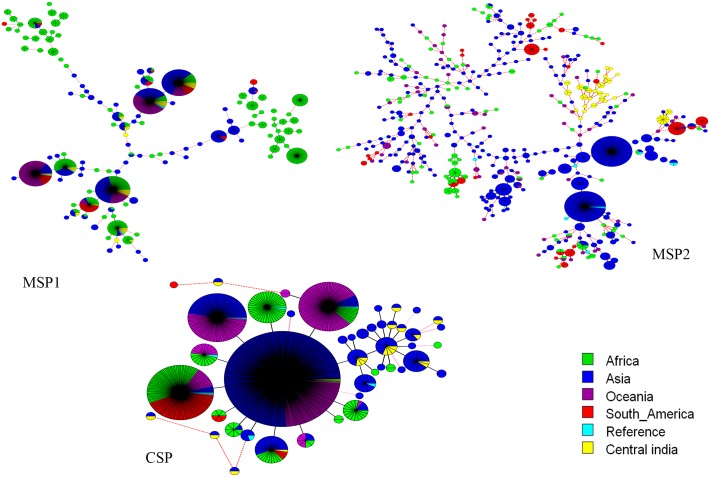
The minimal spanning tree (MST) analysis of Pfmsp1 (1149 sequence), Pfmsp2 (882 sequences) and Pfcsp (985 sequence) from global isolates (Africa, Asia, Oceania, South America and reference sequence) were included and used to construct global population structures (Fig 4, S2 Table). The genetic diversity suggested that the African populations show greater diversity as compared to the other continents. The level of genetic diversity in our study population was very similar to the diversity shown by isolates from other Asian Countries.
Fig 4. Global population structure of P. falciparum msp1, msp2 and csp gene.
A minimal spanning tree (MST) generated using Bio Numerics software version 7.6.1 showing the relationship from worldwide isolates. Each circle represents and individual haplotype and the size of the circle is proportional to the number of isolates belonging to that haplotypes. The line connecting the circle is branch length.
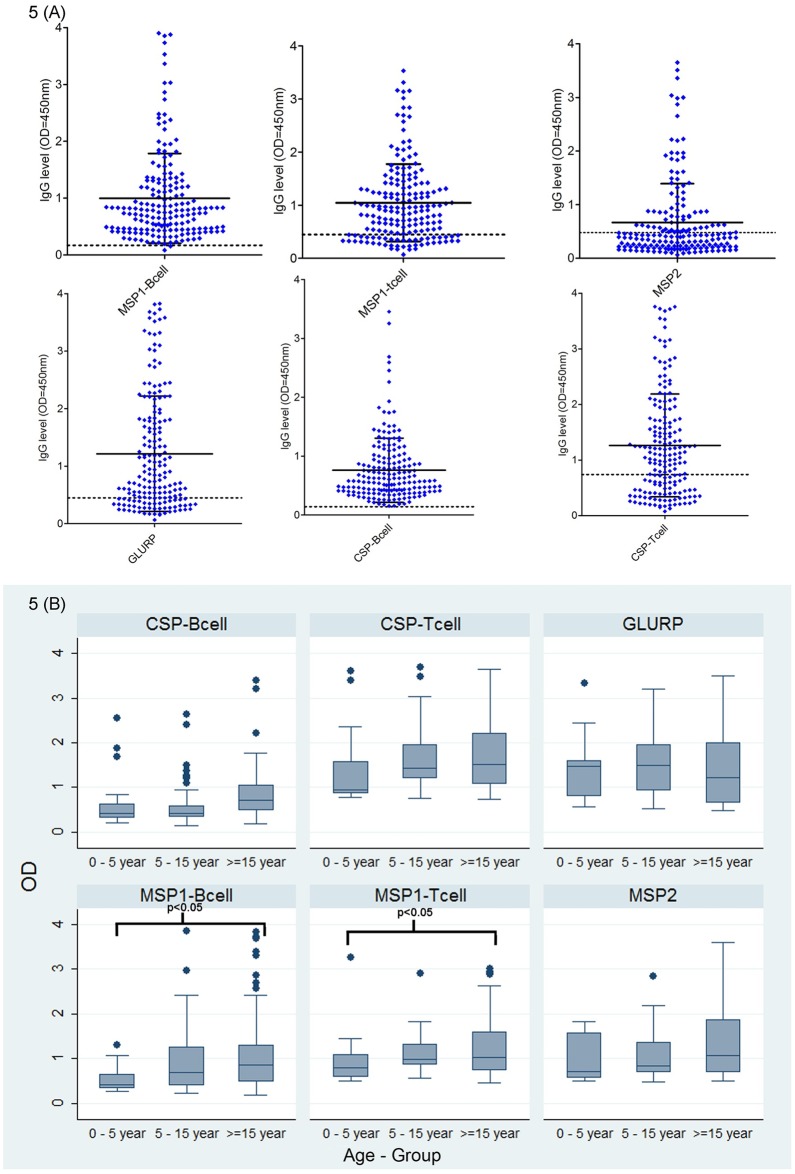
Prevalence and levels of total IgG responses against various P. falciparam antigens / epitopes
The levels of total IgG was assessed against different antigens/epitope in 180 plasma samples of P. falciparum infected patients by Enzyme linked Immunosorbent assay. The overall prevalence of total IgG antibody varied greatly between 37% to 97% to different antigens /epitopes (Fig 5A). The higher level of IgG response was found in B-cell epitopes of pfmsp1 (94%) and pfcsp (97%) antigens while significantly lower levels observed against T-cell epitopes of pfmsp1(71%) and pfcsp (62%) antigens. However, only 37% individuals were found positive for B cell epitope of pfmsp2 antigen. In addition, pfglurp antigen showed 64% positivity. The mean antibody levels presented in Fig 5B illustrate significant differences in the IgG levels between the age groups. Furthermore, when assessed the levels of IgG among different age groups, we observed significantly higher levels of antibodies against B-cell epitopes of pfcsp and pfglurp antigens in the ≥15 years age group than ≤5 year age group (Fig 5B). Further analysis revealed that the IgG antibody response was not associated with the parasite density (S4 Table). Although, samples with low parasite density (500–1000) showed higher antibody response compared to other groups but the difference was not statistically significant (p>0.05). Additionally, the correlation of B-cell epitopes of pfcsp response to NANP repeat showed significantly high positive response where the average NANP repeats was ≥42 repeats (p<0.05).
Fig 5.
(A) Total IgG antibody levels against synthetic peptide of the P. falciparum antigens/epitopes. (B) Levels of total IgG antibody responses among different age groups. (Box plots depict median values with 25th and 75th percentile values represented by the bottom and top edges boxes. Small * indicate that the antibody responses statistically significant differences (* p<0.05) when compared among different age groups.
