Abstract
Context:
Variation in genes that cause maturity-onset diabetes of the young (MODY) has been associated with diabetes incidence and glycemic traits.
Objectives:
This study aimed to determine whether genetic variation in MODY genes leads to differential responses to insulin-sensitizing interventions.
Design and Setting:
This was a secondary analysis of a multicenter, randomized clinical trial, the Diabetes Prevention Program (DPP), involving 27 US academic institutions. We genotyped 22 missense and 221 common variants in the MODY-causing genes in the participants in the DPP.
Participants and Interventions:
The study included 2806 genotyped DPP participants randomized to receive intensive lifestyle intervention (n = 935), metformin (n = 927), or placebo (n = 944).
Main Outcome Measures:
Association of MODY genetic variants with diabetes incidence at a median of 3 years and measures of 1-year β-cell function, insulinogenic index, and oral disposition index. Analyses were stratified by treatment group for significant single-nucleotide polymorphism × treatment interaction (Pint < 0.05). Sequence kernel association tests examined the association between an aggregate of rare missense variants and insulinogenic traits.
Results:
After 1 year, the minor allele of rs3212185 (HNF4A) was associated with improved β-cell function in the metformin and lifestyle groups but not the placebo group; the minor allele of rs6719578 (NEUROD1) was associated with an increase in insulin secretion in the metformin group but not in the placebo and lifestyle groups.
Conclusions:
These results provide evidence that genetic variation among MODY genes may influence response to insulin-sensitizing interventions.
Genetic variation in MODY genes was associated with response to diabetes prevention interventions as measured by β-cell function and diabetes incidence.
Maturity-onset diabetes of the young (MODY) is characterized by a nonketotic form of diabetes mellitus transmitted by an autosomal dominant mode of inheritance that is usually diagnosed before the age of 25 years. HNF4A (MODY1), HNF1A (MODY3), PDX1 (MODY4), HNF1B (MODY5), and NEUROD1 (MODY6) encode transcription factors operational in the pancreatic β-cell, whereas GCK (MODY2) encodes the β-cell glycolytic enzyme glucokinase. Mutations in these genes cause β-cell dysfunction, which leads to the development of MODY (1). More recently, additional genes have been identified as rarer causes of MODY 7 through MODY 13, including KLF11 (2), CEL (3), PAX4 (4), INS (5), BLK (6), ABCC8 (7), and KCNJ11 (8). In contrast to MODY, type 2 diabetes is a polygenic disease and an illness of insulin resistance and relative insulinopenia.
Large candidate gene studies have identified common genetic variants in genes responsible for MODY 1 through MODY 6 that are associated with type 2 diabetes and glycemic traits (9–11). Bonnycastle et al. (9), in a cross-sectional study, showed that genetic variation in GCK, NEUROD1, HNF4A, HNF1A, and HNF1B was nominally associated (P < 0.05) with diabetes risk after considering multiple testing. Winckler et al. (10) did not replicate these findings in their cross-sectional analyses but did show a consistent relationship between an HNF1B variant and diabetes risk in two independent cohorts. A prospective observational study revealed an association between HNF1A and HNF4A common genetic variation and diabetes incidence (11). A cross-sectional study showed no association between groups of 121 rare missense variants identified by sequencing seven MODY genes and early-onset diabetes in a population without particular risks for diabetes (12). A prior study found no association between HNF4A and oral disposition index (DIo) (11), and other studies have primarily examined how GCK variants influence insulin secretion (13–16).
Genome-wide association studies (GWASs) have further confirmed a relationship between MODY genes and diabetes risk (17–20). Genetic variants in HNF4A (19, 20), HNF1A (17, 18, 21), and HNF1B (17, 22) have been associated with diabetes risk in populations of European, Hispanic, South Asian, or East Asian descent at genome-wide statistical significance (P < 5 × 10–8). GWASs have also detected associations of common variants in GCK with fasting glucose (23–25) and hemoglobin A1c (24, 26).
These data show that MODY genes may influence the development of type 2 diabetes and modulate glycemic traits. In our comprehensive analysis of candidate genes in the Diabetes Prevention Program (DPP) (27), a clinical trial that evaluated preventive strategies for type 2 diabetes, we found that rs11868513 (HNF1B) was associated with diabetes incidence only in the placebo group, with both metformin and lifestyle interventions showing significant evidence as effect modifiers; and rs11086926 (HNF4A) was associated with diabetes incidence only in the metformin group, with metformin but not the lifestyle intervention showing significant evidence as an effect modifier (27). Here we build upon these findings by exploring the physiological underpinnings of this association further and investigating how common and rare missense genetic variation in MODY genes may influence insulin secretion in response to insulin-sensitizing interventions.
Methods
The DPP enrolled 3548 US participants from multiple ethnicities with high-risk criteria for diabetes development: overweight, elevated fasting glucose, and impaired glucose tolerance. Of these participants with complete clinical data and consent for genetic testing, 2806 were randomized to placebo (n = 944), metformin 850 mg twice daily (n = 927), or lifestyle intervention (n = 935) with a goal weight loss of ≥7% and ≥150 minutes of physical activity per week; a fourth troglitazone treatment arm was terminated early due to concerns for drug-related hepatotoxicity (28). Participants in this arm (n = 585) were included in genotyping and baseline association analyses, but not included in longitudinal association testing due to early termination (28). Ethical approval was obtained by local human research committees, and all participants signed informed consent forms.
We used Sanger sequencing on an ABI 3730 DNA Analyzer to sequence the exons and splice sites of the six MODY-causing genes (HNF4A, GCK, HNF1A, PDX1, HNF1B, and NEUROD1) in 190 DPP participants. Samples were chosen without regard to subsequent diabetes incidence with relatively equal distribution of males and females (44% male, 56% female) and ethnicity. Sequencing coverage across the MODY 1 through MODY 6 genes was completed in a three-step primer redesign process to maximize coverage of all MODY genes. Forty-three amplicons were designed to sequence exons 1 through 10 and the promoter region in HNF4A, exons 1 through 10 in GCK and HNF1A, exons 1 and 2 in PDX1, 9 exons in HNF1B, and exon 2 in NEUROD1 (exon 1 is untranslated). An overlap of amplicons provided additional genotypes that had polymorphism concordance of 99.5% across all amplicons. The average targeted coverage was 95%.
