ABSTRACT
Most cells in nature are not actively dividing, yet are able to return to the cell cycle given the appropriate environmental signals. There is now ample evidence that quiescent G0 cells are not shut-down but still metabolically and transcriptionally active. Quiescent cells must maintain a basal transcriptional capacity to maintain transcripts and proteins necessary for survival. This implies a tight control over RNA polymerases: RNA pol II for mRNA transcription during G0, but especially RNA pol I and RNA pol III to maintain an appropriate level of structural RNAs, raising the possibility that specific transcriptional control mechanisms evolved in quiescent cells. In accordance with this, we recently discovered that RNA interference is necessary to control RNA polymerase I transcription during G0. While this mini-review focuses on yeast model organisms (Saccharomyces cerevisiae and Schizosaccharomyces pombe), parallels are drawn to other eukaryotes and mammalian systems, in particular stem cells.
KEYWORDS: Dicer, differentiation, dormancy, epigenetics, G0, histone, quiescence, reprogramming, RNA interference, stem cells, transcription
Introduction
Cells only divide under appropriate conditions. In unicellular organisms such as microbes, this is usually linked to the availability of appropriate nutrients in the environment. Outside of the laboratory, these are often limiting. As a result, most microorganisms are present in a non-dividing (hereafter referred to as “G0”) state in nature.1-3 Typical examples of non-dividing microbial cells include “viable but non-culturable” (VBNC) cells4-6 and endospores in bacteria, as well as asexual and sexual spores in fungi. Multi-cellular microbes can also induce large resting structures such as sclerotia and cysts. In multi-cellular organisms possessing different cell types and functions, coordination between cells is important to ensure proper development and requires a tight control on cell proliferation. In the human body, most cells are not actively dividing but either terminally differentiated, such as neurons, or quiescent, such as most stem cells and memory lymphocytes.7-12 Reactivation of quiescent stem cells is essential for tissue regeneration, for example during wound healing.13-14
It is important to note that several types of non-dividing states can exist (Fig 1A). Typically, ‘quiescence’ is defined by metabolic activity and full ability to return to the cell cycle. On the other hand, the absence of metabolic activity is a hallmark of ‘dormancy’, for example in spores. The inability to revert to the cell cycle characterizes post-mitotic cells, and when this inability is irreversibly acquired over-time constitutes a key feature of cellular senescence. For example, progressive loss of this reversibility hinders the self-renewal capability of a stem cell niche, resulting in its depletion.15-20 A consequence of this phenotypic definition is that distinct types—and depths—of quiescence can exist. The existence of different depths of cellular quiescence is illustrated for example in muscular stem cells for which cells can enter a ‘poised’ state in response to certain environmental signals, in which they are more responsive to the signal triggering quiescence-exit. A proposed name for this intermediary state has been ‘G-alert' in stem cells21 and ‘G0(A)' in T lymphocytes.22 This has lead to the proposal of a ‘quiescence cycle' alongside the cell cycle (Fig 1B).21,23 Other examples include the morphological and phenotypic differences in fission yeast between early and late G0 cells,24,25 and the transcriptome differences in human fibroblasts when G0 is induced by different signals.26
Figure 1.
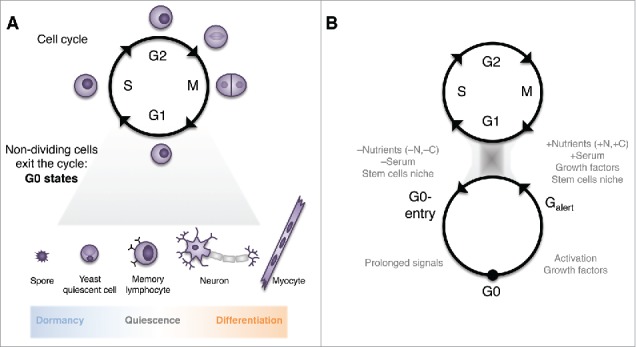
The many facets of cellular quiescence and non-dividing cells. (A) Non-dividing cells exit the cell cycle in response to specific environmental cues to enter different types of G0 states. At one end of the spectrum, microbial spores are metabolically shut-down and dormant; at the other end, cells terminally differentiate in specialized post-mitotic states (such as neurons and myocytes). There is a continuum in terms of reversibility to the cell cycle, metabolic activity and differentiation/specialization. (B) The “quiescence cycle” model proposes a progressive entry into G0, and conversely that in response to environmental signals (such as stem cell activation) G0 cells can enter a poised Galert state facilitating G0-exit.21–23
Controversies
Many efforts in the field have sought a molecular basis underlying quiescence—similarly to the molecular characterization of stem cells and of what constitutes ‘stemness’ (the concomitant presence of ‘self-renewal' and ‘differentiation potential').27,28 In part because of the different types of quiescent cells, there are still important controversies surrounding the definition of cellular quiescence. Some are semantic, such as cancer dormancy—“dormancy” qualifies the cancer and not necessarily the cancer cells underlying the “dormant” phenotype, which are thought to be either quiescent cells, and/or an immune balance between growth of a micro-tumor and the immune system.29 Second, the term “G0” has been used as a synonym of “non-dividing” cells in general, as a synonym for “quiescence,” as well as the notion that cells enter quiescence uniquely from G1. However, in certain conditions cells can enter quiescence from distinct points of the cell cycle30-33 and doing so preferentially in G1 may be a consequence of metabolic slow-down rather than necessity.32 This has led to a controversy over the term “G0” when understood as indicating an exit only from G1.32,34 Third, the choice of the model system and of the signal to induce quiescence has to be considered, as different organisms show different responses to starvation of different nutrients, such as nitrogen-starvation in budding yeast haploids vs. diploids,35 and in fission yeast.24,36 In particular, the use of stationary-phase cultures does not necessarily accurately reflect quiescent cells, as these cultures are heterogeneous and most cells not long-lived.37 Last but not least, it has proven difficult to obtain biomarkers specific for G0 cells.
Markers of cellular quiescence
The first proposed quiescence-specific marker was ‘statin’ in quiescent fibroblasts (not to be confused with the statin class of drugs: statin was later revealed to be lamin A),38-39 but was subsequently found to not be completely specific.40 Nowadays, the most commonly used marker in mammalian cells is pKi-67, which can distinguish between quiescent and proliferating cells; however, pKi-67 is still expressed at low levels in quiescence as it acts in rRNA synthesis.41,42 Other proposed markers in mammalian cells include CDK inhibitors such as p27CDKN1B/Kip1,43-46 which led to a Venus-p27K construct discriminating G0 and G1 cells,47 p21Cip1/Waf1/Sdi1,48 as well as the retinoblastoma (RB) protein (p105, p107, p130) family of cell cycle inhibitors, which regulate the E2F transcription factors,49-52 and the RB yeast analog Whi5.53,54 Quiescent cells have often been defined negatively i.e. what the cell does not do, such as absence of PCNA (i.e., absence of S-phase), and Hoechst 33342+Pyronin Y staining (DNA+RNA content) based on low RNA expression in G0.55 Transcriptomic studies in human fibroblasts,26 and in fission yeast,56,57 have identified potential core ‘quiescent program' genes. This will help identifying better markers for cellular quiescence.
Quiescence: Conserved or specialized?
Despite the importance of quiescence in all organisms, the molecular mechanisms underlying transition between growth and quiescence are not very well understood, nor are the molecular mechanisms of quiescence maintenance over long periods of time. An important question is whether molecular mechanisms underlying cellular quiescence are conserved across evolution. Because states of quiescence can be seen in prokaryotes, unicellular eukaryotes and multicellular organisms, it is likely that some aspects of quiescence are conserved.36 However, it has been proposed that because cycling cells, in any organism, use their energy predominantly to grow and divide, the most likely conserved mechanisms in G0 would be mechanisms of shutting-down growth and division, and that apart from this common aspect, quiescent cells are driven to specialization.58 Further study of quiescent cells in different organisms will help us determine to what extent this may be the case; furthermore, this dual aspect of conservation/specialization highlights the importance of in-depth study of several model organisms.