Discussion
Almost all vaccine candidate antigens present a great challenge for successful development of malaria vaccine due to antigenic diversity in the parasite resulting immune selection in most of the parasite. In this study, we determined the antigenic diversity of sporozoite, merozoite surface proteins, sexual stage antigen and antibody response against these antigens in Indian samples. To the best of our knowledge, this is the first report to analysed genetic polymorphism and antibody responses of five potential vaccine candidates of P. falciparum (msp1, msp2, glurp, csp and pfs25) in India. Although, P. falciparum has been one of the most extensively investigated parasites but there is limited information on genetic diversity of each stage along with antibody response. Present study showed that encoding protein expressed on the surface of the merozoite and sporozoite are more polymorphic than they are expressed during the sexual stage. These results are in agreement with the observation that stage specific surface proteins exhibit high polymorphism when compared with internal antigens [30]. In present study, P. falciparum msp1, msp2 and glurp were highly diverse from central India region in respect of length as well as sequence motifs with prevalence of all the allelic families of msp1 and msp2, which corroborates with earlier reports on Indian isolates [9,10,13,14,31,32]. Block-2 region of pfmsp1 exhibit repetitive tri-nucleotides and appears to be subjected to a rapid intra-genic recombination process. Besides these variability, Cavanaugh et.al. (2004) observed that the antibody against block-2 region of pfmsp1 are also associated with protection from clinical malaria [33]. The high proportion of multiclonal isolation way is accordance with the previous report [9,10]. One investigator also reported that multiple genotypic infections are associated with malaria severity [34] but in the present study, we have enrolled only uncomplicated malaria. Mono-morphic nature of RO33 family of msp1 also show many variants in this study as initially it was considered as limited diversified family [35,36]. The sequence polymorphism in K1 and MAD20 types of block-2 looks initially quite extensive but further analysis towards tripeptide motif, it showed limited (5 to 6) subgroups, which was also reported in Tanzanian isolates [37]. The little evidence from field data is available on the selective effects in the malaria vaccine trial and despite of the extensive polymorphism of pfmsp1, it is considered as one of the good malaria vaccine candidate genes. Although, the block-2 of pfmsp1 is considered highly polymorphic but Cavanagh et al 2004 demonstrated that various construct of block 2 region of pfmsp1 showed good antibody response compared to block 1 region of pfmsp1. It was further confirmed that block 2 region showed protective potential of the immunity [33].