Twenty-two missense single-nucleotide polymorphisms (SNPs) were identified by Sanger sequencing (three with minor allele frequency [MAF] >5% and 19 with MAF <5%) and subsequently genotyped in all DPP participants. DNA was whole-genome amplified with the REPLI-G (Qiagen) kit and purified using Nucleofast (Machery-Nagel). Genotyping was performed by allele-specific primer extension of multiplex amplified products, with detection by matrix-assisted laser desorption ionization–time-of-flight mass spectroscopy on an iPLEX-GOLD Sequenom platform. The genotyping success rate was 99%, and concordance rate between sequence data and Sequenom genotyping was 99.5%. We used publicly available assessment tools, PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2) and SIFT (http://sift.jcvi.org), to predict whether amino acid changes could be detrimental to protein function (29–31).
We used Tagger (32) to select tagging SNPs that would capture (r2 ≥ 0.8) all variations with MAF >1% in European (CEU) and/or African (YRI) HapMap populations in MODY genes. The 221 tag SNPs were genotyped on a custom-designed oligonucleotide pool array with other diabetes-related candidate SNPs. Further details regarding genotyping are described in Jablonski et al. (27).
Glucose (in milligrams per deciliter) and insulin (in units per milliliter) were measured fasting and 30 minutes after a 75-g glucose load at baseline and year 1 (33). We focused on the following two quantitative traits as measurements for insulin secretion: change in the insulinogenic index (InsIndex) and DIo over the first year after randomization. Insulin secretion indices were calculated as follows: InsIndex (U/mL)/(mg/dL) = (30-minute insulin – fasting insulin)/(30-minute glucose – fasting glucose) (34) and DIo (mg/dL)–1 = InsIndex × 1/fasting insulin (35). The DIo is a measure of insulin secretion adjusted for insulin sensitivity. The logged value of InsIndex and DIo was used in the analysis and results were back-transformed. The year 1 change (∆) in InsIndex and log DIo was calculated by subtracting the baseline InsIndex or DIo from the year 1 value. These insulinogenic quantitative measures are negatively associated with diabetes hazard rate in the DPP (35).
Diabetes incidence was determined by a diagnostic fasting or 2-hour glucose after a 75-g oral glucose tolerance test that was confirmed by a second test (fasting plasma glucose ≥126 mg/dL or 2-hour post–oral glucose tolerance test glucose level ≥200 mg/dL) (36).
We examined the 224 SNPs with MAF ≥1% in at least one ethnic group for association with insulin secretion indices and diabetes incidence. We assumed an additive genetic model. All models were adjusted for self-reported ethnicity, age at randomization, sex, and treatment group, with additional adjustment for the respective baseline trait for the association tests examining Δ log InsIndex and Δ log DIo. To examine how the SNP’s effect is influenced by treatment group, the analysis was stratified by treatment group if the treatment group × SNP interaction was significant (P < 0.05). We used an analysis of covariance and proportional hazards model to examine the association between the individual SNPs and insulinogenic traits and diabetes incidence, respectively. For the SNPs that were significantly associated with an insulinogenic trait, we performed an additional analysis using the SNP as a class variable (in two degrees-of-freedom tests), obtaining marginal means of the calculated insulin secretion indices and comparing differences between genotypic groups. Holm procedure was used to adjust P values when testing for differences between treatment groups when the interaction P value was significant for the year 1 change traits.
We examined the association of 224 common SNPs in a prior study (27) with diabetes incidence and reported that rs11868513 (HNF1B) and rs11086926 (HNF4A) were associated with diabetes incidence. To comprehensively examine the association of MODY variants and diabetes and related traits, in this report, we delved deeper into understanding these associations with diabetes incidence, as this was not investigated more closely in the prior study. In this study, we examined diabetes incidence by genotype per treatment group. We compared diabetes incidence for each genotype between the treatment groups using a χ2 test and corrected for multiple testing using stepdown Bonferroni correction.
Three methods were used to examine 19 rare missense SNPs in aggregate with each outcome. First, a genotype risk score (GRS) was calculated by assigning one point per minor allele. In a second examination, we used the combined multivariate and collapsing (CMC) method (37), which coded each participant having at least one allele with an MAF <1% as “present” or no minor alleles “absent.” A third technique used the sequence kernel association test (SKAT), which uses a multiple regression model and allows the variants to have different directions and magnitude of effects (38). SKAT was used only for testing associations with the insulinogenic traits because we were unable to incorporate the time variable into the analysis to assess diabetes incidence.
Additionally, we tested a “damaging” missense variants risk score for association with diabetes incidence composed of p.Val33Ala (C, GCK), p.Pro197His (rs8192556, A, NEUROD1), p.Leu176Ser (C, NEUROD1), p.His314Leu (T, NEUROD1), and p.Ser547Phe (T, HNF1B), which were determined to be probably damaging via bioinformatic analysis.
We chose to highlight SNPs that fulfilled a stringent study-wide significance level of P = 3 × 10–4 as determined by employing a Bonferroni correction for the estimated number of independent tests after taking linkage disequilibrium into account for 224 SNPs (39, 40) based on the HapMap sample’s linkage disequilibrium structure in populations of European, African, and Asian ancestry (CEU, YRI, and CHP/JPT, respectively).
We compiled a list of variants in MODY-related variants from the literature that had been associated with diabetes in prior studies and examined them for association in the DPP with diabetes incidence. Additionally, we reviewed the published literature (88 publications) through December 2014 for prior reports of associations with diabetes and related traits for the missense variants identified in this study.