Quiescence in pathological contexts
Quiescence is also important in pathological contexts, such as cancer, degenerative diseases, and microbial infection. Two hallmarks of cancer are the inability to sustain quiescence, resulting in aberrant proliferation, and the inability to trigger apoptosis to stop these cells. It is therefore especially important to study this question not only from the viewpoint of uncontrolled growth and proliferation, but in the context of unwarranted quiescence-exit.36,58,59 Most cancer-related deaths are caused by secondary tumors after treatment of the primary cancer; one factor in cancer persistence and resurgence is thought to be the presence of quiescent cancer cells, which are less sensitive to antitumor drugs.29,60 An improved understanding of cellular quiescence would therefore permit better targeting of G0 cancerous cells, and develop more specific drugs.61-64
In the context of human pathogens, the formation of quiescent cells contributes to persistence of infection within the body, such as for the bacterial pathogen Mycobacterium tuberculosis3 and for fungal pathogens such as Cryptococcus spp., which can survive in a latent state inside host macrophages,65,66 and Candida albicans.67 Furthermore, the persistence of quiescent cells in the environment is a matter of human health, illustrated by the fact that VBNC cells of pathogens such as Vibrio vulfinicus maintain their infectious capability.3,68–70
Fission yeast is an ideal model for cellular quiescence
Several model organisms are commonly used for the study of cellular quiescence; the most notable are the budding yeast Saccharomyces cerevisiae,71,72 the fission yeast Schizosaccharomyces pombe,36,73 in vitro mammalian cell culture,46,74 the wing cells of Drosophilia melanogaster,75,76 and stem cells.9,77 In fission yeast, quiescence is induced by nitrogen deprivation of a prototrophic heterothallic strain. This results in 2 divisions without growth, resulting in haploid cells with 1c DNA.24 An advantage in using this particular model and signal is that G0-induction is synchroneous,78 all cells retain full viability for extended periods of time,24 and G0-exit is synchroneous,56 more so than when quiescence is brought about by glucose- or phosphate-starvation.36 This improved homogeneity of G0 cells is a big advantage over stationary-phase cultures or in vitro cell cultures, which are a heterogeneous population consisting mostly of cells that are not long-lived.23,37 Furthermore, this model is distinct to that of dormant spores, as nitrogen-deprived fission yeast quiescent cells still require a carbon source for viability.24 Genetic requirements common to several model systems are more likely to be conserved in higher eukaryotes.36
Cellular quiescence is transcriptionally and metabolically active
While the distinction between cellular quiescence and dormancy is not always trivial to assay, these 2 states are conceptually different: the maintenance of metabolic and transcriptional activity in quiescent cells implies the maintenance of an operational transcriptional machinery. In fission yeast, quiescent cells have distinctive transcriptomic signatures.56,57 mRNA and rRNA levels are greatly reduced (to ∼30% and ∼20% respectively) yet display a high diversity of transcripts and of proteins.57 Regulation of RNA pol I and RNA pol III is tightly correlated with growth in all organisms79 and accordingly, we have found that the binding of RNA polymerase I to rDNA is decreased over time in G0, reaching 10% that of G0-entering cells after 8 d ( = 1% of cycling cells in rich medium).80 Conversely, the proportion of H3K9-methylated silent rDNA repeats is increased.80
Quiescent fission yeast cells display an extensive metabolomic change,81,82 reflecting their shift in metabolism and specialization in recycling of nutrients. In this model, the maintenance of quiescence requires glucose24 and accordingly, several of the most abundant transcripts in G0 code for proteins involved in glycolysis,56 reflecting increased oxidative metabolism.57 One of the immediate changes caused by nitrogen-starvation is a reduction in the free aminoacid pool.82 Autophagy is required for quiescence maintenance for recycling of aminoacids83-87 and nucleotides via degradation of RNAs.88,89 Increased catabolism is also reflected by the vast size increase of the vacuole in quiescent cells,24 and many vacuolar genes are essential for quiescence establishment.73 Furthermore, new organelles are formed in quiescent fission yeast and budding yeast cells, such as actin bodies,90 proteasome storage granules91 rapidly re-imported into the nucleus upon quiescence-exit,92 and a quiescent microtubule bundle.93,94
Another indication of the active state of quiescent S. pombe cells is that they are still able to repair DNA damage.95,96 Importantly, transcription itself is a source of damage.97 Transcription of genes exposes single-stranded DNA, which is more prone to several types of damage including cytidine deamination.98 Quiescent cells also retain the ability to repair double-strand DNA breaks and do so preferentially by non-homologous end-joining.95 However, certain DNA lesions may persist in G0 as an increase of DNA repair is seen in several types of G0-exiting cells, such as haematopoietic stem cells99 and fission yeast cells (where repair is detected as Rad52 foci).80,100 In wild type, the proportion of Rad52 foci forming during the first S-phase when cells are exiting quiescence and during the following S-phases are similar, indicating that the level of unrepaired lesions after 48 hours of quiescence and between 2 replications are similar. The number of Rad52 foci dramatically increases in several DNA repair mutants, indicating that spontaneous DNA lesions are efficiently repaired during quiescence.96
Several large-scale screens have been conducted in fission yeast to identify genes important for maintaining viability during cellular quiescence.73,86 Essentiality in dividing cells does not correlate with essentiality in quiescence, and it has been estimated that ∼25% of essential genes are necessary for both (‘super-housekeeping' genes).73 An especially interesting set of genes identified is involved in transcription, such as specific alleles of the RNA pol II C-terminal domain phosphatase Fcp1 and of the common RNA pol I/III subunit AC40,73 and the Krüppel-like family transcription factor Klf1.25
As the characterization of the transcriptome in early G0 cells has allowed identification of several genes essential for quiescence,56 an important future work would be to characterize the transcriptome and proteome after longer times spent in quiescence, as these might help identify factors involved for long-term maintenance rather than early adaptations to the metabolic stress caused by quiescence induction.
Epigenetic mechanisms of quiescence maintenance?
We propose that epigenetic mechanisms are key to maintaining cellular quiescence. Cells in the cell cycle or in quiescence possess the same genotype, yet exhibit distinct phenotypes. It is therefore possible to envision cellular quiescence as a kind of cellular differentation.58 Accordingly, early cloning experiments were greatly enhanced by the induction of fibroblasts into quiescence in advance of nuclear transplantation, leading to the realization of its role as a state of major epigenetic reprogramming.101-103 Accordingly, evidence for essential epigenetic processes in quiescent cells is increasing.
RNA interference controls RNA polymerases
In S. pombe, the key proteins Dicer, Argonaute and RNA-dependent RNA polymerase (respectively Dcr1, Ago1 and Rdp1) are involved in co-transcriptional silencing of pericentromeric repeat transcripts, processing these transcripts into small interfering RNAs (siRNAs) and recruiting the silencing CLRC/Rik1 complex, harboring the H3K9 methyltransferase Clr4SUV39H1. This results in the formation of H3K9me heterochromatin.104-105 Pericentromeric heterochromatin allows proper chromosome segregation in mitosis,104,106 in meiosis107 and in G0-entry mitoses.80 More recently, RNAi has been proposed to also act in post-transcriptional silencing (PTGS) pathways in fission yeast,108 and in control of stress-related genes.109
Transcriptional silencing is a result of the inhibition of RNA polymerase II. In fission yeast heterochromatin, H3K9 is methylated, and binds HP1-like proteins (Swi6 and Chp2). In turn, Swi6HP1 recruits the SHREC complex, containing histone deacetylases,110,111 resulting in hypoacetylation of H3 and H4 tails, limiting RNA pol II recruitment. In S-phase, Swi6HP1 is displaced by H3S10 phosphorylation by Aurora kinase,112 alleviating this silencing; this results in transient transcription of peri-centromeric repeats that will trigger RNAi and heterochromatin formation on both daughter strands. The activity of DNA polymerases and RNA polymerase II on the same template during S-phase requires both processes to be tightly co-regulated. Therefore, we hypothesized that RNAi may act more directly on polymerases, and discovered that RNAi releases RNA pol II from pericentromeric heterochromatin,113 as well as at several highly-transcribed euchromatic loci, tDNAs, and rDNA.114 This sheds new light on why certain specific RNA pol II mutants lose silencing in S. pombe, such as rpb2-m203(N44Y)115 and rpb7-G150D.116
Finding an important role for RNAi at tDNAs and rDNA loci was one of the reasons that prompted us to investigate the response of RNAi mutants in quiescence. Indeed, we found that RNAi mutants have G0-entry defects and lose viability during quiescence maintenance. We found that RNAi releases RNA pol I from rDNA specifically in G0 (Fig 2ABC), and that the G0 defects of RNAi deletion mutants are suppressed by reducing RNA pol I binding (such as in a specific TBP mutant, tbp1-D156Y) or by destabilizing RNA pol I by deleting its non-essential subunit A1280 (Fig 2D). Interestingly, mutants in equivalent subunits in RNA pol I (A12) and in RNA pol II (Rpb9 + TFIIS) specifically suppress, respectively, RNAi quiescence defects and RNAi silencing defects.80,117 This strong parallel buttresses the proposition that RNAi proteins are indeed closely associated to RNA polymerase holocomplexes. Furthermore, the parallel role of A12 and TFIIS in polymerase back-tracking118 may indicate that in the absence of Dicer, this activity becomes detrimental. Pausing of RNA pol II during transcription termination results in polymerase back-tracking, and the 3′ end of the RNA is targeted by the RNA exosome.119 Absence of TFIIS reduces the level of read-through, suggesting that TFIIS and the RNA exosome compete for the RNA 3′ end119 and that in the absence of Dicer, the activity of the RNA exosome becomes essential. The fact that, in the case of RNA pol I, polymerase accumulation in Dicer mutants is not only seen at the 3′ETS region, but over the whole locus (rDNA promoter, 5′ETS and multiple sites in 18S, 5.8S and 28S),80 suggests that Dicer may play a role during elongation in G0 rather than only during termination.