P. falciparum msp2 allelic family are also associated with malaria severity [10] and could be due to super infection of severe cases or strain- specific immunity [38]. It has been demonstrated that anti-msp2 antibodies contribute to the protective immunity [39]. In trial immunization with 3D7 variant of msp2 prevent re-infection with the parasite carrying alleles of the same family [40]. The asexual blood stage antigens i.e. pfmsp1, pfmsp2 and the ring –infected erythrocyte surface antigen (RESA) are combination vaccine known as combination B, which is immunogenic [41,42] and detected a significant immune response against the central variable region of the pfmsp2. In addition, both FC27 and 3D7 family were associated with the extensive polymorphism and immune response. Furthermore, Fluck C et al 2007 demonstrated that family specific dimorphic domain is more likely to account for the pfmsp2 family specific selective effect [43]. In addition, P. falciparum glurp has also been considered an important antigen that playing an important role in the induction of protective immunity [44]. Our results suggest that amino acid sequence type 1,2,3,4 (Table 3) was present more frequently in the conserved part R2 region. Although, the antibody response was found only in 64% samples against the peptide of R0 region. Therefore, this 19 mer peptide of R2 region may be considered as future vaccine candidates either in the combination of other stage-specific antigens or multi-allelic vaccine of some antigens. This finding is consistent with the previous studies [13,44]. R2 repeat region is the immunodominant region and targeted by the human IgG antibody, which is effective in antibody-dependent cellular inhibition (ADCI).
P. falciparum csp antigen is a part of RTS’S vaccine, which showed only partial protection (50%) in the phase- III trial [45], and no clear reason behind the partial efficacy was established. In this study, we observed high degree of amino acid repeats polymorphism in the central repeat region where almost 54% of the samples contained 40 to 44 tetrapeptide (NANP) repeats, which is consistent with our previous findings and other studies [12,46]. In addition to NANP repeats, other minor repeats such as NVDP, NADP and NVNP were also found in the present study, which may be influence the antibody response against pfcsp. A large number of haplotypes observed in the present study is consistent with this region of the circumsporozoite gene being under diversifying selection. The selection of non-vaccine variant of CSP following immunogenicity with RTS, S/AS01 vaccine, which is CSP based was not allele specific [47]. Therefore, this study makes an important understanding the type and distribution of naturally occurring polymorphism in RTS, S vaccine candidate antigens in a population from Chhattisgarh-Central India. Furthermore, as expected, sexual stage antigen pfs25 was found to be conserved and only two site mutated one is Alanine with Glycine and other Valine with Alanine, which is in agreement with previous study [12].
In the present study, agreement with large parasite diversity in these genes may be due to higher malaria transmission intensity as the extent of polymorphism may vary with transmission intensity by applying inter-clonal parasite mating and intragenic recombination [48]. Other factors such as frequent migration within sub-region, gametocyte reservoir and antimalarial drug resistance may have intensified the level of P. falciparum diversity. Therefore, the understanding the population structure and diversity of P. falciparum has implication for vaccine, drug and epidemiological studies including monitoring of malaria [49,50].