Results
Twenty-two missense variants were examined in this study, of which eight are unique. Three of the missense SNPs were common (overall MAF >5%), whereas 19 had MAF <5%. p.Val33Ala (GCK), p.Pro197His (rs8192556, NEUROD1), p.Leu176Ser (NEUROD1), p.His314Leu (NEUROD1), and p.Ser547Phe (HNF1B) were determined to be damaging to protein function consistently across two bioinformatics tools, PolyPhen-2 and SIFT (Table 1). The allele frequencies and number of individuals carrying each genotype by ethnicity for the 22 missense SNPs are provided in Table 1 and Supplemental Table 1 (173.4KB, xlsx) . A literature review of the missense SNPs for association with diabetes and related traits is in Supplemental Table 2 (173.4KB, xlsx) .
Table 1.
Bioinformatic Assessment of 22 Missense Variants
| Residue Change and Codon Position | Chromosome, Position (HG19) | MAF | Allele Change (Major to Minor) | rs Number | PolyPhen-2/SIFT |
|---|---|---|---|---|---|
| GCK | |||||
| p.Ala11Thr | 7, 44228522 | 0.0027 | GCC-aCC | rs116093166 | Probably damaging/tolerated |
| p.Thr396Sera | 7, 44185162 | 0.0006 | ACC-AgC | N/A | Benign/damaging |
| p.Glu272Alaa | 7, 44187297 | 0.0003 | GAG-GcG | N/A | Possibly damaging/tolerated |
| p.Val33Alaa,b | 7, 44193010 | 0.0002 | GTG-GcG | N/A | Possibly damaging/damaging |
| HNF4A | |||||
| p.Thr139Ile | 20, 43042364 | 0.0239 | ACT-AtT | rs1800961 | Benign/tolerated |
| NEUROD1 | |||||
| p.Thr45Alab | 2, 182543455 | 0.3237 | GCC-aCC | rs1801262 | Benign/tolerated |
| p.Pro197Hisb | 2, 182542998 | 0.0185 | CCT-CaT | rs8192556 | Possibly damaging/damaging |
| p.Val239Ileb | 2, 182542873 | 0.0003 | GTC-aTC | rs145050582 | Benign/tolerated |
| p.Leu176Sera,b | 2, 182543061 | 0.0002 | TTA-TcA | N/A | Possibly damaging/damaging |
| p.His314Leua,b | 2, 182542647 | 0.0006 | CAC-CtC | N/A | Possibly damaging/damaging |
| HNF1A | |||||
| p.Ser487Asnb | 12, 121435427 | 0.2993 | AGC-AaC | rs2464196 | Benign/tolerated |
| p.Ala98Valb | 12, 121416864 | 0.0239 | GCC-GtC | rs1800574 | Benign/tolerated |
| p.Gly574Serb | 12, 121437382 | 0.0081 | GGC-aGC | rs1169305 | Benign/tolerated |
| p.Arg583Glnb | 12, 121437410 | 0.0009 | CGG-CaG | rs137853242 | Benign/tolerated |
| p.Pro894Serb | 12, 121432124 | 0.0003 | CCA-tCA | rs151256267 | Benign/tolerated |
| p.Ile27Leub | 12, 121416650 | 0.2927 | ATC-cTC | rs1169288 | Benign/tolerated |
| p.Gly554Arg | 12, 121437322 | 0.0005 | GGG-aGG | N/A | Probably damaging/tolerated |
| HNF1B | |||||
| p.Gly370Serb | 17, 36070609 | 0.0009 | GGC-aGC | rs113042313 | Benign/tolerated |
| p.Asn228Lys | 17, 33363607 | 0.0042 | AAC-AAg | N/A | Benign/damaging |
| p.Ser547Phea,b | 17, 36059095 | 0.0002 | TCT-TtT | N/A | Probably damaging/damaging |
| p.Thr436Sera,b | 17, 36064957 | 0.0002 | ACA-tCA | N/A | Benign/tolerated |
| p.His332Glna | 17, 36091635 | 0.0005 | CAC-CAg | N/A | Probably damaging/tolerated |
Major allele denoted by underline. We used the publicly available assessment tools PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2) and SIFT (http://sift.jcvi.org) to predict if amino acid changes could be detrimental to protein function (29–31). PolyPhen-2 predictions were based on the Hum Div testing model. This model was compiled from all damaging alleles with known effects on the molecular function causing human Mendelian diseases, present in the UniProtKB database, together with differences between human proteins and their closely related mammalian homologs, assumed to be nondamaging.
Abbreviation: N/A, not available.
Indicates a unique variant.
SNPs were consistent for both bioinformatics tools.
Individual variants and aggregate scores tested for association with baseline insulin secretion traits are shown in Table 2 and Supplemental Table 3 (173.4KB, xlsx) .
Table 2.
