Abstract
The dentate gyrus (DG) is thought to perform pattern separation on inputs received from the entorhinal cortex, such that the DG forms distinct representations of different input patterns. Neuronal responses, however, are known to be variable, and that variability has the potential to confuse the representations of different inputs, thereby hindering the pattern separation function. This variability can be especially problematic for tissues like the DG, in which the responses can persist for tens of seconds following stimulation: the long response duration allows for variability from many different sources to accumulate. To understand how the DG can robustly encode different input patterns, we investigated a recently-developed in vitro hippocampal dentate gyrus preparation that generates persistent responses to transient electrical stimulation. For 10–20s after stimulation, the responses are indicative of the pattern of stimulation that was applied, even though the responses exhibit significant trial-to-trial variability. Analyzing the dynamical trajectories of the evoked responses, we found that, following stimulation, the neural responses follow distinct paths through the space of possible neural activations, with a different path associated with each stimulation pattern. The neural responses’ trial-to-trial variability shifts the responses along these paths rather than between them, maintaining the separability of the input patterns. Manipulations that redistributed the variability more isotropically over the space of possible neural activations impeded the pattern separation function. Consequently, we conclude that the confinement of neuronal variability to these one-dimensional paths mitigates the impacts of variability on pattern encoding and thus may be an important aspect of the DG’s ability to robustly encode input patterns.
Keywords: Persistence, Pattern Separation, Dentate Gyrus, Intracellular, Slice, Variability
Introduction
The hippocampal dentate gyrus is thought to perform pattern separation on the inputs it receives from the entorhinal cortex [Leutgeb et al., 2007; Myers and Scharfman, 2009]. In other words, the dentate should yield distinct responses even when presented with overlapping stimulus patterns [O’Reilly and McClelland, 1994]. Confounding this function is the fact that, even over repeat presentations of the same stimulus, neural activities tend to be highly variable [Britten et al., 1993; Softky and Koch, 1993; Faisal et al., 2008]. This variability within the representation can confuse the representations of different input patterns, leading us to wonder how the dentate can robustly encode these patterns.
The issue of robust representation in the presence of variability has been relatively well-studied in the peripheral sensory systems [Faisal et al., 2008; Hu et al., 2014; Aver beck et al., 2006; Romo et al., 2003; Cayco-Gajic et al., 2015; Shamir, 2014; da Silveira and Berry, 2014], where we have a rapidly-deepening understanding of how robust population codes can be constructed from variable single-cell responses. While similarly high levels of variability are observed in the “deeper” cortical structures, our understanding of how those systems form robust representations is relatively poor. The problem of robustness is especially important in the context of persistent mnemonic (symbolic) representations — involved in functions like working memory — where the long duration of the representation means that there is ample time for noise from different sources to accumulate.
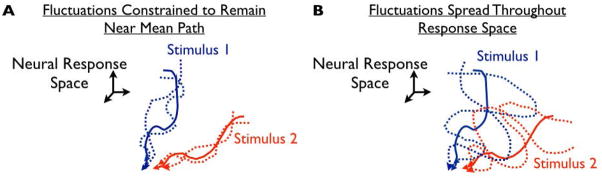
To address the questions of robust sustained pattern encoding, we exploited a recently-developed in vitro hippocampal dentate gyrus preparation that exhibits sustained responses to electrical stimulation that last for more than 20s after the stimulus is turned off [Hyde and Strowbridge 2012]. Importantly, no pharmacological manipulations are needed to trigger these persistent activities: they are innate properties of the tissue. We previously [Hyde and Strowbridge 2012] demonstrated that these responses, when averaged over the duration of the response, could be decoded on a trial-by-trial basis to reveal the pattern of stimulation that was applied. Does the same separability persists on shorter time scales — so that, at any epoch, the stimulation patterns can be distinguished based on the current neural activations? Or are the patterns only separable after lengthy integration of the neural responses, that can “average away” the variability? For example, in Fig. 1A, the responses to different stimuli overlap relatively little on an epoch-by-epoch and trial-by-trial basis, and so they could be decoded in short time windows to recover the applied stimulus. For contrast, in Fig. 1B, there are epochs at which the responses to different stimuli overlap significantly, and so the responses could not be accurately decoded, on an epoch-by epoch and / or trial-by-trial basis, to recover the stimulus identity.
Fig. 1. Constraining fluctuations near stimulus-evoked response trajectories may yield robust representations.
In the cartoons (A and B), we consider the space of all possible neural responses. Each axis in the space is the response of a given neuron, and the dimensionality of the space is equal to the number of neurons in the population. Within this space, we consider the dynamical trajectories of the evoked neural responses. The cartoons show both the mean response trajectories (solid line), and two example trajectories — observed on different trials (dashed lines) — generated in response to 2 different stimuli. In (A), the variability is structured such that responses to each stimulus remain close to the mean trajectory. For contrast, in (B), the trial-by-trial fluctuations are more spread out from the mean trajectory. Accordingly, the responses to different stimuli in (B) overlap more — and thus the stimulus identity is more ambiguously encoded in the neural activities.
Herein, we find that the stimulation patterns can be distinguished, epoch-by-epoch, based on the neural activations on individual trials, for up to 20s following stimulation. Next, we identify the structure of the neural responses that maintains the separability of input patterns over time. Given that a brain that required long integration times to identify input patterns would lead to slow behavioral reactions, the neural response structures we identified — that allow for pattern encoding using short integration windows — could be important to the hippocampus’ (and thus the brain’s) reaction speed.