Figure 2.
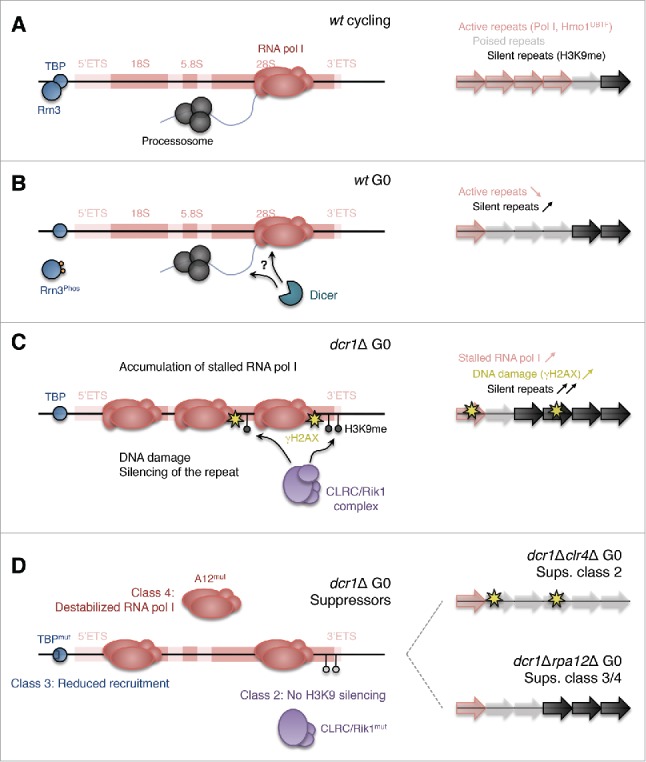
Model for the novel essential role for Dicer in RNA polymerase I release in G0. (A) In wild-type cycling cells, RNA pol I transcribes the rDNA repeats. (B) Wild-type G0 cells lower the recruitment of RNA pol I to rDNA, in part via phosphorylation of the Rrn3 initiation factor, shifting the ratio of active vs. silent rDNA repeats. Dicer contributes to RNA pol I release, although it is still unknown whether this occurs directly at the level of rRNA or via RNA pol I itself. (C) Dicer mutants in G0 are defective in RNA pol I release, resulting in accumulation of stalled RNA pol I, DNA damage, and the recruitment of the silencing CLRC/Rik1 complex at the repeat, causing a hyper-silencing of rDNA repeats via H3K9 methylation. (D) The Dicer defect is suppressed by mutants in the H3K9 methylation pathway (class 2: such as dcr1Δclr4Δ), by reducing RNA pol I transcription initiation (class 3: such as dcr1Δtbp1-D156Y), or by destabilizing RNA pol I itself (class 4: such as dcr1Δrpa12Δ).80 (Note: class 1 suppressors are not represented, and concern chromosomal segregation during G0-entry).
One difference is that Dicer's requirement in RNA pol II release from rDNA is independent of its catalytic activity and of Argonaute,114 while all RNAi proteins are required for RNA pol I release in G0, including Dicer's catalytic activity.80 Whether this reflects a difference in different RNA polymerase compositions or cell cycle stage would be an interesting aspect to investigate. Presumably, Dicer acts on a RNA template; and the genetic requirement for catalytic activity and RNAi proteins in this novel mechanism also points to specific small RNAs (sRNAs) being involved. However, we have not found Dicer-dependent novel sRNAs in G0.80 What is the molecular target of Dicer in G0 cells? One possibility is that the rRNA itself is targeted by Dicer, potentially in a torpedo-like mechanism as used by the RNAse III Rnt1.120 In Candida albicans, the Dicer ortholog (although closer to the RNT1 family121) cleaves the 28S rRNA at the 3′ETS and the 3′ tail of U4 snRNA,122 but we did not detect signatures of this activity in our S. pombe sRNA-seq80 (and unpublished observations). Another possibility is that Dicer may act indirectly on the rRNA, potentially through single-cleavage of a specific non-coding RNA (ncRNA) in G0, in a manner reminiscent of pRNA-mediated123 and long ncRNA-mediated124,125 rDNA silencing in mammals. Interestingly, the few genomic loci that are upregulated in Dicer mutants in G0 include several long ncRNAs of unknown function (unpublished observations); testing whether any of these ncRNAs plays any function in rDNA silencing is an important next step. Finally, similarly to its function controlling RNA pol I and RNA pol II, we hypothesize that RNAi may release RNA pol III through its C11 subunit at specific genomic loci.
Histone marks in cellular quiescence
Dicer mutants cause over-activation of the CLRC/Rik1 silencing complex at the rDNA resulting in unchecked accumulation of H3K9 methylation (Fig 2C).80 H3K9me causes cell death, as H3K9me mutants rescue RNAi mutants specifically in G0. In fact, our suppressor screen of G0 viability loss of dcr1Δ uncovered a large number of mutants involved in heterochromatin formation, and may potentially be used to uncover novel silencing factors. In addition to CLRC/Rik1 complex mutants (rik1-V449G, rik1-K812*, rik1-A875P, rik1-T942K, clr4-R126*, clr4-C317F, clr4-Y451*, raf2-G37V), we obtained Swi6HP1 mutants (swi6-T278K, swi6-W293*).80 In S. pombe, the CLRC/Rik1 complex associates with DNA polymerase ε (Cdc20) through its replication targeting subunit Raf2, and several cdc20 mutants lose silencing126,127; further, we uncovered a novel cdc20–1 mutant required for Dicer mutant cells to enter G0 (unpublished observation). Proper regulation of H3K9me in G0 is important: wild-type cells show a physiologic increase of H3K9me at the rDNA to adjust the active/silent rDNA repeat ratio. Complete loss of H3K9me results in a mild defect in quiescence maintenance (although much milder than defects resulting from H3K9me over-accumulation).80
Several other histone marks are thought to play an important role in quiescence maintenance, belonging to several classes: (i) marks to silence proliferation-genes, such as lack of histone H3 and H4 acetylation; (ii) bivalent marks at ‘poised’ genes, presumably for efficient G0-exit given the appropriate signal. Silencing marks cover a large part of the genome, and are thought to be an important factor in chromatin compaction, a hallmark of G0 cells in most organisms.24,128,129 In budding yeast, condensin,129 the H1-like Hho1 protein and H4K16 deacetylation130-133 are thought to participate in this process; several quiescence-specific transcription factors (Xbp1 and Stb3) have been shown to recruit the class I histone deacetylase (HDAC) Rpd3 to half of gene promoters,134,135 and to rDNA.136 However, Schizosaccharomyces spp. do not have Rpd3 orthologs but only the Clr6 and Hos2 lineages of class I HDACs,137 and their genomes do not encode H1 orthologs. The contribution of condensin to G0 chromatin compaction has not yet been investigated in S. pombe. Quiescent naive T cells also display chromatin compaction by condensin.138 Another mark of interest is H4K20 methylation, which is found at facultative heterochromatin in quiescent muscle stem cells,139 in primary fibroblasts,140 in serum-deprived NIH 3T3 cells,125 and accumulates in quiescent brine shrimp encysted embryos.141 H4K20 methylation is present in S. pombe, associates with histone turn-over rate at certain genes142 and DNA repair via Crb253BP1 recruitment,143,144 but has not yet been investigated in cellular quiescence. In S. pombe, it has been proposed that H3K9 methylation contributes to general silencing in G0.145 The second class of marks believed to be pivotal for cellular quiescence is thought to signal a ‘bivalent’ state. Indeed quiescent ES cells display bivalent marks (H3K4 methylation and H3K27 methylation) at the promoters of many genes important for lineage differentiation.146,147 S. cerevisiae quiescent cells retain high levels of active marks such as H3K4me3, H3K36me3 and H3K79me3,148 and H3K36 methylation by ASH1/Trithorax is important for quiescence and self-renewal of haematopoietic stem cells.18 The contribution of these histone marks to quiescence maintenance has not yet been investigated in fission yeast.