MODY SNPs Associated With Log Baseline InsIndex at P < 3 × 10–4 and Their Association With Log Baseline DIo and Insulin Sensitivity (Log 1/FI)
| Gene | SNP | Alleles (Effect/Other) | Log Baseline InsIndex | Log Baseline DIo | Log Baseline 1/FI | Diabetes Incidence | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P Value | Genotype (n) | Adjusted Means (95% CI) | β (SE) | P Value | β (SE) | P Value | HR (95% CI) | P Value | |||
| NEUROD1 | rs11884960 | A/G (MAF = 2.2%) | 0.084 (0.023) | 3 × 10–4 | GG (3291) | 1.200 (1.149–1.252) | 0.065 (0.034) | 0.06 | –0.065 (0.044) | 0.14 | 0.71 (0.48–1.07) | 0.10 |
| AG (135) | 1.502 (1.332–1.681) | |||||||||||
| AA (9) | 1.501 (0.940–2.177) | |||||||||||
| PDX1 | rs4769581 | T/C (MAF = 39%) | 0.027 (0.007) | 2 × 10–4 | CC (1267) | 1.149 (1.086–1.213) | 0.026 (0.0105) | 0.01 | –0.005 (0.0134) | 0.71 | 1.0 (0.88–1.13) | 1.0 |
| TC (1666) | 1.223 (1.164–1.283) | |||||||||||
| TT (506) | 1.317 (1.230–1.407) | |||||||||||
| HNF1B | rs3110638 | A/G (MAF = 0.83%) | 0.145 (0.038) | 1 × 10–4 | GG (3376) | 1.933 (0.342–4.689) | 0.182 (0.0557) | 0.001 | 0.034 (0.0716) | 0.64 | 0.86 (0.45–1.6) | 0.64 |
| AG (55) | 1.691 (1.426–1.977) | |||||||||||
| AA (1) | 1.207 (1.156–1.259) | |||||||||||
The underlined alleles designate the minor allele. The adjusted means are back-transformed for InsIndex. Supplemental Table 3 (173.4KB, xlsx) shows the association of all the SNPs examined and baseline insulinogenic traits.
Abbreviations: FI, fasting insulin; HR, hazard ratio; β (SE), β estimate (standard error) of the association of each minor allele with the respective trait.
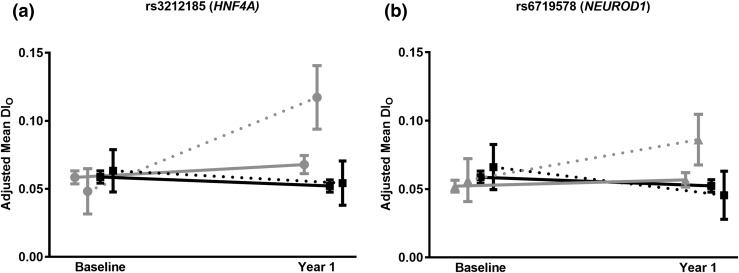
Two SNPs were found to have a significant SNP × treatment interaction and, after stratifying by treatment group, reached the study-wide level of significance for association with Δ log DIo in either the lifestyle or metformin groups. SNP rs3212185 (HNF4A), which was predominant in African-American participants (MAF 0.08), had an SNP × lifestyle vs placebo Pinteraction = 0.0002. The C allele (minor) of rs3212185 was associated with an increase in log DIo in response to the lifestyle intervention over 1 year that was absent in the placebo group and attenuated in the metformin group. A consistent relationship was noted for association with Δ log InsIndex, though at a weaker level of statistical significance (Table 3). Heterozygotes at rs3212185 had an increase in mean DIo from baseline (0.0481, 95% confidence interval [CI]: 0.031 to 0.065) to 1 year (0.117, 95% CI: 0.094 to 0.141) in the lifestyle group, compared with heterozygotes in the placebo group and TT homozygotes in the placebo and lifestyle groups, where DIo remained unchanged [Fig. 1(a)].
Table 3.
Genetic Variation in HNF4A and NEUROD1 Influences the Improvement in Insulin Secretion in One of the Treatment Groups
| Gene | SNP | Trait | Alleles |
SNP × Treatment Interaction |
Placebo |
Metformin |
Lifestyle |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| (Effect/Other) | P Value | β (SE) | P Value | β (SE) | P Value | β (SE) | P value | |||
| HNF4A | ||||||||||
| Significant interaction for SNP × lifestyle (vs placebo) | ||||||||||
| rs3212185 | Δ log InsIndex | C/T (MAF = 1.7%) | 0.01 | –0.005 (0.200) | 0.98 | 0.493 (0.167) | 0.003 | 0.396 (0.173) | 0.02 | |
| Δ log DIo | 0.0002 | –0.001 (0.008) | 0.88 | 0.017 (0.007) | 0.03 | 0.043 (0.011) | 2 × 10–4 | |||
| NEUROD1 | ||||||||||
| Significant interaction for SNP × metformin (vs placebo) | ||||||||||
| rs6719578 | Δ log InsIndex | C/G (MAF = 1.2%) | 0.03 | 0.028 (0.233) | 0.90 | 0.525 (0.229) | 0.02 | –0.092 (0.210) | 0.66 | |
| Δlog DIo | 0.002 | –0.003 (0.010) | 0.79 | 0.042 (0.010) | 1 × 10–4 | 0.005 (0.014) | 0.72 | |||
The underlined allele is the minor allele.
Abbreviation: β (SE), β estimate (standard error) of the association of each minor allele with the respective trait.
Figure 1.
SNPs in HNF4A and NEUROD1 were associated with a significant SNP × treatment interaction and insulin secretion as measured by DIo. This figure displays the adjusted means at baseline and year 1 of these SNPs comparing the two groups that demonstrated the interaction. The minor homozygotes are not shown because, due to low frequency, they were not present in all of the treatment groups. (a) rs3212185 × lifestyle (vs placebo) interaction, P value = 0.0002. TC heterozygotes (gray circle and dotted line) at SNP rs3212185 showed improved DIo after 1 year of lifestyle intervention, whereas the TT (gray circle and solid line) homozygotes had no response to lifestyle intervention. The TT and TC genotypes in the placebo group (black square) did not change in DIo. (b) rs6719578 × metformin (vs placebo) interaction, P value = 0.002. GC heterozygotes (gray triangle and dotted line) at SNP rs6719578 had improved DIo after 1 year of metformin intervention, whereas the GG (gray triangle and solid line) homozygotes did not respond to metformin intervention. Participants with the GG and GC (black square) genotypes in the placebo group did not change DIo.