The blood stage (msp1, msp2 and glurp) and liver stage (csp) vaccine candidates have been reported to have B cell and T cell recognition domain. In the present investigation, we determined the IgG antibody response against synthetic peptides representing the conserved region of B cell & T cell of msp1, csp and B cell of msp2, glurp among patients infected with P. falciparum in Central India, where malaria is endemic [51]. In this study, majority of the plasma samples reacted against the B cell epitope of pfmsp1 and pfcsp despite its short amino acid sequence. In addition, the level of IgG antibody responses varies in different antigens/epitopes, which is in agreement with the previous study where T cell response was significantly lower than B-cell response, suggesting that the peptide is immunogenic [52]. However, several other studies have found difference in the immune response to these antigens may be due the different structure of the epitopes [53]. The existence of allele specific antibodies in P. falciparum infected samples has been demonstrated, however their role in protection was not established [52]. Furthermore, a negative association between antibody response to pfmsp2 and risk of malaria was observed, suggesting a possible protective role for anti-msp2 antibody in natural infection [54]. Although we found a low level of antibody response against pfmsp2. Several studies have demonstrated that antibodies against both repeat and non-repeat region of glurp can inhibit parasite growth in-vitro [55]. In the present study we also observed higher level of IgG antibody response against glurp antigen, suggest partial protective response and it should be considered as future vaccine candidate antigen. The age specific antibody levels against B-cell epitopes of pfcsp and pfglurp antigens showed a positive association with increasing age suggesting the role of intrinsic age-related factors in immune maturation. The world wide dynamics of P. falciparum populations is complex and distribution of different parasite strains differ from region to region and evolves over time possibly because of immune selection. Therefore, a study from a range of different settings are required to show how the vaccine candidates dependent to the different alleles of the vaccine candidate antigens.
The strengths of this study is the comprehensive use of sensitive and standardized assays, rendering our findings amenable to a detailed analysis of genetic polymorphisms as well as antibody response in P. falciparum vaccine candidate antigens. In the present study, we aimed for such analysis to be relevant for epidemiologic investigations, malaria transmission dynamics and vaccine design.
The limitation of present study is that we could not assess the genetic polymorphism of P. falciparum csp T-cell (Th2 and Th3) epitopes. However, the present study has robustly genotypes the P. falciparum merozoite, sporozoite surface and sexual stage protein to obtain a conclusive genetic diversity data set for this region. It has been proposed that diversity in the repeat region might have evolved as an immune evasion mechanism by the parasite in which the host mount the protective immune response against this region while the parasite escapes into the liver [56].
Conclusion
This is a first extensive study that contributes in understanding the type and distribution of naturally evolved genetic polymorphism in the P. falciparum vaccine candidate antigens from Chhattisgarh, Central India, where malaria is endemic. There is probably some association between malaria hyper endemicity and the parasite mixture as well as the number of alleles present in the area. This data would be helpful in the future malaria vaccine trial in India and to monitor changes in the parasite population. Furthermore, the characterization of the pattern of antibody response to malarial antigens/epitopes may also be useful in monitoring the effect of malaria vaccine. The simultaneous presence of antibody response, in our population, to more than one antigen is indicative of a lower frequency of malarial infection and suggests that a successful malaria vaccine should contain multiple antigens. Therefore, similar study should be carried out on a large scale to cover the entire country to map the different endemic regions. Finally, the global analysis of allelic variants reported in this study would be helpful in the identifying the predominant allele in the region and may aid in designing region specific vaccine and would pave the way for an effective malaria intervention measures.
Supporting information
(DOCX)
(XLSX)
(DOCX)
(DOCX)
(TIF)
Acknowledgments
This publication was made possible by NPRP grant [NPRP 5–098–3–021] from the Qatar National Research Fund (a member of Qatar Foundation). Disclaimer: The statements made herein are solely the responsibility of the author[s]. We thank all the study participants and their relatives for providing the informed consent.