More specifically, we found that, following stimulation, the activities of neural populations in our hippocampal preparation tend to travel along distinct paths in the space of possible neural activations, with a different path for each applied stimulus. This observation suggests that the stimulus identity is most persistently reflected in the identity of the path on which the neural responses lie, rather than any specific pattern of neural activities over the population. The variability in the neural responses tends to spread the response trajectories out along the appropriate paths, rather than between them. Our results show that this structure makes the neural representation of input patterns more robust because the noise tends not to push the responses towards the “other” paths, corresponding to stimuli other than the one that was presented (for illustration, compare Figs. 1AB).
Materials and Methods
Experimental Set-up
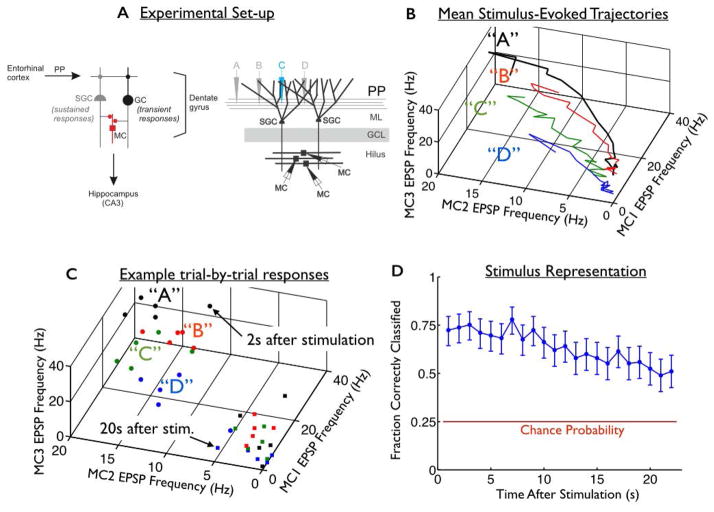
Herein, we revisit the data from a previous study [Hyde and Strowbridge, 2012]. The preparation consists of horizontal slices of rat brain that pass through the hippocampal formation. In each slice, we identified the perforant path (PP: which provides inputs to the hippocampus from the entorhinal cortex), and implanted an array of stimulating electrodes into the PP (Fig. 2A). We then transiently stimulated the PP with brief (200 μs) shocks from one of four stimulating electrodes, and thereafter we recorded intracellularly from triplets of mossy cells (MCs) in the dentate gyrus [Scharfman and Schwartzkroin, 1988]. Hilar mossy cells have extensive dendritic arbors, and therefore, they broadly sample the population of upstream granule cells and semilunar granule cells (Fig. 2A) [Larimer and Strowbridge, 2010; Williams et al., 2007]. For our analyses, we extracted the rates of excitatory post-synaptic potentials (EPSPs) received by our three mossy cells, measured over 1-second-long intervals at different times post-stimulation (from 1s up to >20s post-stimulation).
Fig. 2. Persistent mnemonic representations formed by variable neural activities.
(A) (Left) Diagram of excitatory synaptic connections within the dentate gyrus. Perforant path (PP) input from entorhinal cortical neurons excites both granule cells (GC) and semilunar granule cells (SGCs). Both cell types are excitatory and project toward the CA3 subfield of the hippocampus. While GCs respond transiently to PP inputs, SGCs can fire persistently in response to brief synaptic input. Axon collaterals of both GCs and SGCs synapse on mossy cells (MC) in the dentate hilus. (Right) To probe the structure of the dentate’s persistent representations, we implanted an array of four stimulating electrodes (labelled “A”, “B”, “C”, and “D”) into the perforant path. We transiently stimulated the PP using one stimulating electrode at a time, and then recorded from downstream mossy cells. The mossy cells receive persistent synaptic inputs from semilunar granule cells, in the molecular layer (ML) of the dentate gyrus. The granule cell layer (GCL) is also shown. (B) Mean EPSP frequency response trajectories of the 3 recorded mossy cells, in response to four different stimuli (labelled “A”, “B”, “C”, and “D”). (C) Responses to the 4 stimuli on each of 5 trials are shown. The circles are data recorded 2s after stimulation, whereas the squares are 20s post-stimulation. As time passes after stimulation, the responses to different stimuli drift towards the origin and blend together, hindering the representation. (D) (blue curve) Accuracy of the mnemonic representation, quantified by fraction of correctly classified responses, as a function of when they occurred after the stimulus offset. Error bars are 95% confidence intervals, using data from all 9 slice experiments. Red horizontal line represents chance performance.
Each stimulus was applied between 3 and 5 times, and a total of 4 different stimuli were applied to each slice. Owing to the long duration of the evoked responses, there was a 210s delay between subsequent stimulation events, to allow the tissue to return to its resting state. The amount of time per trial (210s), and the limited (~1h) duration of the slice experiments restricted the number of repeats of each stimulus. The experiment and associated analyses were repeated on 9 different slices. The data from each slice were analyzed separately, and the results reported herein are average quantities (averaged over those 9 slices).