SNP rs6719578 (NEUROD1) had an SNP × metformin vs placebo Pinteraction = 0.002. The C allele (minor) of rs6719578 was associated with a positive Δ log DIo among the metformin-treated participants during the first year that was not seen in the placebo or lifestyle groups. The C allele was similarly associated with Δ log InsIndex among the metformin group at a lower level of statistical significance (P = 0.02). Heterozygotes had an increase in mean DIo from baseline (0.057, 95% CI: 0.041 to 0.072) to 1 year (0.086, 95% CI: 0.07 to 0.10) in the metformin group, whereas DIo in heterozygotes in the placebo group and in GG homozygotes in the placebo and metformin groups remained largely flat, if not trending downward [Fig. 1(b)].
All the individual variants and aggregate scores tested for association with Δ log InsIndex and Δ log DIo are shown in Supplemental Tables 4 and 5 (173.4KB, xlsx) .
None of the common missense variants nor the GRS and CMC scores of the 19 rare missense variants were associated with change in the insulinogenic traits over the first year. The SKAT analysis revealed a nominally significant association between the aggregate of the 19 uncommon missense variants and change in InsIndex (P = 0.03) but no change in DIo (P = 0.6) (Supplemental Table 6 (173.4KB, xlsx) ).
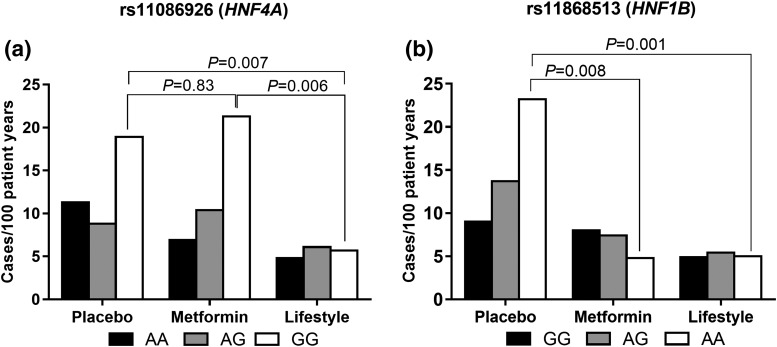
We have shown previously that SNPs rs11086926 (HNF4A) and rs11868513 (HNF1B) were associated with diabetes incidence in one of the treatment groups (27), with significant genotype × treatment interactions. To investigate this further, we examined diabetes incidence by genotype within each treatment group (Fig. 2). GG homozygotes at rs11086926 in the lifestyle group were less likely to develop diabetes compared with persons with the same genotype in the placebo (P = 0.02) and metformin (P = 0.02) groups, with no difference between the metformin and placebo groups [Fig. 2(a)]. AA homozygotes at rs11868513 had the highest diabetes incidence in the placebo group but a dramatic response to metformin (P = 0.008) and lifestyle (P = 0.002) therapy. AG heterozygotes had lower diabetes incidence rates than AA homozygotes, but their rates were also lowered both by lifestyle (P < 0.001) and metformin (P = 0.002). Diabetes incidence rates were lowered in GG homozygotes by the lifestyle intervention (P < 0.001) but not significantly by metformin (P = 0.2) compared with placebo [Fig. 2(b)].
Figure 2.
Diabetes diagnoses per 100 patient-years for (a) rs11086926 and (b) rs11868513 for each genotype group within each treatment arm. (a) The GG homozygotes of rs11086926 had a differential response to lifestyle and metformin (rs11086926 × metformin vs placebo, Pinteraction = 0.002). The GG carriers in the lifestyle group had a significantly lower diabetes incidence than did carriers of the same genotype in the placebo (P = 0.007) and metformin (P = 0.006) groups. These GG homozygotes had no response to metformin therapy compared with placebo (P = 0.8). (b) There was a significant treatment × genotype interaction for rs1186513. rs11868513 × metformin vs placeb, Pinteraction = 0.0007, and rs11868513 × lifestyle vs placebo Pinteraction = 0.03. The AA genotype of rs11868513, despite having a higher diabetes risk in the placebo group, had a dramatic response to metformin (P = 0.008) and lifestyle (P = 0.001) therapy. The AG genotype also demonstrated a decrease to both lifestyle (P < 0.001) and metformin (P = 0.002) therapy compared with placebo. The GG genotype carriers showed a decrease in diabetes incidence to lifestyle intervention (P < 0.001) and no response to metformin (P = 0.2) compared with placebo.
Neither the missense variant GRS nor CMC burden scores were associated with diabetes incidence. Association results for diabetes incidence in the DPP for variants in MODY genes previously associated with type 2 diabetes are shown in Table 4.
Table 4.