Data Availability
The authors have reported all the findings in the manuscript and the molecular data of all four Plasmodium species (sequence data) is submitted to the National Center for Biotechnology Information (NCBI) data base which is public domain and the Gen Bank accession numbers are (KY425824 -KY426012). This accession no. is already given in the manuscript. The patient information sheet is available with the director NIRTH in institutional computer. This contains unique identification code and patient personal details such as name, age, home address etc. If anyone wants to look the data set or want to use the data they can contact to the Director of the Institute: Dr. Neeru Singh, Director, National Institute for Research in Tribal Health (NIRTH), NIRTH (ICMR) Campus, Nagpur Road, Garha, Jabalpur - 482003, Madhya Pradesh, India. Phone: (Office) +91-761-2672239. Email: neeru.singh@gmail.com. Website: www.nirth.res.in.
Funding Statement
This work was supported by Qatar National Research Fund, NPRP 5 - 098 - 3 – 021.
References
- 1.World Malaria Report 2015. World Health Organization, Geneva. 2015. http://www.who.int/malaria/publications/world-malaria-report-2015/report/en/
- 2.Malaria situation in India. NVDCP. http://www.nvbdcp.gov.in/Doc/malaria-situation-Oct16.pdf
- 3.Krishna S, Bharti PK, Chandel HS, Ahmad A, Kumar R, Singh PP, et al. Detection of Mixed Infections with Plasmodium spp. by PCR, India, 2014. Emerg Infect Dis. 2015;21:1853–1857. doi: 10.3201/eid2110.150678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma RK, Thakor HG, Saha KB, Sonal GS, Dhariwal AC, Singh N. Malaria situation in India with special reference to tribal areas. Indian J Med Res. 2015;141:537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crompton PD, Pierce SK, Miller LH. Advances and challenges in malaria vaccine development. J Clin Invest. 2010;120:4168–4178. doi: 10.1172/JCI44423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plowe CV, Alonso P, Hoffman SL. The potential role of vaccines in the elimination of falciparum malaria and the eventual eradication of malaria. J Infect Dis. 2009;200:1646–1649. doi: 10.1086/646613 [DOI] [PubMed] [Google Scholar]
- 7.Conway DJ. Natural selection on polymorphic malaria antigens and the search for a vaccine. Parasitology Today. 1997;13:26–29. [DOI] [PubMed] [Google Scholar]
- 8.Escalante AA, Lal AA, Ayala FJ. Genetic Polymorphism and Natural Selection in the Malaria Parasite Plasmodium falciparum. Genetics. 1998;149:189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bharti PK, Shukla MM, Sharma YD, Singh N. Genetic diversity in the block 2 region of the merozoite surface protein-1 of Plasmodium falciparum in central India. Malar J. 2012;11:78 doi: 10.1186/1475-2875-11-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranjit MR, Das A, Das BP, Das BN, Dash BP, Chhotray GP. Distribution of Plasmodium falciparum genotypes in clinically mild and severe malaria cases in Orissa, India. Trans R Soc Trop Med Hyg. 2005;99:389–395. doi: 10.1016/j.trstmh.2004.09.010 [DOI] [PubMed] [Google Scholar]
- 11.Mahajan RC, Farooq U, Dubey ML, Malla N. Genetic polymorphism in Plasmodium falciparum vaccine candidate antigens. Indian JPatholMicrobiol. 2005;48:429–438. [PubMed] [Google Scholar]
- 12.Escalante AA, Grebert HM, Isea R, Goldman IF, Basco L, Magris M, et al. A study of genetic diversity in the gene encoding the circumsporozoite protein (CSP) of Plasmodium falciparum from different transmission areas—XVI. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2002;125:83–90. [DOI] [PubMed] [Google Scholar]