An important question is whether specific novel histone marks are present during cellular quiescence. To our knowledge, this has not yet been investigated; while mass-spectrometry of histone post-translational modifications have allowed the identification of many novel marks of unknown function,149-153 including in primarily non-dividing tissues such as the mouse brain,154 such studies are usually conducted, in yeast, on growing cells.150,155,156 An alternative, complementary approach would be to generate a library of histone mutants157,158 to assay the effect of each aminoacid on viability specifically during cellular quiescence. Toward this goal, we have designed a system for mutating H3 in S. pombe,80 more physiological than previous systems159 and therefore amenable to G0 study as well as making partial loss-of-function mutants. This approach has the potential to uncover novel histone marks of biological interest.
Conclusion
The importance of quiescence makes it likely not only that many molecular mechanisms will be discovered for its establishment and regulation, but also that many of these basic mechanisms may be evolutionarily conserved.36 Importantly, the essentiality of genes is usually determined using laboratory growth conditions, yet in nature, the prevalence of non-dividing states stresses the importance to identify genes essential specifically in quiescence (and genes essential for both growth and quiescence, often termed “super-housekeeping” genes73). Because of the reversible nature of quiescence and because it can be considered a kind of cellular differentiation, we think that epigenetic mechanisms may be key factors controlling maintenance of quiescence and rewiring its transcriptional program. Preliminary data suggests that indeed, other key chromatin genes are specifically essential in G0 in addition to key RNAi proteins80 (and unpublished observations). In fact, it is important to test familiar mechanisms in quiescence as it appears—as is the case with RNAi—that quiescence can uncover novel mechanisms and functions that are important or essential. This also applies for numerous genes that are apparently devoid of function. Indeed mutants affected in growth are not necessarily affected in quiescence and vice versa.73,86 Because of its advantages in studying cellular quiescence as well as in studying epigenetic pathways, we think that the fission yeast S. pombe is poised to continue to be a model system of choice. Accordingly, a lot of resources for fission yeast G0 have been developed recently, such as the G0 transcriptome56,57 proteome,57 metabolome.81 Gene replacement in yeast also allows the use of molecular tools such as G0 over-expressing promoters,80,160 and G0-shutoff systems.
Advances in fundamental research in cellular quiescence will have consequences in several fields relevant to human health, such as stem cell biology and cancer biology. For example, a strategy that has been successful in developing novel anti-cancer drugs is to identify negative epistatis (synthetic lethality) networks.161-163 Large-scale negative epistasis screens can be conducted on yeast before being tested on human cells, allowing more interactions to be tested164; importantly, there is good conservation of negative epistatic interactions. Using this type of approach to screen for synthetic lethality specific to quiescence would therefore be a promising approach to target quiescent cancer cells, an essential step toward effective treatment and avoiding relapse. We conclude that the investigation of cellular quiescence therefore opens not only a deeper understanding of fundamental biology, but also new avenues in medicine.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the S. pombe community for discussion. We thank the S. pombe database Pombase, as its resources are an important contribution to work in our laboratories. We apologize to authors whose work was not cited due to space constraints.
References
- 1.Lewis DL, Gattie DK. The ecology of quiescent microbes. ASM News 1991; 57:27-32 [Google Scholar]
- 2.Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol 2006; 4:113-20; PMID:16415927; https://dx.doi.org/ 10.1038/nrmicro1340 [DOI] [PubMed] [Google Scholar]
- 3.Rittershaus ES, Baek SH, Sassetti CM. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe 2013; 13:643-51; PMID:23768489; https://dx.doi.org/ 10.1016/j.chom.2013.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol 1982; 8:313-23; PMID:24226049; https://dx.doi.org/ 10.1007/BF02010671 [DOI] [PubMed] [Google Scholar]
- 5.Roszak DB, Colwell RR. Survival strategies of bacteria in the natural environment. Microbiol Rev 1987; 51:365-79; PMID:3312987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 2010; 34:415-25; PMID:20059548; https://dx.doi.org/ 10.1111/j.1574-6976.2009.00200.x [DOI] [PubMed] [Google Scholar]
- 7.Suda T, Arai F, Hirao A. Hematopoietic stem cells and their niche. Trends Immunol 2005; 26:426-33; PMID:15979407; https://dx.doi.org/ 10.1016/j.it.2005.06.006 [DOI] [PubMed] [Google Scholar]
- 8.Glauche I, Moore K, Thielecke L, Horn K, Loeffler M, Roeder I. Stem cell proliferation and quiescence—two side of the same coin. PLoS Comput Biol 2009; 5:e1000447; PMID:19629161; https://dx.doi.org/ 10.1371/journal.pcbi.1000447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 2013; 14:329-40; PMID:23698583; https://dx.doi.org/ 10.1038/nrm3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 2014; 82:545-59; PMID:24811379; https://dx.doi.org/ 10.1016/j.neuron.2014.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hija A, Salpeter S, Klochendler A, Grimsby J, Brandeis M, Glaser B, Dor Y. G0-G1 transition and the restriction point in pancreatic β-cells in vivo. Diabetes 2014; 63:578-84; PMID:24130333; https://dx.doi.org/ 10.2337/db12-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okhrimenko A, Grün JR, Westendorf K, Fang Z, Reinke S, von Roth P, Wassilew G, Kühl AA, Kudernatsch R, Demski S, et al.. Human memory T cells from the bone marrow are resting and maintain long-lasting systemic memory. Proc Natl Acad Sci U S A 2014; 111(25):9229-34; PMID:24927527; https://dx.doi.org/ 10.1073/pnas.1318731111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng T, Frank DB, Kadzik RS, Morley MP, Rathi KS, Wang T, Zhou S, Cheng L, Lu MM, Morrisey EE. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 2015; 526:578-82; PMID:26436454; https://dx.doi.org/ 10.1038/nature14984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rumman M, Dhawan J, Kassem M. Quiescence in adult stem cells: biological significance and relevance to tissue regeneration. Stem Cells 2015; 33:2903-12; PMID:26075660; https://dx.doi.org/ 10.1002/stem.2056 [DOI] [PubMed] [Google Scholar]
- 15.Thorén LA, Liuba K, Bryder D, Nygren JM, Jensen CT, Qian H, Antonchuk J, Jacobsen SE. Kit regulates maintenance of quiescent hematopoietic stem cells. J Immunol 2008; 180:2045-53; PMID:18250409; https://doi.org/ 10.4049/jimmunol.180.4.2045 [DOI] [PubMed] [Google Scholar]
- 16.Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature 2012; 490:355-60; PMID:23023126; https://dx.doi.org/ 10.1038/nature11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodgers JT, Rando TA. Sprouting a new take on stem cell aging. EMBO J 2012; 31:4103-5; PMID:23042560; https://dx.doi.org/ 10.1038/emboj.2012.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones M, Chase J, Brinkmeier M, Xu J, Weinberg DN, Schira J, Friedman A, Malek S, Grembecka J, Cierpicki T, et al.. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J Clin Invest 2015; 125:2007-20; PMID:25866973; https://dx.doi.org/ 10.1172/JCI78124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavallucci V, Fidaleo M, Pani G. Neural stem cells and nutrients: poised between quiescence and exhaustion. Trends Endocrinol Metab 2016; 27:756-69; PMID:27387597; https://dx.doi.org/ 10.1016/j.tem.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 20.Yue F, Bi P, Wang C, Shan T, Nie Y, Ratliff TL, Gavin TP, Kuang S. Pten is necessary for the quiescence and maintenance of adult muscle stem cells. Nat Commun 2017; 8:14328; PMID:28094257; https://dx.doi.org/ 10.1038/ncomms14328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR, et al.. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 2014; 510:393-6; PMID:24870234; https://dx.doi.org/ 10.1038/nature13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lea NC, Orr SJ, Stoeber K, Williams GH, Lam EW, Ibrahim MA, Mufti GJ, Thomas NS. Commitment point during G0–>G1 that controls entry into the cell cycle. Mol Cell Biol 2013; 23:2351-61; PMID:12640120; https://doi.org/ 10.1128/MCB.23.7.2351-2361.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhawan J, Laxman S. Decoding the stem cell quiescence cycle—lessons from yeast for regenerative biology. J Cell Sci 2015; 128:4467-74; PMID:26672015; https://dx.doi.org/ 10.1242/jcs.177758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su SS, Tanaka Y, Samejima I, Tanaka K, Yanagida M. A nitrogen starvation-induced dormant G0 state in fission yeast: the establishment from uncommitted G1 state and its delay for return to proliferation. J Cell Sci 1996; 109:1347-57; PMID:8799823 [DOI] [PubMed] [Google Scholar]