In vivo, stimuli will not necessary arrive in isolation and well-separated in time: randomly-timed barrages of stimuli are more likely. In our recordings, however, the stimuli were intentionally well-separated in time, so that the network synaptic activity assayed by intracellular recordings could return to its baseline activity level between stimuli. For contrast, if the stimuli were more closely spaced, and the activity levels did not return to the resting state between stimuli, then contextual effects (as in [Hyde and Strowbridge, 2012]) would impact the neural activity patterns. Our experimental protocol removed this confound, allowing us to cleanly investigate the neural activity patterns responsible for encoding the stimulation position.
Decoding the Mossy Cell Responses
To quantify the representation of the applied stimulation patterns by the hilar neural activities, we sought to estimate the applied stimulus given the resultant neural activities. To do so, we used the standard k-nearest neighbors (KNN) algorithm, as follows. For each experiment, we went through the recorded data points one-by-one, and tried to guess which stimulus generated the recorded response (each data point is the mean EPSP rates of the 3 cells in a given 1-s-long window, on a given trial, of a given stimulus). For each such “test” data point, we computed the K nearest-neighboring data points from that experiment (smallest Euclidean distance). In other words, among all time epochs, trials, and stimuli, we found those data points most similar to the test point. Next, we took a majority vote over the stimulus labels from those K neighbors, and used that as our guess for the stimulus that generated the “test” data point. We used K=5, and verified that other choices of K yield very similar results. After recording the guesses for all recorded data points, we computed the fraction of such guesses that were correct in a given epoch, and averaged the results over all 9 slice experiments.
We emphasize that the test point is not used in building the classifier (i.e., the test data point is not allowed to be its own neighbor). However, one potential concern with our approach is that the response data points used in building the classifier include those recorded on the same trial as the test point (albeit at different epochs). Data points recorded on the same trial may be atypically similar, posing a potential confound to our analysis. One possible way to address this concern would be to modify the KNN analysis so that, in classifying the test point, the classifier uses only responses recorded on trials other than the one from which the test point was taken. This approach, however, is problematic because the number of trials is small (3–5), and so removing a whole trial makes the classifier perform very poorly (regardless of the correlations between data points).
However, we still wanted to verify that our classification result is not confounded by the autocorrelation (within each trial). To do this, we generated surrogate data via a method (described below) that is guaranteed to yield independence of neighboring time points, even within the same trial. To do this, we did the following:
For each experiment, we went through the data stimulus-by-stimulus and epoch-by-epoch. Thus, for each stimulus / epoch, we extracted the EPSP rates of the 3 cells on the 3–5 different trials.
We computed the mean and covariance (over trials) of these data points.
We generated surrogate data by drawing Gaussian random variables with the same mean and covariance as the actual data. For consistency with the analysis of the actual data, and with the “random rotation” analysis (described below), we generated the same number of surrogate trials as there were in the actual data.
We repeated steps 1–3 independently for each epoch. Accordingly, the different epochs on a given surrogate trial are no more correlated than are data points from different surrogate trials.
We repeated this surrogate-data generation procedure for all different stimuli from a given experiment, and performed the KNN analysis as described above.
We repeated steps 1–5 for all 9 slice experiments, and averaged the results.
This analysis shows very similar classification rates (not shown) as did the analysis done on the “actual” data, giving us added confidence that the KNN performance is not confounded by autocorrelation within trials.
Measuring Distances From Mean Trajectories
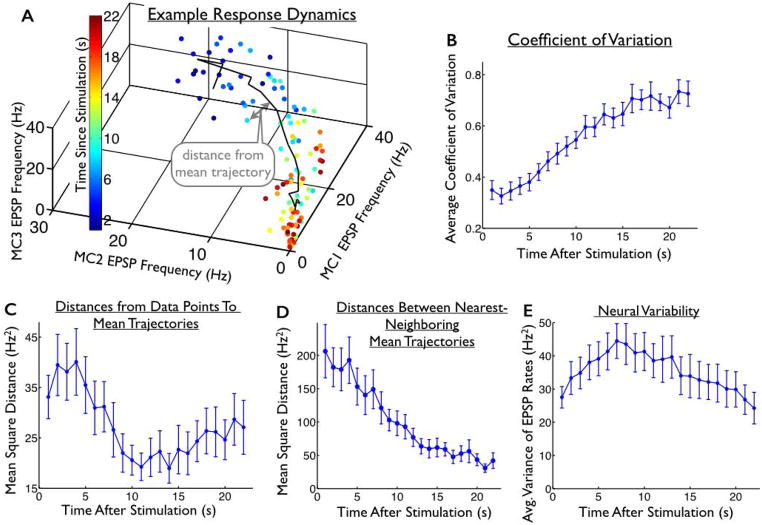
To understand where (in the space of possible neural responses) the trial-to-trial variability is most concentrated, we measured the deviations of responses on individual trials from the mean stimulus-evoked trajectories. This measurement is described schematically in Fig. 3A.
Fig. 3. Structure of the trial-to-trial variability in the evoked responses.