- 13.Kumar D, Dhiman S, Rabha B, Goswami D, Deka M, Singh L, et al. Genetic polymorphism and amino acid sequence variation in Plasmodium falciparum GLURP R2 repeat region in Assam, India, at an interval of five years. Malar J. 2014;13:450 doi: 10.1186/1475-2875-13-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi H, Valecha N, Verma A, Kaul A, Mallick PK, Shalini S, et al. Genetic structure of Plasmodium falciparum field isolates in eastern and north-eastern India. Malar J. 2007;6:60 doi: 10.1186/1475-2875-6-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z, Xiao L, Branch OH, Kariuki S, Nahlen BL, Lal AA. Antibody responses to repetitive epitopes of the circumsporozoite protein, liver stage antigen-1, and merozoite surface protein-2 in infants residing in a Plasmodium falciparum-hyperendemic area of western Kenya. XIII. Asembo Bay Cohort Project. The Am J Trop Med Hyg. 2002;66:7–12. [DOI] [PubMed] [Google Scholar]
- 16.Taylor RR, Smith DB, Robinson VJ, McBride JS, Riley EM. Human antibody response to Plasmodium falciparum merozoite surface protein 2 is serogroup specific and predominantly of the immunoglobulin G3 subclass. Infect Immun. 1995;63:4382–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubois P, Druilhe P, Arriat D, Jendoubi M, Jouin H. Changes in recognition of Plasmodium falciparum antigens by human sera depending on previous malaria exposure. Ann Inst Pasteur Immunol. 1987;138:383–396. [DOI] [PubMed] [Google Scholar]
- 18.White NJ. Malaria In: Garden C, editor.Manson’s tropical diseases. London: WB Saunders Co; 1996. pp. 1087–1164. [Google Scholar]
- 19.Felger I, Tavul L, Kabintik S, Marshall V, Genton B, Alpers M, et al. Plasmodium falciparum: Extensive Polymorphism in Merozoite Surface Antigen 2 Alleles in an Area with Endemic Malaria in Papua New Guinea. Exp Parasitol. 1994;79:106–116. doi: 10.1006/expr.1994.1070 [DOI] [PubMed] [Google Scholar]
- 20.Singh J, Bhattacharji LM. Rapid Staining of Malarial Parasites by a Water Soluble Stain. Ind Med Gaz. 1944;79:102–104. [PMC free article] [PubMed] [Google Scholar]
- 21.Diagnosis and treatment of malaria.NVDCP.2013. http://www.nvbdcp.gov.in/Doc/Diagnosis-Treatment-Malaria-2013.pdf.
- 22.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nei M. Molecular Evolutionary Genetics. 2ndedNew York:Columbia University. Press;1987 [Google Scholar]
- 25.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992;9:678–687. [DOI] [PubMed] [Google Scholar]
- 26.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Bio Evol. 1986;3:418–426. [DOI] [PubMed] [Google Scholar]
- 27.Jukes TH, Cantor CR. Evolution of Protein Molecules. Munro HN, editor: Academy Press; 1969. [Google Scholar]
- 28.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 29.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCutchan TF, de la Cruz VF, Good MF, Wellems TE. Antigenic diversity in Plasmodium falciparum. ProgAllergy. 1988;41:173–192. [DOI] [PubMed] [Google Scholar]
- 31.Okoyeh JN, Pillai CR, Chitnis CE. Plasmodium falciparum Field Isolates Commonly Use Erythrocyte Invasion Pathways That Are Independent of Sialic Acid Residues of Glycophorin A. Infect Immun. 1999;67:5784–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baruah S, Lourembam SD, Sawian CE, Baruah I, Goswami D. Temporal and spatial variation in MSP1 clonal composition of Plasmodium falciparum in districts of Assam, Northeast India. Infect Genet and Evol. 2009;9:853–859. [DOI] [PubMed] [Google Scholar]
- 33.Cavanagh DR, Dodoo D, Hviid L, Kurtzhals JA, Theander TG, Akanmori BD, et al. Antibodies to the N-terminal block 2 of Plasmodium falciparum merozoite surface protein 1 are associated with protection against clinical malaria. Infect Immun. 2004;72:6492–6502 doi: 10.1128/IAI.72.11.6492-6502.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rout R, Mohapatra BN, Kar SK, Ranjit M. Genetic complexity and transmissibility of Plasmodium falciparum parasites causing severe malaria in central-east coast India. Trop Biomed. 2009;26:165–172. [PubMed] [Google Scholar]