- 25.Shimanuki M, Uehara L, Pluskal T, Yoshida T, Kokubu A, Kawasaki Y, Yanagida M. Klf1, a C2H2 zinc finger-transcription factor, is required for cell wall maintenance during long-term quiescence in differentiated G0 phase. PLoS One 2013; 8:e78545; PMID:24167631; https://dx.doi.org/ 10.1371/journal.pone.0078545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol 2006; 4:e83; PMID:16509772; https://dx.doi.org/ 10.1371/journal.pbio.0040083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikkers H, Frisén J. Deconstructing stemness. EMBO J 2015; 24:2715-9; PMID:16037819; https://dx.doi.org/ 10.1038/sj.emboj.7600749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanza R, Atala A, Eds. Essentials of stem cell biology. 3rd ed. Academic Press, San Diego: 2014; https://doi.org/ 10.1016/c2012-0-06957-8 [DOI] [Google Scholar]
- 29.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007; 7:834-46; PMID:17957189; https://dx.doi.org/ 10.1038/nrc2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei W, Nurse P, Broek D. Yeast cells can enter a quiescent state through G1, S, G2 or M phase of the cell cycle. Cancer Res 1993; 53:1867-70; PMID:8467507 [PubMed] [Google Scholar]
- 31.Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J Cell Biol 2011; 192:949-57; PMID:21402786; https://dx.doi.org/ 10.1083/jcb.201009028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daignan-Fornier B, Sagot I. Proliferation/quiescence: the controversial “aller-retour.” Cell Div 2011; 6:10; PMID:21554667; https://dx.doi.org/ 10.1186/1747-1028-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR. Yeast cells can access distinct quiescent states. Genes Dev 2011; 25:336-49; PMID:21289062; https://dx.doi.org/ 10.1101/gad.2011311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper S. Reappraisal of serum starvation, the restriction point, G0, and G1 phase arrest points. FASEB J 2003; 17:333-40; PMID:12631573; https://dx.doi.org/ 10.1096/fj.02-0352rev [DOI] [PubMed] [Google Scholar]
- 35.Madhani H. From a to α: yeast as a model for cellular differentiation. Cold Spring Harbor Press, Cold Spring Harbor. 2007. Print [Google Scholar]
- 36.Yanagida M. Cellular quiescence: are controlling genes conserved? Trends Cell Biol 2009; 19:705-15; PMID:19833516; https://dx.doi.org/ 10.1016/j.tcb.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 37.Allen C, Büttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo, et al.. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol 2006; 174:89-100; PMID:16818721; https://dx.doi.org/ 10.1083/jcb.200604072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang E. Rapid disappearance of statin, a nonproliferating and senescent cell-specific protein, upon reentering the process of cell cycling. J Cell Biol 1985; 101:1695-701; PMID:3902853; https://doi.org/ 10.1083/jcb.101.5.1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coates PJ, Hobbs RC, Crocker J, Rowlands DC, Murray P, Quinlan R, Hall PA. Identification of the antigen recognized by the monoclonal antibody BU31 as lamins A and C. J Pathol 1996; 178:21-9; PMID:8778310; https://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 40.Ansari B, Dover R, Gillmore CP, Hall PA. Expression of the nuclear membrane protein statin in cycling cells. J Pathol 1993; 169:391-6; PMID:8501536; https://dx.doi.org/ 10.1002/path.1711690402 [DOI] [PubMed] [Google Scholar]
- 41.Bullwinkel J, Baron-Lühr B, Lüdemann A, Wohlenberg C, Gerdes J, Scholzen T. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J Cell Physiol 2006; 206:624-35; PMID:16206250; https://dx.doi.org/ 10.1002/jcp.20494 [DOI] [PubMed] [Google Scholar]
- 42.Rahmanzadeh R, Hüttmann G, Gerdes J, Scholzen T. Chromophore-assisted light inactivation of pKi-67 leads to inhibition of ribosomal RNA synthesis. Cell Prolif 2007; 40:422-30; PMID:17531085; https://dx.doi.org/ 10.1111/j.1365-2184.2007.00433.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivard N, L'Allemain G, Bartek J, Pouysségur J. Abrogation of p27Kip1 by cDNA antisense suppresses quiescence (G0 state) in fibroblasts. J Biol Chem 1996; 271:18337-41; PMID:8702474; https://doi.org/ 10.1074/jbc.271.31.18337 [DOI] [PubMed] [Google Scholar]
- 44.Bresnahan WA, Boldogh I, Ma T, Albrecht T, Thompson EA. Cyclin E/Cdk2 activity is controlled by different mechanisms in the G0 and G1 phases of the cell cycle. Cell Growth Differ 1996; 7:1283-90; PMID:8891332 [PubMed] [Google Scholar]
- 45.Mellor HR, Ferguson DJ, Callaghan R. A model of quiescent tumour microregions for evaluating multicellular resistance to chemotherapeutic drugs. Br J Cancer 2005; 93:302-9; PMID:16052217; https://dx.doi.org/ 10.1038/sj.bjc.6602710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salmenperä P, Karhemo PR, Räsänen K, Laakkonen P, Vaheri A. Fibroblast spheroids as a model to study sustained fibroblast quiescence and their crosstalk with tumor cells. Exp Cell Res 2016; 345:17-24; PMID:27177832; https://dx.doi.org/ 10.1016/j.yexcr.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 47.Oki T, Nishimura K, Kitaura J, Togami K, Maehara A, Izawa K, Sakaue-Sawano A, Niida A, Miyano S, Aburatani H, et al.. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci Rep 2014; 4:4012; PMID:24500246; https://dx.doi.org/ 10.1038/srep04012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakanishi M, Adami GR, Robetorye RS, Noda A, Venable SF, Dimitrov D, Pereira-Smith OM, Smith JR. Exit from G0 and entry into the cell cycle of cells expressing p21Sdi1 antisense RNA. Proc Natl Acad Sci U S A 1995; 92:4352-6; PMID:7753810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas NS, Burke LC, Bybee A, Linch DC. The phosphorylation state of the retinoblastoma (RB) protein in G0/G1 is dependent on growth status. Oncogene 1991; 6:317-22; PMID:2000225 [PubMed] [Google Scholar]
- 50.Sun A, Bagella L, Tutton S, Romano G, Giordano A. From G0 to S phase: a view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J Cell Biochem 2007; 102:1400-4; PMID:17979151; https://dx.doi.org/ 10.1002/jcb.21609 [DOI] [PubMed] [Google Scholar]
- 51.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol 2008; 10:476-82; PMID:18364697; https://dx.doi.org/ 10.1038/ncb1711 [DOI] [PubMed] [Google Scholar]
- 52.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A 2012; 81:552-66; PMID:22693166; https://dx.doi.org/ 10.1002/cyto.a.22075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charvin G, Oikonomou C, Siggia ED, Cross FR. Origin of irreversibility of cell cycle start in budding yeast. PLoS Biol 2010; 8:e1000284; PMID:20087409; https://dx.doi.org/ 10.1371/journal.pbio.1000284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Travesa A, Kalashnikova TI, de Bruin RA, Cass SR, Chahwan C, Lee DE, Lowndes NF, Wittenberg C. Repression of G1/S transcription is mediated via interaction of the GTB motifs of Nrm1 and Whi5 with Swi6. Mol Cell Biol 2013; 33:1476-86; PMID:23382076; https://dx.doi.org/ 10.1128/MCB.01333-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim KH, Sederstrom JM. Assaying cell cycle status using flow cytometry. Curr Protoc Mol Biol 2015; 111:28.6.1-11; PMID:26131851; https://dx.doi.org/ 10.1002/0471142727.mb2806s111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimanuki M, Chung SY, Chikashige Y, Kawasaki Y, Uehara L, Tsutsumi C, Hatanaka M, Hiraoka Y, Nagao K, Yanagida M. Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells 2007; 12:677-92; PMID:17535257; https://dx.doi.org/ 10.1111/j.1365-2443.2007.01079.x [DOI] [PubMed] [Google Scholar]
- 57.Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bähler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 2012; 151:671-83; PMID:23101633; https://dx.doi.org/ 10.1016/j.cell.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Farrell PH. Quiescence: early evolutionary origins and universality do not imply uniformity. Philos Trans R Soc Lond B Sci 2011; 366:3498-507; PMID:22084377; https://dx.doi.org/ 10.1098/rstb.2011.0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer 2001; 1:222-31; PMID:11902577; https://dx.doi.org/ 10.1038/35106065 [DOI] [PubMed] [Google Scholar]