(A) Responses of one mossy cell triplet to one location of stimulation. Each dot represents the mean EPSP frequencies in a 1s window on a given trial; 5 trials are shown. The colors of the dots indicate the time since stimulation. Black line represents the mean trajectory, averaged over trials. (B) Average coefficient of variation (standard deviation divided by mean) of the neural responses, as a function of when they were observed. (C) For each data point, we measured the distance to the nearest point on the mean trajectory. The mean of these distances (averaged over trials) is shown as a function of time since stimulation. During the first ~12s of the responses, these distances decrease. (D) For comparison, we also show the mean squared distances between the mean trajectories and their nearest-neighboring trajectory. (E) Average (over cells and stimuli) of the trial-to-trial variance in neural responses, as a function of time since stimulation. The (average) level of variability increases over the first ~10s of the responses, then decreases. Thus, during the first ~10s of the responses, the overall response variability increases (BE) while the distances from the data points to the mean trajectories decrease (C), underlining the fact that response variability is not evenly spread over the space of possible neural activities. Error bars in all panels represent the standard error of the mean (S.E.M.) over all 4 stimuli and 9 slice experiments.
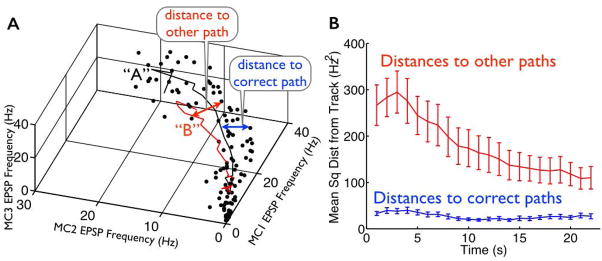
To carry out this measurement, we computed the mean trajectory following each stimulus by averaging over all responses to that stimulus within each epoch (mean trajectories shown in Figs. 2B and 3A). We then interpolated between these (22) data points to “fill in the curve”, and measured the Euclidean distance from each data point to the nearest point on the mean trajectory, to yield the data shown in Fig. 3C. Similarly, for the results in Fig. 6B we measured the distances from each response data point to the nearest point on the mean trajectories associated with stimuli other than the one that generated the responses.
Fig. 6. Responses are typically far from the “wrong” trajectories.
(A) Mean response trajectories of one mossy cell triplet following two locations of stimulation (“A” and “B”, corresponding to black and red solid lines, respectively). Also shown are the mean EPSP frequencies in 1s windows on individual trials in which stimulus “A” was presented; 5 such trials are shown (black dots). The schematic arrows indicate the measurement of distances from these response data points to the nearest points on the two mean trajectory curves. (B) (blue curve; same as Fig. 3C) We computed the distance from each recorded response to the nearest point on the mean trajectory associated with the applied stimulus. For comparison, we also computed the distances from the recorded responses to the nearest points on the mean trajectories associated with stimuli other than the one that was actually applied (red curve: mean +/− S.E.M. over the 9 slice experiments and 4 stimuli). For example, if the response was evoked by stimulus “A”, we measured the (average) distance to the mean trajectories evoked by stimuli “B”, “C”, and “D”. The distances to the mean trajectories evoked by “other” stimuli (red curve) are typically much larger than the distances to the mean trajectories evoked by the “correct” stimulus (blue curve).
Measuring Distances Between Mean Trajectories
To estimate the distances between the mean stimulus-evoked trajectories (Fig. 3D), we took each trajectory and, for each epoch, we found the nearest (in Euclidean distance) of the other mean trajectories. We then recorded the distance between these nearest-neighboring trajectories at this epoch. This procedure was repeated for all epochs, to trace out the distance vs time curve: data shown are averaged over all stimuli and experiments.
Measuring Average Levels of Variability
To estimate the overall level of trial-to-trial variability in the neural activities (Fig. 3E), we took all responses of a given cell to a given stimulus, in a given post-stimulation epoch. We then computed the trial-to-trial variance of these responses, and average that quantity over all 3 cells, to quantify the overall levels of trial-to-trial variability. To compute the coefficient of variation (Fig. 3B), we divided the square root of each cell’s variance by its mean response (again, for each epoch and stimulus) and averaged those values over all cells.
Perturbing the Structure of the Response Variability
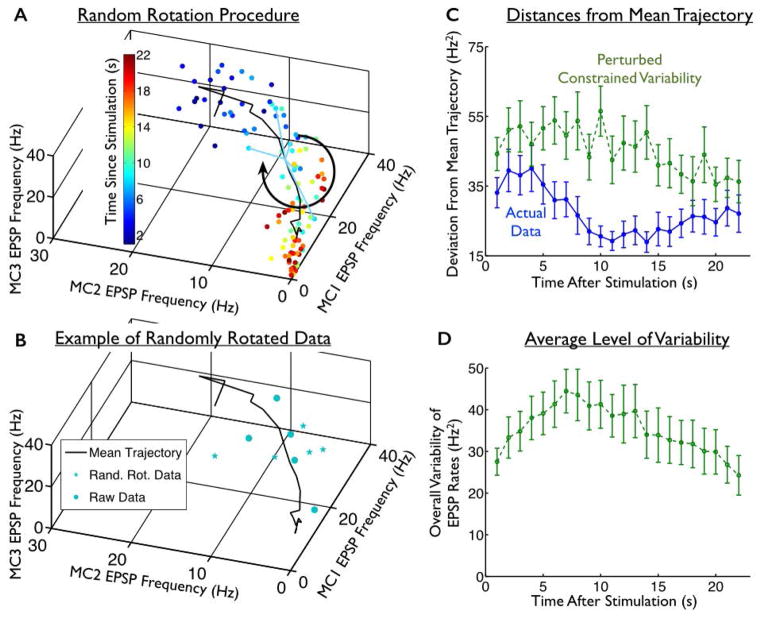
To construct surrogate data in which the “confined” structure of the trial-to-trial variability is interrupted, we did the following. For each stimulus, and each post-stimulation epoch, we computed the mean response (over all trials), and subtracted that from the responses on each trial. This yielded the residuals, which are 3-dimensional vectors describing the trial-to-trial fluctuations in the population responses. We then randomly rotated these vectors in the 3-dimensional space, and added these rotated residuals to the mean responses (Fig. 4AB).