Replication of Previously Reported Type 2 Diabetes–Associated MODY Genetic Variants for Association With Diabetes Incidence in the DPP
| SNP | Effect/Other | Gene | ORa (95% CI) | Proxy (Effect/Other)b | r2 (CEU) | Diabetes HR (95% CI) | P Value |
|---|---|---|---|---|---|---|---|
| rs2244164 (9) | C/T | GCK | 0.81 (0.72–0.92) | rs2080033 (C/T) | 1 | 1.05 (0.93–1.18) | 0.42 |
| rs1169288 (11) | T/G | HNF1A | 1.20 (1.10–1.3)a | 0.99 (0.87–1.12) | 0.89 | ||
| rs2701175 (9) | C/A | HNF1A | 1.34 (1.06–1.68) | rs1169288 (G/T) | 0.6 | 1.01 (0.89–1.15) | 0.89 |
| rs2071190 (9) | A/T | HNF1A | 2.08 (1.30–3.31) | 1.10 (0.96–1.27) | 0.15 | ||
| rs7305618a (18) | C/T | HNF1A | 1.14 (1.09–1.20) | 0.98 (0.86–1.12) | 0.80 | ||
| rs7957197a (17) | T/A | HNF1A | 1.07 (1.05–1.10) | rs7305618 (G/A) | 0.57 | 0.98 (0.86–1.12) | 0.80 |
| rs11263755 (10) | A/G | HNF1B | 1.13 (1.04–1.23) | 0.92 (0.79–1.05) | 0.22 | ||
| rs2285741 (10) | A/G | HNF1B | 0.94 (0.88–1.00) | 0.89 (0.79–1.01) | 0.06 | ||
| rs2689 (10) | A/T | HNF1B | 1.09 (1.02–1.17) | 1.08 (0.96–1.22) | 0.19 | ||
| rs3110641 (10) | C/T | HNF1B | 1.10 (1.04–1.17) | 0.96 (0.85–1.1) | 0.57 | ||
| rs3094513 (10) | T/C | HNF1B | 1.08 (1.01–1.16) | rs3110640 (C/T) | 0.93 | 1.09 (0.97–1.23) | 0.14 |
| rs757210 (10) | A/G | HNF1B | 1.12 (1.07–1.18) | 1.13 (1.01–1.27) | 0.04 | ||
| rs12450628 (9) | T/C | HNF1B | 1.63 (1.20–2.23) | rs2411153 (C/G) | 0.68 | 0.89 (0.80–1.00) | 0.06 |
| rs1008284 (9) | A/G | HNF1B | 0.53 (0.37–0.75) | 1.07 (0.94–1.22) | 0.32 | ||
| rs4430796a (17) | G/A | HNF1B | 1.14 (1.08–1.20) | 1.08 (0.96–1.21) | 0.17 | ||
| rs4810424 (11) | C /G | HNF4A | 1.30 (1.00–1.60)c | rs1884614 (A/G) | 0.95 | 1.04 (0.90–1.20) | 0.58 |
| rs3212198 (11) | C/T | HNF4A | 1.10 (1.00–1.20)c | 0.96 (0.86–1.08) | 0.54 | ||
| rs6103716 (9) | C/A | HNF4A | 1.26 (1.11–1.44) | 1.07 (0.95–1.21) | 0.25 | ||
| rs6017317a (20) | G/T | HNF4A | 1.09 (1.07–1.12) | rs1884614 (A/G) | 0.7 | 1.04 (0.90–1.20) | 0.58 |
| rs4812829a (19) | A/G | HNF4A | 1.09 (1.06–1.12) | rs1884614 (A/G) | 1 | 1.04 (0.90–1.20) | 0.58 |
| rs3916026 (9) | C/G | NEUROD1 | 0.73 (0.61–0.87) | 1.00 (0.88–1.12) | 0.94 | ||
| rs2297316d (9) | A/G | PDX1 | 0.77 (0.64–0.92) | — | — |
The article origin for each SNP is in parentheses to the right of the SNP.
Abbreviations: HR, hazard ratio; OR, odds ratio.
SNPs found to be associated with type 2 diabetes in GWASs.
Proxy alleles are consistent with the major or minor effect allele in the original study.
The effect represents a hazard ratio.
rs2297316 did not have an adequate proxy (r2 > 0.5) genotyped in the DPP.
Discussion
We have examined genetic variation among six canonical MODY genes for association with insulinogenic traits and diabetes incidence in response to diabetes preventive interventions. Our key findings show that genetic variation within HNF4A, HNF1B, and NEUROD1 is associated with a differential response to lifestyle and/or metformin interventions in insulinogenic traits and diabetes development. Our study supports prior findings demonstrating that variation in genes where pathogenic mutations are known to cause MODY contribute to the risk of diabetes and variation in insulinogenic traits (9–11, 17–20, 22). We furthermore demonstrated how the influence of variation in MODY genes is modified by insulin-sensitizing interventions.
Among common variants in MODY genes previously associated with type 2 diabetes from candidate gene studies and GWASs (listed in Table 4), we replicated rs757210 in HNF1B for association with diabetes incidence (10). We reported an association between rs11868513 in HNF1B and diabetes incidence among the placebo group participants, but rs11868513 and rs757210 are not in linkage disequilibrium (r2 < 0.009) and appear to be exerting independent effects on diabetes incidence.
In a comprehensive literature review of the missense variants identified among the DPP participants (summarized in Supplemental Table 2 (173.4KB, xlsx) ), we illustrate how these variants have been associated with diabetes and related traits in prior literature. Three low-frequency variants [p.Thr45Ala (NEUROD1), p.Ser487Asn (HNF1A), and p.Ile27Leu (HNF1A)] have been studied most comprehensively previously. p.Thr45Ala (rs1801262) has been associated with β-cell dysfunction in both type 1 and type 2 diabetes (41, 42) and a faster deterioration in β-cell function (43) in Asians. The DPP may be underpowered to detect this association in participants with Asian ancestry. We confirmed findings for association of Ser487Asn (rs2464196) with baseline DIo (P = 0.02) (11, 44), although this finding was not seen in other reports where a variety of tools were used to measure β-cell function (45–47). Last, we confirmed prior findings of the association of Ile27Leu (rs1169288) with β-cell function (InsIndex, P = 0.006) (11, 44, 48).
In contrast to prior case-control, cross-sectional, or observational prospective studies, the DPP randomized design allowed us to characterize how these genetic variants might influence response to diabetes preventive interventions. Because DPP participants were phenotyped at regular intervals for glycemia-related traits during the study intervention, we were able to assess β-cell function during the first year in response to treatment. This is of particular relevance to MODY genes, as the primary defect in the monogenic disease is impaired insulin secretion (1). Two SNPs (rs3212185 [HNF4A] and rs671978 [NEUROD1]) showed significant interactions with the lifestyle or metformin interventions, respectively, and reached study-wide statistical significance for association with change in insulin secretion after stratification in one of the treatment groups. At both SNPs, heterozygote minor allele carriers showed an improvement in insulin secretion as measured by DIo during the first year compared with the major allele homozygotes, an effect that was not seen in the placebo arm.