- 60.Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol 2011; pii:396076:1-11; PMID:20936110; https://dx.doi.org/ 10.1155/2011/396076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cavnar SP, Rickelmann AD, Meguiar KF, Xiao A, Dosch J, Leung BM, Cai Lesher-Perez S, Chitta S, Luker KE, Takayama S, et al.. Modeling selective elimination of quiescent cancer cells from bone marrow. Neoplasia 2015; 17:625-33; PMID:26408255; https://dx.doi.org/ 10.1016/j.neo.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X, de Milito A, Olofsson MH, Gullbo J, D'Arcy P, Linder S. Targeting mitochondrial function to treat quiescent tumor cells in solid tumors. Int J Mol Sci 2015; 16:27313-26; PMID:26580606; https://dx.doi.org/ 10.3390/ijms161126020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xi Z, Yao M, Li Y, Xie C, Holst J, Liu T, Cai S, Lao Y, Tan H, Xu HX, et al.. Guttiferone K impedes cell cycle re-entry of quiescent prostate cancer cells via stabilization of FBXW7 and subsequent c-MYC degradation. Cell Death Dis 2016; 7:e2252; PMID:27253416; https://dx.doi.org/ 10.1038/cddis.2016.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai J, Miller MA, Everetts NJ, Wang X, Li P, Li Y, Xu JH, Yao G. Elimination of quiescent slow-cycling cells via reducing quiescence depth by natural compounds purified from Ganoderma lucidum. Oncotarget 2017; 8(8):13770-13781; PMID:28099150; https://dx.doi.org/ 10.18632/oncotarget.14634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.García-Rodas R, Zaragoza O. Catch me if you can: phagocytosis and killing avoidance by Cryptococcus neoformans. FEMS Immunol Med Microbiol 2012; 64:147-61; PMID:22029633; https://dx.doi.org/ 10.1111/j.1574-695X.2011.00871.x [DOI] [PubMed] [Google Scholar]
- 66.Coelho C, Bocca AL, Casadevall A. The intracellular life of Cryptococcus neoformans. Annu Rev Pathol 2014; 9:219-38; PMID:24050625; https://dx.doi.org/ 10.1146/annurev-pathol-012513-104653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du H, Guan G, Li X, Gulati M, Tao L, Cao C, Johnson AD, Nobile CJ, Huang G. N-acetylglucosamine-induced cell death in Candida albicans and its implications for adaptive mechanisms of nutrient sensing in yeasts. MBio 2015; 6:e01376-15; PMID:26350972; https://dx.doi.org/ 10.1128/mBio.01376-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oliver JD, Bockian R. In vivo resuscitation, and virulence towards mice, of viable but nonculturable cells of Vibrio vulnificus. Appl Environ Microbiol 1995; 61:2620-3; PMID:7618873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol 2014; 5:258; PMID:24917854; https://dx.doi.org/ 10.3389/fmicb.2014.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding T, Suo Y, Xiang Q, Zhao X, Chen S, Ye X, Liu D. Significance of viable but nonculturable Escherichia coli: induction, detection, and control. J Microbiol Biotechnol 2016; 27:417–28; PMID:27974728; https://dx.doi.org/ 10.4014/jmb.1609.09063 [DOI] [PubMed] [Google Scholar]
- 71.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M “Sleeping beauty:” quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 2004; 68:187-206; PMID:15187181; https://dx.doi.org/ 10.1128/MMBR.68.2.187-206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Virgilio C. The essence of yeast quiescence. FEMS Microbiol Rev 2012; 36:306-39; PMID:21658086; https://dx.doi.org/ 10.1111/j.1574-6976.2011.00287.x [DOI] [PubMed] [Google Scholar]
- 73.Sajiki K, Hatanaka M, Nakamura T, Takeda K, Shimanuki M, Yoshida T, Hanyu Y, Hayashi T, Nakaseko Y, Yanagida M. Genetic control of cellular quiescence in S. pombe. J Cell Sci 2009; 122:1418-29; PMID:19366728; https://dx.doi.org/ 10.1242/jcs.046466 [DOI] [PubMed] [Google Scholar]
- 74.Yao G. Modelling mammalian cellular quiescence. Interface Focus 2014; 4:20130074; PMID:24904737; https://dx.doi.org/ 10.1098/rsfs.2013.0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun D, Buttitta L. Protein phosphatase 2A promotes the transition to G0 during terminal differentiation in Drosophila. Development 2015; 142:3033-45; PMID:26253406; https://dx.doi.org/ 10.1242/dev.120824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo Y, Flegel K, Kumar J, McKay DJ, Buttitta LA. Ecdysone signaling induces two phases of cell cycle exit in Drosophila cells. Biol Open 2016; 5:1648-61; PMID:27737823; https://dx.doi.org/ 10.1242/bio.017525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li L, Bhatia R. Stem cell quiescence. Clin Cancer Res 2011; 17:4936-41; PMID:21593194; https://dx.doi.org/ 10.1158/1078-0432.CCR-10-1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yanagida M, Ikai N, Shimanuki M, Sajiki K. Nutrient limitations alter cell division control and chromosome segregation through growth-related kinases and phosphatases. Philos Trans R Soc Lond B Biol Sci 2011; 366:3508-20; PMID:22084378; https://dx.doi.org/ 10.1098/rstb.2011.0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klein J, Grummt I. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc Natl Acad Sci U S A 1999; 96:6096-101; PMID:10339547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roche B, Arcangioli B, Martienssen RA. RNA interference is essential for cellular quiescence. Science 2016; 354:piiaah5651; PMID:27738016; https://doi.org/ 10.1126/science.aah5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pluskal T, Hayashi T, Saitoh S, Fujisawa A, Yanagida M. Specific biomarkers for stochastic division patterns and starvation-induced quiescence under limited glucose levels in fission yeast. FEBS J 2011; 278:1299-315; PMID:21306563; https://dx.doi.org/ 10.1111/j.1742-4658.2011.08050.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sajiki K, Pluskal T, Shimanuki M, Yanagida M. Metabolomic analysis of fission yeast at the onset of nitrogen starvation. Metabolites 2013; 3:1118-29; PMID:24958269; https://dx.doi.org/ 10.3390/metabo3041118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kohda TA, Tanaka K, Konomi M, Sato M, Osumi M, Yamamoto M. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells 2007; 12:155-70; PMID:17295836; https://dx.doi.org/ 10.1111/j.1365-2443.2007.01041.x [DOI] [PubMed] [Google Scholar]
- 84.Markaki M, Tavernarakis N. Metabolic control by target of rapamycin and autophagy during ageing—a mini-review. Gerontology 2013; 59:340-8; PMID:23594965; https://dx.doi.org/ 10.1159/000348599 [DOI] [PubMed] [Google Scholar]
- 85.An Z, Tassa A, Thomas C, Zhong R, Xiao G, Fotedar R, Tu BP, Klionsky DJ, Levine B. Autophagy is required for G1/G0 quiescence in response to nitrogen starvation in Saccharomyces cerevisiae. Autophagy 2014; 10:1702-11; PMID:25126732; https://dx.doi.org/ 10.4161/auto.32122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sideri T, Rallis C, Bitton DA, Lages BM, Suo F, Rodríguez-López M, Du LL, Bähler J. Parallel profiling of fission yeast deletion mutants for proliferation and for lifespan during long-term quiescence. G3 (Bethesda) 2014; 5:145-55; PMID:25452419; https://dx.doi.org/ 10.1534/g3.114.014415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Müller M, Schmidt O, Angelova M, Faseri K, Weys S, Kremser L, Pfaffenwimmer T, Dalik T, Kraft C, Trajanoski Z, et al.. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. Elife 2015; 4:e07736; PMID:25902403; https://dx.doi.org/ 10.7554/eLife.07736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang H, Kawamata T, Horie T, Tsugawa H, Nakayama Y, Ohsumi Y, Fukusaki E. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J 2015; 34:154-68; PMID:25468960; https://dx.doi.org/ 10.15252/embj.201489083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Welter E, Elazar Z. Autophagy mediates nonselective RNA degradation in starving yeast. EMBO J 2015; 34:131-3; PMID:25492883; https://dx.doi.org/ 10.15252/embj.201490621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sagot I, Pinson B, Salin B, Daignan-Fornier B. Actin bodies in yeast quiescent cells: an immediately available actin reserve? Mol Biol Cell 2006; 17:4645-55; PMID:16914523; https://dx.doi.org/ 10.1091/mbc.E06-04-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laporte D, Salin B, Daignan-Fornier B, Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol 2008; 181:737-45; PMID:18504300; https://dx.doi.org/ 10.1083/jcb.200711154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yedidi RS, Fatehi AK, Enenkel C. Proteasome dynamics between proliferation and quiescence stages of Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol 2016; 51:497-512; PMID:27677933; https://dx.doi.org/ 10.1080/10409238.2016.1230087 [DOI] [PubMed] [Google Scholar]