- 35.Aubouy A, Migot-Nabias F, Deloron P. Polymorphism in two merozoite surface proteins of Plasmodium falciparum isolates from Gabon. Malar J. 2003;2:12 doi: 10.1186/1475-2875-2-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peyerl-Hoffmann G, Jelinek T, Kilian A, Kabagambe G, Metzger WG, von Sonnenburg F. Genetic diversity of Plasmodium falciparum and its relationship to parasite density in an area with different malaria endemicities in West Uganda. Trop Med Int Health. 2001;6:607–613. [DOI] [PubMed] [Google Scholar]
- 37.Jiang G, Daubenberger C, Huber W, Matile H, Tanner M, Pluschke G. Sequence diversity of the merozoite surface protein 1 of Plasmodium falciparum in clinical isolates from the Kilombero District, Tanzania. Acta tropica. 2000;74:51–61. [DOI] [PubMed] [Google Scholar]
- 38.Eisen DP, Saul A, Fryauff DJ, Reeder JC, Coppel RL. Alterations in Plasmodium falciparum genotypes during sequential infections suggest the presence of strain specific immunity. Am J Trop Med Hyg. 2002;67:8–16. [DOI] [PubMed] [Google Scholar]
- 39.Franks S, Baton L, Tetteh K, Tongren E, Dewin D, Akanmori BD, et al. Genetic Diversity and Antigenic Polymorphism in Plasmodium falciparum: Extensive Serological Cross-Reactivity between Allelic Variants of Merozoite Surface Protein 2. Infect Immun. 2003;71:3485–3495. doi: 10.1128/IAI.71.6.3485-3495.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genton B, Betuela I, Felger I, Al-Yaman F, Anders RF, Saul A, et al. A Recombinant Blood-Stage Malaria Vaccine Reduces Plasmodium falciparum Density and Exerts Selective Pressure on Parasite Populations in a Phase 1-2b Trial in Papua New Guinea. J Infect Dis. 2002;185:820–827. doi: 10.1086/339342 [DOI] [PubMed] [Google Scholar]
- 41.Genton B, Al-Yaman F, Betuela I, Anders RF, Saul A, Baea K, et al. Safety and immunogenicity of a three-component blood-stage malaria vaccine (MSP1, MSP2, RESA) against Plasmodium falciparum in Papua New Guinean children. Vaccine. 2003;22:30–41. [DOI] [PubMed] [Google Scholar]
- 42.Sallenave-Sales S, Faria CP, Zalis MG, Daniel-Ribeiro CT, Ferreira-da-Cruz Mde F. Merozoite surface protein 2 allelic variation influences the specific antibody response during acute malaria in individuals from a Brazilian endemic area. Memorias do Instituto Oswaldo Cruz. 2007;102:421–424. [DOI] [PubMed] [Google Scholar]
- 43.Fluck C, Schopflin S, Smith T, Genton B, Alpers MP, Beck HP, et al. Effect of the malaria vaccine Combination B on merozoite surface antigen 2 diversity. Infect Genet Evol: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2007;7:44–51. doi: 10.1016/j.meegid.2006.03.006 [DOI] [PubMed] [Google Scholar]
- 44.Borre MB, Dziegiel M, Hogh B, Petersen E, Rieneck K, Riley E, et al. Primary structure and localization of a conserved immunogenic Plasmodium falciparum glutamate rich protein (GLURP) expressed in both the preerythrocytic and erythrocytic stages of the vertebrate life cycle. Mol Biochem Parasitol. 1991;49:119–131. [DOI] [PubMed] [Google Scholar]
- 45.Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BG, Kabwende AL, et al. A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N Eng J Med. 2012;367:2284–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeeshan M, Alam MT, Vinayak S, Bora H, Tyagi RK, Alam MS, et al. Genetic variation in the Plasmodium falciparum circumsporozoite protein in India and its relevance to RTS,S malaria vaccine. PLoS One. 2012;7:e43430 doi: 10.1371/journal.pone.0043430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waitumbi JN, Anyona SB, Hunja CW, Kifude CM, Polhemus ME, et al. Impact of RTS,S/AS02(A) and RTS,S/AS01(B) on genotypes of P. falciparum in adults participating in a malaria vaccine clinical trial. PLoS One. 2009;4: e7849 doi: 10.1371/journal.pone.0007849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babiker HA, Abdel-Muhsin AA, Hamad A, Mackinnon MJ, Hill WG, Walliker D. Population dynamics of Plasmodium falciparum in an unstable malaria area of eastern Sudan. Parasitology. 2000;120:105–111. [DOI] [PubMed] [Google Scholar]