SNP rs3212185 (HNF4A) and rs671978 (NEUROD1) are in genes that are crucial to β-cell function. HNF4A and NEUROD1 are transcription factors that are involved in endocrine pancreatic development and regulate insulin gene transcription (1). Insulin sensitivity is known to modulate insulin secretion. In the DPP, the lifestyle intervention improved insulin sensitivity and enhanced β-cell function (35). The metformin intervention had a similar attenuated result. We have shown that genetic variation in HNF4A and NEUROD1 modifies the response to interventions designed to improve insulin resistance [metformin and lifestyle (diet, exercise, and weight loss)]. The major homozygotes at rs3212185 and rs671978 appear resistant to improvement in β-cell function by lifestyle and metformin therapy, respectively, whereas minor allele carriers have enhanced β-cell function (1). These findings underscore that interventions that improve insulin resistance may contribute to improvement in β-cell function among certain genotype carriers in genes known to impair insulin secretion. It is notable that rs3212185 is associated with glycemic traits among predominantly African ancestry carriers, where HNF4A variation has not been studied in detail despite the higher type 2 diabetes risk in this population (49).
SNP rs11086926 showed a significant interaction with metformin on diabetes incidence (27). Specifically, minor allele homozygotes at rs11086926 appear to be resistant to metformin therapy but were responsive to lifestyle intervention. Perhaps the minor allele carriers are resistant to metformin’s action in decreasing insulin resistance at the liver, where HNF4A is also highly expressed (49). This variant has been shown to be nominally associated with type 2 diabetes in GWASs of Hispanic ancestry (50).
SNP rs11868513 showed a significant interaction with metformin and lifestyle interventions (27). The metformin and lifestyle interventions ablated the increased diabetes risk seen among the A allele carriers in the placebo group. Furthermore, AA carriers had a substantial decrease in diabetes incidence with metformin therapy and lifestyle intervention. These results illustrate that metformin and lifestyle interventions reduce the risk of diabetes of AA carriers to the levels of GG and AG carriers.
We used three methods to investigate the contribution of an aggregate of 19 rare missense variants on diabetes incidence and insulinogenic traits. Similar to the study by Flannick et al. (12) in an unselected population, MODY missense variants did not appear to influence diabetes incidence in our high-risk population. Using SKAT, a method that does not assume that rare variants influence the phenotype in the same direction, we found a nominally significant association between the aggregate of 19 missense variant’s baseline InsIndex and DIo and the Δ InsIndex over the first year. Because SKAT provides a P value but no effect estimate, we would need to test each variant individually to determine which SNP(s) may be driving the association and the direction of effect. We are not powered to test these rare variants individually in the DPP but, using publically available databases for type 2 diabetes, we noted that p.Val33Ala (GCK), p.Thr139Ile (HNF4A), p.Ala98Val (HNF1A), and p.Ile27Leu (HNF1A) have been associated with type 2 diabetes at nominal to locus-wide levels of significance (50), which supports the suggested association with the aggregate of missense variants and insulinogenic traits. Nonetheless, large cohorts genotyped for the missense variants examined here will have the power to further explore the individual contribution of rare missense variants in the MODY genes on diabetes incidence and insulinogenic traits.
Some limitations of our study should be emphasized. First, as our sequencing efforts started prior to the introduction of next-generation sequencing techniques, we only sequenced six MODY genes in 190 participants and so did not comprehensively catalog all rare variation in our sample of 3442 individuals. Second, because sample sizes in clinical trials are finite, power to detect associations with rare variants is limited. Third, we have not examined newly discovered MODY genes. Fourth, we have not been able to replicate our results, as suitable venues to do so are not available for this unique clinical trial; thus, though we have tried to be circumspect in the selection of statistical thresholds, the findings reported here should be considered hypothesis generating. Some of these limitations will be overcome with the deployment of an exome array in the DPP, which will allow for a more thorough evaluation of low-frequency variants across all MODY genes.
In summary, select MODY gene variants annotated to HNF4A, HNF1B, and NEUROD1 are associated with response to insulin-sensitizing interventions on either diabetes incidence and/or insulinogenic traits. These results provide evidence that genetic variation among MODY genes influences response to insulin-sensitizing interventions. These data underscore the need for further genotype-directed studies to determine whether carriers of the aforementioned gene variants respond differently to insulin-sensitizing and, moreover, insulin secretagogue therapy.
Acknowledgments
We thank the commitment and dedication of the participants of the DPP and the investigators in the DPP Research Group. A complete list of centers, investigators, and staff can be found in the Supplemental Appendix (28KB, docx) .
Acknowledgments
This work was supported by a subcontract from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (NIH/NIDDK) Grant U01 DK048489 at the DPP Data Coordinating Center in George Washington University (to J.C.F.); National Research Service Award Institutional Training Grant T32 DK007028-35 to Massachusetts General Hospital (to L.K.B.); the Endocrine Society’s Lilly Scholar’s Award; an NIH Loan Repayment Award, NIDDK 1 L30 DK089944-01; a Distinguished Clinical Scientist Award from the Doris Duke Charitable Foundation (to D.A.); and a NorthShore University HealthSystem Auxiliary Research Scholar Award.
Acknowledgments
Author contributions: L.K.B. formulated the analysis plan, cleaned sequencing/genotyping data, interpreted results, and wrote the manuscript under the guidance of J.C.F. K.A.J., and L.T. formulated the analysis plan, carried out the analyses, and wrote and edited the manuscript. A.S.W. and Y.-C.C. assembled manuscript tables and reviewed the manuscript. J.B.M. performed the sequencing and genotyping of the samples and reviewed the manuscript. A.R.S., D.A.E., A.K.M., D.D., P.W.F., S.E.K., T.I.P., W.C.K., and D.A. reviewed the analysis plan, contributed to discussion, and reviewed and/or edited the manuscript. J.C.F. is the guarantor of this manuscript.