- 93.Laporte D, Courtout F, Salin B, Ceschin J, Sagot I. An array of nuclear microtubules reorganizes the budding yeast nucleus during quiescence. J Cell Biol 2013; 203:585-94; PMID:14147429; https://dx.doi.org/ 10.1083/jcb.201306075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Laporte D, Courtout F, Pinson B, Dompierre J, Salin B, Brocard L, Sagot I. A stable microtubule array drives fission yeast polarity reestablishment upon quiescence exit. J Cell Biol 2015; 210:99-113; PMID:26124291; https://dx.doi.org/ 10.1083/jcb.201502025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mochida S, Yanagida M. Distinct modes of DNA damage response in S. pombe G0 and vegetative cells. Genes Cells 2006; 11:13-27; PMID:16371129; https://dx.doi.org/ 10.1111/j.1365-2443.2005.00917.x [DOI] [PubMed] [Google Scholar]
- 96.Gangloff S, Arcangioli B. DNA repair and mutations during quiescence in yeast. FEMS Yeast Res 2017; PMID:28087675; https://dx.doi.org/ 10.1093/femsyr/fox002 [DOI] [PubMed] [Google Scholar]
- 97.Kim N, Jinks-Robertson S. Transcription as a source of genomic instability. Nat Rev Genet 2012; 13:204-14; PMID:22330764; https//dx.doi.org/ 10.1038/nrg3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993; 362:709-15; PMID:8469282; https://dx.doi.org/ 10.1038/362709a0 [DOI] [PubMed] [Google Scholar]
- 99.Walter D, Lier A, Geiselhart A, Thalheimer FB, Huntscha S, Sobotta MC, Moehrle B, Brocks D, Bayindir I, Kaschutnig P, et al.. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 2015; 520:549-52; PMID:25707806; https://dx.doi.org/ 10.1038/nature14131 [DOI] [PubMed] [Google Scholar]
- 100.Ben Hassine S, Arcangioli B. Tdp1 protects against oxidative DNA damage in non-dividing fission yeast. EMBO J 2009; 28:632-40; PMID:19197239; https://dx.doi.org/ 10.1038/emboj.2009.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature 1996; 380:64-6; PMID:8598906; https://dx.doi.org/ 10.1038/380064a0 [DOI] [PubMed] [Google Scholar]
- 102.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature 1997; 385:810-3; PMID:9039911; https://dx.doi.org/ 10.1038/385810a0 [DOI] [PubMed] [Google Scholar]
- 103.Kallingappa PK, Turner PM, Eichenlaub MP, Green AL, Oback FC, Chibnall AM, Wells DN, Oback B. Quiescence loosens epigenetic constraints in bovine somatic cells and improves their reprogramming into totipotency. Biol Reprod 2016; 95:16; PMID:27281704; https://dx.doi.org/ 10.1095/biolreprod.115.137109 [DOI] [PubMed] [Google Scholar]
- 104.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002; 297:1833-7; PMID:12193640; https://dx.doi.org/ 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 105.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 2013; 14:100-12; PMID:23329111; https://dx.doi.org/ 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. RNA interference is required for normal centromere function in fission yeast. Chromosome Res 2003; 11:137-46; PMID:12733640; https://doi.org/ 10.1023/A:1022815931524 [DOI] [PubMed] [Google Scholar]
- 107.Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A 2003; 100:193-8; PMID:12509501; https://dx.doi.org/ 10.1073/pnas.232688099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smialowska A, Djupedal I, Wang J, Kylsten P, Swoboda P, Ekwall K. RNAi mediates post-transcriptional repression of gene expression in fission yeast Schizosaccharomyces pombe. Biochem Biophys Res Commun 2014; 444:254-9; PMID:24462781; https://dx.doi.org/ 10.1016/j.bbrc.2014.01.057 [DOI] [PubMed] [Google Scholar]
- 109.Woolcock KJ, Stunnenberg R, Gaidatzis D, Hotz HR, Emmerth S, Barraud P, Bühler M. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev 2012; 27:683-92; PMID:22431512; https://dx.doi.org/ 10.1101/gad.186866.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 2007; 128:491-504; PMID:17289569; https://dx.doi.org/ 10.1016/j.cell.2006.12.035 [DOI] [PubMed] [Google Scholar]
- 111.Shimada A, Dohke K, Sadaie M, Shinmyozu K, Nakayama J, Urano T, Murakami Y. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev 2009; 23:18-23; PMID:19136623; https://dx.doi.org/ 10.1101/gad.1708009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 2008; 18:490-5; PMID:18394897; https://dx.doi.org/ 10.1016/j.cub.2008.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marín L, Chang AY, Goto D, et al.. RNAi promotes heterochromatic silencing through replication-coupled release of RNA pol II. Nature 2011; 479:135-8; PMID:22002604; https://dx.doi.org/ 10.1038/nature10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Castel SE, Ren J, Bhattacharjee S, Chang AY, Sánchez M, Valbuena A, Antequera F, Martienssen RA. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 2014; 159:572-83; PMID:25417108; https://dx.doi.org/ 10.1016/j.cell.2014.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 2005; 309:467-9; PMID:15947136; https://dx.doi.org/ 10.1126/science.1114955 [DOI] [PubMed] [Google Scholar]
- 116.Djupedal I, Portoso M, Spåhr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 2005; 19:2301-6; PMID:16204182; https://dx.doi.org/ 10.1101/gad.344205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 2011; 18:1132-8; PMID:21892171; https://dx.doi.org/ 10.1038/nsmb.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lisica A, Engel C, Jahnel M, É Roldán, Galburt EA, Cramer P, Grill SW. Mechanisms of backtrack recovery by RNA polymerases I and II. Proc Natl Acad Sci U S A 2016; 113(11):2946-51; PMID:26929337; https://dx.doi.org/ 10.1073/pnas.1517011113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lemay JF, Larochelle M, Marguerat S, Atkinson S, Bähler J, Bachand F. The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat Struct Mol Biol 2014; 21:919-26; PMID:25240800; https://dx.doi.org/ 10.1038/nsmb.2893 [DOI] [PubMed] [Google Scholar]
- 120.Braglia P, Kawauchi J, Proudfoot NJ. Co-transcriptional RNA cleavage provides a failsafe termination mechanism for yeast RNA polymerase I. Nucleid Acids Res 2011; 39:1439-48; PMID:20972219; https://dx.doi.org/ 10.1093/nar/gkq894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bernstein DA, Vyas VK, Fink GR. Genes come and go: the evolutionary path of budding yeast RNase III enzymes. RNA Biol 2012; 9:1123-8; PMID:23018782; https://dx.doi.org/ 10.4161/rna.21360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bernstein DA, Vyas VK, Weinberg DE, Drinnenberg IA, Bartel DP, Fink GR. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. Proc Natl Acad Sci U S A 2012; 109:523-8; PMID:22173636; https://dx.doi.org/ 10.1073/pnas.1118859109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wehner S, Dörrich AK, Ciba P, Wilde A, Marz M. pRNA: NoRC-associated RNA of rRNA operons. RNA Biol 2014; 11:3-9; PMID:24440945; https://dx.doi.org/ 10.4161/rna.27448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bierhoff H, Schmitz K, Maass F, Ye J, Grummt I. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb Symp Quant Biol 2010; 75:357-64; PMID:21502405; https://dx.doi.org/ 10.1101/sqb.2010.75.060 [DOI] [PubMed] [Google Scholar]
- 125.Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. Quiescence-induced lncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 2014; 54:675-82; PMID:24768537; https://dx.doi.org/ 10.1016/j.molcel.2014.03.032 [DOI] [PubMed] [Google Scholar]
- 126.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature 2011; 475:244-8; PMID:21725325; https://dx.doi.org/ 10.1038/nature10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gonzalez M, Li F. DNA replication, RNAi and epigenetic inheritance. Epigenetics 2012; 7:14-9; PMID:22207359; https://dx.doi.org/ 10.4161/epi.7.1.18545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Piñon R. Folded chromosomes in non-cycling yeast cells: evidence for a characteristic g0 form. Chromosoma 1978; 67:263-74; PMID:359278; https://doi.org/ 10.1007/BF02569039 [DOI] [PubMed] [Google Scholar]