- 49.Chenet SM, Taylor JE, Blair S, Zuluaga L, Escalante AA. Longitudinal analysis of Plasmodium falciparum genetic variation in Turbo, Colombia: implications for malaria control and elimination. Malar J. 2015;14:363 doi: 10.1186/s12936-015-0887-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffing SM, Mixson-Hayden T, Sridaran S, Alam MT, McCollum AM, Cabezas C, et al. South American Plasmodium falciparum after the Malaria Eradication Era: Clonal Population Expansion and Survival of the Fittest Hybrids. PLoS One. 2011;6:e23486 doi: 10.1371/journal.pone.0023486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jain V, Basak S, Bhandari S, Bharti PK, Thomas T, Singh MP, et al. Burden of complicated malaria in a densely forested Bastar region of Chhattisgarh State (Central India). PLoS One. 2014;9:e115266 doi: 10.1371/journal.pone.0115266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mamillapalli A, Sunil S, Diwan SS, Sharma SK, Tyagi PK, Adak T, et al. Polymorphism and epitope sharing between the alleles of merozoite surface protein-1 of Plasmodium falciparum among Indian isolates. Malar J. 2007;6:95 doi: 10.1186/1475-2875-6-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marsh K, Hayes RH, Carson DC, Otoo L, Shenton F, Byass P, et al. Anti-sporozoite antibodies and immunity to malaria in a rural Gambian population. Trans R Soc Trop Med Hyg. 1988;82:532–537. [DOI] [PubMed] [Google Scholar]
- 54.Al-Yaman F, Genton B, Anders RF, Falk M, Triglia T, Lewis D, et al. Relationship Between Humoral Response to Plasmodium falciparum Merozoite Surface Antigen-2 and Malaria Morbidity in a Highly Endemic Area of Papua New Guinea. Am J Trop Med Hyg. 1994;51:593–602. [DOI] [PubMed] [Google Scholar]
- 55.Theisen M, Dodoo D, Toure-Balde A, Soe S, Corradin G, Koram KK, et al. Selection of Glutamate-Rich Protein Long Synthetic Peptides for Vaccine Development: Antigenicity and Relationship with Clinical Protection and Immunogenicity. Infect Immun. 2001;69:5223–5229. doi: 10.1128/IAI.69.9.5223-5229.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schofield L. The circumsporozoite protein of Plasmodium: a mechanism of immune evasion by the malaria parasite? Bull World Health Organ. 1990;68(Suppl):66–73. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLSX)
(DOCX)
(DOCX)
(TIF)
Data Availability Statement
The authors have reported all the findings in the manuscript and the molecular data of all four Plasmodium species (sequence data) is submitted to the National Center for Biotechnology Information (NCBI) data base which is public domain and the Gen Bank accession numbers are (KY425824 -KY426012). This accession no. is already given in the manuscript. The patient information sheet is available with the director NIRTH in institutional computer. This contains unique identification code and patient personal details such as name, age, home address etc. If anyone wants to look the data set or want to use the data they can contact to the Director of the Institute: Dr. Neeru Singh, Director, National Institute for Research in Tribal Health (NIRTH), NIRTH (ICMR) Campus, Nagpur Road, Garha, Jabalpur - 482003, Madhya Pradesh, India. Phone: (Office) +91-761-2672239. Email: neeru.singh@gmail.com. Website: www.nirth.res.in.