Fig. 4. Perturbed constrained variability.
(A) We constructed surrogate data in which the mean trajectories evoked by each stimulus (black solid curve; data shown are for one stimulus), and the overall levels of trial-to-trial variability, matched the recorded data, but the variability was distributed relatively isotropically over the space of neural responses. To do this, we took the constellations of points recorded at each epoch, and rotated them in a random direction about the mean response recorded at that epoch; this is shown schematically for the teal data points, corresponding to responses recorded 8s post-stimulation. (B) The raw data points observed 8s post-stimulation (teal circles) are shown alongside an example of the randomly-rotated data points (stars) from that same epoch. (C) For these perturbed surrogate data, we computed the distance from each response to the mean response trajectory associated with the applied stimulus. We then averaged these over all stimuli and experiments, and show those distances as a function of time post-stimulation (green curve). This is similar to the measurement shown schematically in Fig. 3A. For comparison, we also show the same quantity for the actual experimental data, in which the structure of the fluctuations was left intact (blue curve; same data as in Fig. 3C). (D). As in Fig. 3E, we measured the average trial-to-trial variability in the surrogate responses (with perturbed trial-to-trial variability) as a function of time. By construction, this level of variability is identical to that which was displayed by the actual (unperturbed) responses (in Fig. 3E). Error bars in all panels represent the standard error of the mean (S.E.M.) over all 4 stimuli and 9 slice experiments.
To randomly rotate the residual vectors — thereby redistributing trial-to-trial variability in random directions in the response space — we generated random rotation matrices. We then multiplied the residual vectors by these rotation matrices. A different random rotation was applied for each post-stimulation epoch.
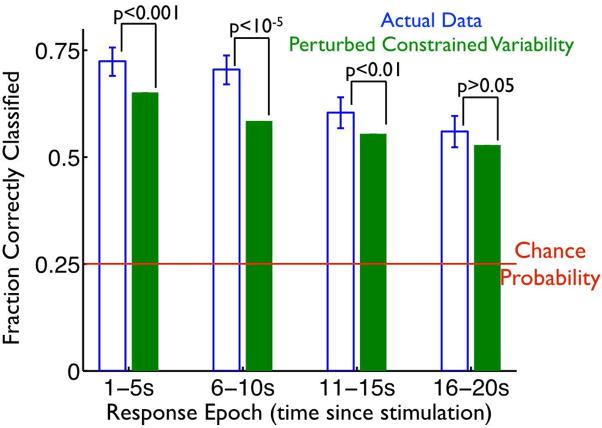
For the results in Fig. 5, that compare the decoding performance on the surrogate vs. actual data, we estimated the decoding performance with our KNN algorithm for each random rotation, and averaged the result over 10,000 surrogate datasets, each of which had different random rotations.
Fig. 5. Variability constrained to lie near mean trajectories enhances pattern separability in the dentate gyrus.
We used our KNN classifier to identify the stimulus responsible for the neural responses (as in Fig. 2D). The classifier was applied to both the actual experimental data (blue bars), and the surrogate data (green bars) in which the structure of the trial-to-trial variability was perturbed (as in Fig. 4AB), reducing the tendency of fluctuations to lie along / near the mean stimulus-evoked trajectories. The classification was performed on the mean EPSP frequencies measured in 1s windows post-stimulation, and the bar heights indicate the fraction of successfully classified responses, in 4 different post-stimulation epochs. Error bars indicate 95% confidence intervals. To generate the green bars, we repeated the random rotation procedure 10,000 times, yielding enough surrogate trials that the error bars associated with the performance values are vanishingly small. At each epoch, we show the p-values for comparisons between the performance obtained with the raw, and the surrogate, data; these p-values come from binomial tests.
Results
In vitro persistent mnemonic representations
Herein, we revisit the data from a previous study [Hyde and Strowbridge, 2012]. The preparation consists of horizontal slices of rat brain, which pass through the hippocampal formation. In each slice, we identified the perforant path (PP: which provides inputs to the hippocampus from the entorhinal cortex), and implanted an array of stimulating electrodes into the PP (Fig. 2A). We then transiently stimulated the PP with brief (200 μs) shocks from one of four stimulating electrodes, and thereafter we recorded intracellularly from triplets of mossy cells (MCs) in the dentate gyrus. As in [Hyde and Strowbridge, 2012], we extracted from these intracellullar recordings the rates of excitatory post-synaptic potentials (EPSPs) received by our three mossy cells. Hilar mossy cells have extensive dendritic arbors, and therefore, they broadly sample the population of upstream granule cells and semilunar granule cells (Fig. 2A) [Larimer and Strowbridge, 2010; Williams et al., 2007]. For our analyses, we considered the three MCs’ EPSP rates over 1-second-long intervals at different times post-stimulation (from 1s up to >20s post-stimulation). Each stimulus was repeated 3–5 times, and the same procedure was performed on 9 different slices.