- 129.Rutledge MT, Russo M, Belton JM, Dekker J, Broach JR. The yeast genome undergoes significant topological reorganization in quiescence. Nucleid Acids Res 2015; 43:8299-313; PMID:26202961; https://dx.doi.org/ 10.1093/nar/gkv723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schäfer G, McEvoy CR, Patterton HG. The Saccharomyces cerevisiae linker histone Hho1p is essential for chromatin compaction in stationary phase and is displaced by transcription. Proc Natl Acad Sci U S A 2008; 105:14838-43; PMID:18799740; https://dx.doi.org/ 10.1073/pnas.0806337105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu Q, Kuzmiak H, Zou Y, Olsen L, Defossez PA, Bi X. Saccharomyces cerevisiae linker histone Hho1p functionally interacts with core histone H4 and negatively regulates the establishment of transcriptionally silent chromatin. J Biol Chem 2009; 284:740-50; PMID:19017647; https://dx.doi.org/ 10.1074/jbc.M806274200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ngubo M, Kemp G, Patterton HG. Nano-electrospray tandem mass spectrometric analysis of the acetylation state of histones H3 and H4 in stationary phase in Saccharomyces cerevisiae. BMC Biochem 2011; 12:34; PMID:21726436; https://dx.doi.org/ 10.1186/1471-2091-12-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Laporte D, Courtout F, Tollis S, Sagot I. Quiescent Saccharomyces cerevisiae forms telomere hyperclusters at the nuclear membrane vicinity through a multifaceted mechanism involving Esc1, the Sir complex, and chromatin condensation. Mol Biol Cell 2016; 27:1875-84; PMID:27122604; https://dx.doi.org/ 10.1091/mbc.E16-01-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McKnight JN, Boerma JW, Breeden LL, Tsukiyama T. Global promoter targeting of a conserved lysine deacetylase for transcriptional shutoff during quiescence entry. Mol Cell 2015; 59:732-43; PMID:26300265; https://dx.doi.org/ 10.1016/j.molcel.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miles S, Breeden LL. A common strategy for initiating the transition from proliferation to quiescence. Curr Genet 2016; 63:179-86; PMID:27544284; https://dx.doi.org/ 10.1007/s00294-016-0640-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sandmeier JJ, French S, Osheim Y, Cheung WL, Gallo CM, Beyer AL, Smith JS. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J 2002; 21:4959-68; PMID:12234935; https://doi.org/ 10.1093/emboj/cdf498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nishida H, Matsumoto T, Kondo S, Hamamoto M, Yoshikawa H. The early diverging ascomycetous budding yeast Saitoella complicata has three histone deacetylases belonging to the Clr6, Hos2 and Rpd3 lineages. J Gen Appl Microbiol 2014; 60:7-12; PMID:24646756; https://doi.org/ 10.2323/jgam.60.7 [DOI] [PubMed] [Google Scholar]
- 138.Rawlings JS, Gatzka M, Thomas PG, Ihle JN. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J 2011; 30:263-76; PMID:21169989; https://dx.doi.org/ 10.1038/emboj.2010.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Boonsanay V, Zhang T, Georgieva A, Kostin S, Qi H, Yuan X, Zhou Y, Braun T. Regulation of skeletal muscle stem cell quiescence by Suv4-20h1-dependent facultative heterochromatin formation. 2016; 18:229-42; PMID:26669898; https://dx.doi.org/ 10.1016/j.stem.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 140.Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA. H4K20 methylation regulates quiescence and chromatin compaction. Mol Biol Cell 2013; 24:3025-37; PMID:23924899; https://dx.doi.org/ 10.1091/mbc.E12-07-0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dai L, Ye S, Li HW, Chen DF, Wang HL, Jia SN, Lin C, Yang JS, Yang F, Nagasawa H, et al.. SETD4 regulates cell quiescence and catalyzes the trimethylation of H4K20 during diapause formation of Artemia. Mol Cell Biol 2017; 37:7 e00453-16; PMID:28031330; https://doi.org/ 10.1128/MCB.00453-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Svensson JP, Shukla M, Menendez-Benito V, Norman-Alexsson U, Audergon P, Sinha I, Tanny JC, Allshire RC, Ekwall K. A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin. Genome Res 2015; 25:872-83; PMID:25778913; https://dx.doi.org/ 10.1101/gr.188870.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysing 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004; 119:603-14; PMID:15550243; https://dx.doi.org/ 10.1016/j.cell.2004.11.009 [DOI] [PubMed] [Google Scholar]
- 144.Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR 3rd, Jia S. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell 2009; 33:428-37; PMID:19250904; https://dx.doi.org/ 10.1016/j.molcel.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Joh RI, Khanduja JS, Calvo IA, Mistry M, Palmieri CM, Savol AJ, Ho Sui SJ, Sadreyev RI, Aryee MJ, Motamedi M. Survival in quiescence requires the euchromatic deployment of Clr4/SUV39H by Argonaute-associated small RNAs. Mol Cell 2016; 64:1088-101; PMID:27984744; https://dx.doi.org/ 10.1016/j.molcel.2016.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315-26; PMID:16630819; https://dx.doi.org/ 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 147.Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 2013; 4:189-204; PMID:23810552; https://dx.doi.org/ 10.1016/j.celrep.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Young CP, Hillyer C, Hokamp K, Fitzpatrick DJ, Konstantinov NK, Welty JS, Ness SA, Werner-Washburne M, Fleming AB, Osley MA. Distinct histone methylation and transcription profiles are established during the development of cellular quiescence in yeast. BMC Genomics 2017; 18:107; PMID:28122508; https://dx.doi.org/ 10.1186/s12864-017-3509-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Brumbaugh J, Phanstiel D, Coon JJ. Unraveling the histone's potential: a proteomics perspective. Epigenetics 2008; 3:254-7; PMID:18849650; https://doi.org/ 10.4161/epi.3.5.7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang K, Chen Y, Zhang Z, Zhao Y. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. J Proteome Res 2008; 8:900-6; PMID:19113941; https://dx.doi.org/ 10.1021/pr8005155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Arnaudo AM, Garcia BA. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin 2013; 6:24; PMID:23916056; https://dx.doi.org/ 10.1186/1756-8935-6-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu YM, Du JY, Lau AT. Posttranslational modifications of human histone H3: an update. Proteomics 2014; 14:2047-60; PMID:25044606; https://dx.doi.org/ 10.1002/pmic.201300435 [DOI] [PubMed] [Google Scholar]
- 153.Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol 2015; 7:a025064; PMID:26330523; https://dx.doi.org/ 10.1101/cshperspect.a025064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou M, Paša-Tolić L, Stenoien DL. Profiling of histone post-translational modifications in mouse brain with high-resolution top-down mass spectrometry. J Proteome Res 2017; 16:599-608; PMID:28001079; https://dx.doi.org/ 10.1021/acs.jproteome.6b00694 [DOI] [PubMed] [Google Scholar]
- 155.inha I, Buchanan L, Rönnerblad M, Bonilla C, Durand-Dubief M, Shevchenko A, Grunstein M, Stewart AF, Ekwall K. Genome-wide mapping of histone modifications and mass spectrometry reveal H4 acetylation bias and H3K36 methylation at gene promoters in fission yeast. Epigenomics 2010; 2:377-93; PMID:22121899; https://dx.doi.org/ 10.2217/epi.10.18 [DOI] [PubMed] [Google Scholar]
- 156.Mews P, Zee BM, Liu S, Donahue G, Garcia BA, Berger SL. Histone methylation has dynamics distinct from those of histone acetylation in cell cycle reentry from quiescence. Mol Cell Biol 2014; 34:3968-80; PMID:25154414; https://dx.doi.org/ 10.1128/MCB.00763-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Matsubara K, Sano N, Umehara T, Horikoshi M. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 2007; 12:13-33; PMID:17212652; https://dx.doi.org/ 10.1111/j.1365-2443.2007.01031.x [DOI] [PubMed] [Google Scholar]
- 158.Kawashima S, Nakabayashi Y, Matsubara K, Sano N, Enomoto T, Tanaka K, Seki M, Horikoshi M. Global analysis of core histones reveals nucleosomal surfaces required for chromosome bi-orientation. EMBO J 2011; 30:3353-67; PMID:21772248; https://dx.doi.org/ 10.1038/emboj.2011.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mellone BG, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr Biol 2003; 13:1748-57; PMID:14561399; https://doi.org/ 10.1016/j.cub.2003.09.031 [DOI] [PubMed] [Google Scholar]
- 160.Watt S, Mata J, López-Maury L, Marguerat S, Burns G, Bähler J. urg1: a uracil-regulatable promoter system for fission yeast with short induction and repression times. PLoS One 2008; 3:e1428; PMID:18197241; https://dx.doi.org/ 10.1371/journal.pone.0001428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chan DA, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat Rev Drug Discov 2011; 10:351-64; PMID:21532565; https://dx.doi.org/ 10.1038/nrd3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Weidle UH, Maisel D, Eick D. Synthetic lethality-based targets for discovery of new cancer therapeutics. Cancer Genomics Proteomics 2011; 8:159-71; PMID:21737609 [PubMed] [Google Scholar]
- 163.Thompson JM, Nguyen QH, Singh M, Razorenova OV. Approaches to identifying synthetic lethal interactions in cancer. Yale J Biol Med 2015; 88:145-55; PMID:26029013 [PMC free article] [PubMed] [Google Scholar]
- 164.Srivas R, Shen JP, Yang CC, Sun SM, Li J, Gross AM, Jensen J, Licon K, Bojorquez-Gomez A, Klepper K, et al.. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol Cell 2016; 63:514-25; PMID:27453043; https://dx.doi.org/ 10.1016/j.molcel.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]