Mnemonic representations in the presence of noise
To confirm that our preparation does indeed form persistent mnemonic (symbolic) representations, we attempted to decode the mossy cell EPSP rates to identify which of the four stimulating electrodes (labelled “A”, “B”, “C”, and “D”) was used to stimulate the PP. For the decoding, we used the k-nearest neighbors (KNN) algorithm, which is a simplified form of maximum likelihood estimation that works well with even modest amounts of data. The KNN algorithm takes a given response data point, and then identifies the most similar (smallest Euclidean distance) of the other data points. The algorithm then asks which stimulus was responsible for the majority of the nearest-neighboring response points, and uses that as the guess for the stimulus that caused the response under consideration. We then recorded this guess for each response data point, and computed the fraction of those guesses that correctly identified the stimulus that was applied to the PP. All classifications were performed using a procedure in which the classifier was constructed without including the test point. When we repeated our analysis on data with shuffled stimulus labels, the correct classification rate was approximately 25% for estimating which of four different (roughly equiprobable) stimuli was presented — this value is what we use to define “chance” performance.
For tasks with no fixed delay period, it is important that the representation can be readout at any time post-stimulation. We thus repeated our decoding analysis on the responses observed at different post-stimulation epochs. The data points used in building the classifier included those recorded at all epochs. We observed that the responses can identify the applied stimulus at levels well above chance for more than 20s after the stimulus is received (Fig. 2D, blue curve, p<10−6 for all epochs, based on a binomial test, based on EPSP frequencies in 1s-long windows).
The KNN analysis presented herein improves upon the linear discriminant analysis (LDA) we used in [Hyde and Strowbridge, 2012] because it does not assume that the responses to different stimuli will be linearly separable, and because it was applied to the neural responses in shorter time windows, thereby showing that the representation can be read-out relatively quickly, without requiring long integration times.
The sustained nature of the representation is interesting because the responses change dramatically over the post-stimulation period (Fig. 2B; mean trajectories following each different stimulus from one of our 9 slice experiments), and because the level of trial-to-trial variability in the neural responses — which corrupts the stimulus representation — increases significantly following stimulation (Fig. 3E, p<0.05 for comparisons of overall variability at t=1 s and t=12 s post-stimulation, based on a two-sided paired t-test; and Fig. 3B, p<10−6 for comparisons of coefficient of variation at t=1 s and t=12 s post-stimulation, based on a two-sided paired t-test).
Variability is unevenly spread out over the neural response space
We observed (above) that the neural responses evolve dramatically, and show a significant increase in variability during the period following stimulation, but can nevertheless be decoded on a trial-by-trial (and epoch-by-epoch) basis to identify the applied stimulus. Based on theoretical studies about signal vs noise in the nervous system [Hu et al., 2014; Averbeck et al., 2006; da Silveira and Berry, 2013], we postulated that the neural responses are confined to relatively non-overlapping regions of the neural response space, with a different region corresponding to each applied stimulus. In other words, the set of responses to stimulus “A” might lie in a thin cylindrical region around the black curve (mean trajectory) in Fig. 2B, whereas the set of responses to stimulus “C” might lie in another cylindrical region around the green curve in Fig. 2B, and so on. The thinness of these cylinders reduces the extent to which they can overlap. For contrast, if the trial-by-trial responses were relatively spread out away from the mean trajectories — corresponding to “fat” cylinders — this would allow the responses to different stimuli to overlap more, thereby hindering the representation of distinct stimulation patterns (see, for examples, Figs. 1AB).
To investigate this hypothesis, we took each response to stimulus “A” (measured at different epochs post-stimulation), and measured the Euclidean distance from those data points to the nearest point on the mean trajectory that the responses follow after the application of stimulus “A” (Fig. 3A): this effectively measures the radius of the “cylinder” described above. We then repeated this analysis on the responses to the other stimuli, in all cases measuring the distance from each data point to the nearest point on the mean trajectory following the applied stimulus (Fig. 3C). A comparison of Figs. 3BCE shows that, for the first ~10–15s of the neural responses, the overall variability in the responses increases (Figs. 3BE), but that variability is not evenly distributed in all directions in the response space; indeed, the spread of trajectories away from the mean trajectory actually decreases during that period (Fig. 3C, p<10−3 for comparisons of distance from mean trajectory at t=1 s and t=12 s post-stimulation, based on a two-sided paired t-test). This indicates that the increasing variability (Figs. 3BE) is not evenly spread out over the neural response space; the variability is structured such that the neural responses are constrained to lie relatively near the mean stimulus-evoked trajectories.
For comparison with the levels of variability in the neural responses, we also show the distances between each of the mean trajectories, and their nearest-neighboring mean trajectory (Fig. 3D). These distances decrease over time. Accordingly, if the response data points did not converge to the mean trajectories over time, the responses to different stimuli would quickly blend together, destroying the representation. However, the responses do converge towards the mean trajectories (Fig. 3C), and hence the representational accuracy stays relatively high (Fig. 2D).
Having variability constrained to lie near the mean trajectories imparts robustness to the representation
We have seen that, despite significant variability (Fig. 3BE), the neural responses can be decoded epoch-by-epoch to identify the applied stimulation pattern (Fig. 2D). Is the robustness of the representation a result of the fact that the variability is unevenly spread over different parts of the neural response space, such that the neural responses are constrained to lie near the mean stimulus-evoked trajectories (Fig. 3C)?
To address this question, we constructed surrogate data in which the mean trajectories, and the levels of trial-to-trial variability in the neural responses were the same as in our experimental data, but the “constrained” structure of the trial-to-trial variability was disrupted by randomly rotating the constellations of data points obtained at each epoch about the mean response trajectory (Fig. 4AB; see Methods for details). A different random rotation was applied at each epoch.
This procedure yielded surrogate data that have the same mean trajectories and the same overall levels of variability as in the original data (compare Fig. 3E and Fig. 4D, which are identical by construction). However, the variability in the surrogate data is spread out in random directions in the response space, and not necessarily constrained to be near the mean trajectory. As a result, the surrogate data do not show the same decrease over time in the distances from their mean trajectories as do the actual experimental data (Fig. 4C, p < 0.05 at all epochs for comparisons of the distances to the mean trajectories in the surrogate data and the actual data, based on a paired two-sided t-test). Thus, our random rotation procedure maintained the mean response trajectories, and the overall levels of trial-to-trial variability, but successfully disrupted the confined nature of the neural variability.
We could thus compare the stimulus representation formed by the actual, and the surrogate data, to test the impact of the confined response variability on the population code. To do this, we repeated our KNN classification on the surrogate responses, and observed that they were significantly less decodable to identify the location of applied stimulation than were the real neural responses (Fig. 5; blue bars are for the experimentally observed “raw” responses, green bars are for artificially perturbed surrogate responses. Green and blue bars are different, with p<0.01, for the epochs of 1–5s post-stimulation, 6–10s post-stimulation, and 11–15s post-stimulation, based on a binomial test, and green bars are below blue ones for all epochs). This effect was strongest in the first 10 or 15 s post-stimulation, which is the period in which response variability is the largest (Fig. 3E). Our results indicate that the structure of the trial-to-trial variability in the hilar populations, wherein that variability is constrained to lie near the mean stimulus-evoked trajectories, increases the robustness of mnemonic representations by reducing the similarity between responses to different stimuli.
While the stimulus decoding performance obtained with the raw responses, and that obtained with the surrogate responses, are statistically significantly different, that difference is — depending on the epoch — somewhat modest in magnitude. This is because the mean trajectories are fairly well-separated in the space of neural responses. Accordingly, the responses to a particular stimulus (say, “A”) are typically quite far from the paths associated with the other stimuli (“B”, “C”, and “D” for this example) (Fig. 6B). Consequently, an increase in the distances of responses from the paths associated with the applied stimulus, caused by the perturbation to the structure of trial-to-trial variability (as in Fig. 4C) can have a modest impact on stimulus decodability. For contrast, were the mean trajectories closer together, the impact could be much larger. We thus expect that the phenomenon we identified — of variability constrained to lie near the appropriate response “paths” — could have a larger impact on pattern separation in situations where there are more different patterns to store and / or separate. In that situation, the paths associated with different patterns will be — on average — closer together, and so the importance of responses staying close to the correct paths will be magnified.
Discussion
Herein, we report that the trial-to-trial variability of stimulus-specific persistent responses recorded in populations of hilar neurons is constrained to lie near the mean stimulus-evoked trajectories. This structure significantly facilitated decoding of which stimulation pattern was presented, and may thus be an important aspect of the dentate’s ability to separately encode different patterns of applied stimuli [Leutgeb et al., 2007; Myers and Scharfman, 2009; O’Reilly and McClelland, 1994]. Moreover, because neural systems typically display high levels of variability [Faisal et al., 2008; Hu et al., 2014; Averbeck et al., 2006; Romo et al., 2003], and the “confined” structure provides robustness against this “noise”, our results may have implications for other preparations.
In the DG, the consistent average response trajectories we defined likely reflect the triggering of plateau potentials in similar subgroups of semilunar granule cells (SGC’s) on each trial [Larimer and Strowbridge 2010; Williams et al., 2007]. Response variability, in turn, may arise from trial-to-trial differences in the durations of these plateau potentials. This mechanism would force population responses in downstream hilar neurons to follow similar trajectories through a high dimensional neural response space, albeit with different velocities on each trial, depending on when the plateau potentials begin to decay in each presynaptic SGC. Accordingly, though there may be significant trial-to-trial variability in the neural responses, those responses would nonetheless all lie near the same mean trajectory (as we observed herein).
In addition to enhancing the ability to accurately decode population responses at one specific time point (e.g., at the offset of a visual cue in a working memory task, as in [Funahashi et al., 1993]), the response structure we identified in the DG activities enables time-independent decoding of population responses. In particular, the stimulus is persistently encoded in the identity of the path on (or near) which the responses lie. This finding suggests novel time-invariant mechanisms for decoding transient inputs, and predicts that biological systems that decode time invariant memories may take advantage of these structures. It may be possible that downstream neural structures perform “path-based” decoding rather than decoding based on static patterns of activity. Given the importance of persistent representations, formed by dynamical neural activities [Druckmann and Chklovskii, 2012], we suspect that similar representational structures might be quite ubiquitous within the nervous system.
Relatedly, there are many prior reports of neural responses being constrained to low-dimensional regions of the space of possible neural responses [Ganguli et al., 2008; Seung, 1996; Wimmer et al., 2005; Yoon et al., 2013]. The current study adds to this literature the observation that trial-to-trial variability (in addition to mean neural responses) in the dentate is also confined to low-dimensional subspaces. In vivo, these low-dimensional neural-representational structures have the potential to reflect the animal’s memories and / or decision-making, and thus their identification in awake behaving animals could yield major advances in connecting brain function to behavior.
Supplementary Material
Acknowledgments
Funding:
Grant sponsor: NSF; Grant numbers: DMS-1208027 and DMS-1056125
Grant Sponsor: NIH; Grant Number: R01-DC04285
References
- Averbeck BB, Latham P, Pouget A. Neural populations, population coding, and computation. Nat Rev Neurosci. 2006;7:358–366. doi: 10.1038/nrn1888. [DOI] [PubMed] [Google Scholar]
- Britten K, Shadlen M, Newsome W, Movshon J. Responses of neurons in macaque MT to stochastic motion signals. Visual Neurosci. 1993;10:1157–1169. doi: 10.1017/s0952523800010269. [DOI] [PubMed] [Google Scholar]
- Cayco-Gajic NA, Zylberberg J, Shea-Brown E. Triplet correlations among similarly tuned cells impact population coding. Front Comput Neurosci. 2015;9:57. doi: 10.3389/fncom.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckmann S, Chklovskii DB. Neuronal circuits underlying persistent representations despite time varying activity. Curr Biol. 2012;22:2095–2103. doi: 10.1016/j.cub.2012.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisal A, Selen L, Wolpert D. Noise in the nervous system. Nat Rev Neurosci. 2008;9:292–303. doi: 10.1038/nrn2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi S, Chafee MV, Goldman-Rakic PS. Prefrontal neuronal activity in rhesus monkeys performing a delayed anti-saccade task. Nature. 1993;365:753–756. doi: 10.1038/365753a0. [DOI] [PubMed] [Google Scholar]
- Ganguli S, Bisley JW, Roitman JD, Shadlen MN, Goldberg ME, Miller KD. One-dimensional dynamics of attention and decision making in LIP. Neuron. 2008;58:15–25. doi: 10.1016/j.neuron.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zylberberg J, Shea-Brown E. The sign rule and beyond: boundary effects, flexibility, and noise correlations in neural population codes. PLoS Comput Biol. 2014;10:e1003469. doi: 10.1371/journal.pcbi.1003469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde RA, Strowbridge BW. Mnemonic representations of transient stimuli and temporal sequences in the rodent hippocampus in vitro. Nat Neurosci. 2012;15:1430–1438. doi: 10.1038/nn.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer P, Strowbridge BW. Representing information in cell assemblies: persistent activity mediated by semilunar granule cells. Nat Neurosci. 2010;13:213–222. doi: 10.1038/nn.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Moser SB, Moser EI. Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science. 2007;16:961–966. doi: 10.1126/science.1135801. [DOI] [PubMed] [Google Scholar]
- Myers CE, Scharfman HE. A role for hilar cells in pattern separation in the dentate gyrus: a computational approach. Hippocampus. 2009;19:321–327. doi: 10.1002/hipo.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly RC, McClelland JL. Hippocampal conjunctive encoding, storage, and recall: Avoiding a trade-off. Hippocampus. 1994;4:661–682. doi: 10.1002/hipo.450040605. [DOI] [PubMed] [Google Scholar]
- Romo R, Hernandez A, Zainos A, Salinas E. Correlated neuronal discharges that increase coding efficiency during perceptual discrimination. Neuron. 2003;38:649–657. doi: 10.1016/s0896-6273(03)00287-3. [DOI] [PubMed] [Google Scholar]
- Scharfman HE, Schwartzkroin PA. Electrophysiology of morphologically identified mossy cells of the dentate hilus recorded in ginea pig hippocampal slices. J Neurosci. 1988;8:3812–3821. doi: 10.1523/JNEUROSCI.08-10-03812.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung HS. How the brain keeps the eyes still. Proc Natl Acad Sci USA. 1996;93:13339–13344. doi: 10.1073/pnas.93.23.13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir M. Emerging principles of population coding: in search for the neural code. Curr Opin Neurobiol. 2014;25:140–148. doi: 10.1016/j.conb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- da Silveira RA, Berry MJ. High-fidelity coding with correlated neurons. PLoS Comput Biol. 2014;10:e1003970. doi: 10.1371/journal.pcbi.1003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Softky W, Koch C. The highly irregular firing of cortical cells is inconsistent with temporal integration of random EPSP’s. J Neurosci. 1993;13:334–350. doi: 10.1523/JNEUROSCI.13-01-00334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PA, Larimer P, Gao Y, Strowbridge BW. Semilunar granule cells: glutamatergic neurons in the rat dentate gyrus with axon collaterals in the inner molecular layer. J Neurosci. 2007;27:13756–13761. doi: 10.1523/JNEUROSCI.4053-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer K, Nykamp DQ, Constantinidis C, Compte A. Bump attractor dynamics in prefrontal cortex explains behavioral precision in spatial working memory. Nat Neurosci. 2014;17:431–439. doi: 10.1038/nn.3645. [DOI] [PubMed] [Google Scholar]
- Yoon K, Buice MA, Barry C, Hayman R, Burgess N, Fiete IR. Specific evidence of low-dimensional continuous attractor dynamics in grid cells. Nat Neurosci. 2013;16:1077–1084. doi: 10.1038/nn.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.