Disclosure Summary: L.K.B. is a principal investigator on clinical trials, a paid advisory board member, and speaker for Novo Nordisk. P.W.F. is a paid advisory board member for Sanofi Aventis and Eli Lilly. J.C.F. received consulting honoraria from Merck and Boehringer-Ingelheim. The remaining authors have nothing to disclose
Footnotes
- CI
- confidence interval
- CMC
- combined multivariate and collapsing
- DIo
- oral disposition index
- DPP
- Diabetes Prevention Program
- GRS
- genotype risk score
- GWAS
- genome-wide association study
- InsIndex
- insulinogenic index
- MAF
- minor allele frequency
- MODY
- maturity-onset diabetes of the young
- SKAT
- sequence kernel association test
- SNP
- single-nucleotide polymorphism.
References
- 1.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345(13):971–980. [DOI] [PubMed] [Google Scholar]
- 2.Neve B, Fernandez-Zapico ME, Ashkenazi-Katalan V, Dina C, Hamid YH, Joly E, Vaillant E, Benmezroua Y, Durand E, Bakaher N, Delannoy V, Vaxillaire M, Cook T, Dallinga-Thie GM, Jansen H, Charles MA, Clément K, Galan P, Hercberg S, Helbecque N, Charpentier G, Prentki M, Hansen T, Pedersen O, Urrutia R, Melloul D, Froguel P. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic β cell function. Proc Natl Acad Sci USA. 2005;102(13):4807–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torsvik J, Johansson S, Johansen A, Ek J, Minton J, Raeder H, Ellard S, Hattersley A, Pedersen O, Hansen T, Molven A, Njølstad PR. Mutations in the VNTR of the carboxyl-ester lipase gene (CEL) are a rare cause of monogenic diabetes. Hum Genet. 2010;127(1):55–64. [DOI] [PubMed] [Google Scholar]
- 4.Plengvidhya N, Kooptiwut S, Songtawee N, Doi A, Furuta H, Nishi M, Nanjo K, Tantibhedhyangkul W, Boonyasrisawat W, Yenchitsomanus PT, Doria A, Banchuin N. PAX4 mutations in Thais with maturity onset diabetes of the young. J Clin Endocrinol Metab. 2007;92(7):2821–2826. [DOI] [PubMed] [Google Scholar]
- 5.Edghill EL, Flanagan SE, Patch AM, Boustred C, Parrish A, Shields B, Shepherd MH, Hussain K, Kapoor RR, Malecki M, MacDonald MJ, Støy J, Steiner DF, Philipson LH, Bell GI, Hattersley AT, Ellard S; Neonatal Diabetes International Collaborative Group . Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes. 2008;57(4):1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SH, Ma X, Weremowicz S, Ercolino T, Powers C, Mlynarski W, Bashan KA, Warram JH, Mychaleckyj J, Rich SS, Krolewski AS, Doria A. Identification of a locus for maturity-onset diabetes of the young on chromosome 8p23. Diabetes. 2004;53(5):1375–1384. [DOI] [PubMed] [Google Scholar]
- 7.Bowman P, Flanagan SE, Edghill EL, Damhuis A, Shepherd MH, Paisey R, Hattersley AT, Ellard S. Heterozygous ABCC8 mutations are a cause of MODY. Diabetologia. 2012;55(1):123–127. [DOI] [PubMed] [Google Scholar]
- 8.Bonnefond A, Philippe J, Durand E, Dechaume A, Huyvaert M, Montagne L, Marre M, Balkau B, Fajardy I, Vambergue A, Vatin V, Delplanque J, Le Guilcher D, De Graeve F, Lecoeur C, Sand O, Vaxillaire M, Froguel P. Whole-exome sequencing and high throughput genotyping identified KCNJ11 as the thirteenth MODY gene. PLoS One. 2012;7(6):e37423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnycastle LL, Willer CJ, Conneely KN, Jackson AU, Burrill CP, Watanabe RM, Chines PS, Narisu N, Scott LJ, Enloe ST, Swift AJ, Duren WL, Stringham HM, Erdos MR, Riebow NL, Buchanan TA, Valle TT, Tuomilehto J, Bergman RN, Mohlke KL, Boehnke M, Collins FS. Common variants in maturity-onset diabetes of the young genes contribute to risk of type 2 diabetes in Finns. Diabetes. 2006;55(9):2534–2540. [DOI] [PubMed] [Google Scholar]
- 10.Winckler W, Weedon MN, Graham RR, McCarroll SA, Purcell S, Almgren P, Tuomi T, Gaudet D, Boström KB, Walker M, Hitman G, Hattersley AT, McCarthy MI, Ardlie KG, Hirschhorn JN, Daly MJ, Frayling TM, Groop L, Altshuler D. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56(3):685–693. [DOI] [PubMed] [Google Scholar]
- 11.Holmkvist J, Almgren P, Lyssenko V, Lindgren CM, Eriksson KF, Isomaa B, Tuomi T, Nilsson P, Groop L. Common variants in maturity-onset diabetes of the young genes and future risk of type 2 diabetes. Diabetes. 2008;57(6):1738–1744. [DOI] [PubMed] [Google Scholar]
- 12.Flannick J, Beer NL, Bick AG, Agarwala V, Molnes J, Gupta N, Burtt NP, Florez JC, Meigs JB, Taylor H, Lyssenko V, Irgens H, Fox E, Burslem F, Johansson S, Brosnan MJ, Trimmer JK, Newton-Cheh C, Tuomi T, Molven A, Wilson JG, O’Donnell CJ, Kathiresan S, Hirschhorn JN, Njølstad PR, Rolph T, Seidman JG, Gabriel S, Cox DR, Seidman CE, Groop L, Altshuler D. Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat Genet. 2013;45(11):1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rissanen J, Saarinen L, Heikkinen S, Kekäläinen P, Mykkänen L, Kuusisto J, Deeb SS, Laakso M. Glucokinase gene islet promoter region variant (G-->A) at nucleotide -30 is not associated with reduced insulin secretion in Finns. Diabetes Care. 1998;21(7):1194–1197. [DOI] [PubMed] [Google Scholar]
- 14.Zaidi FK, Wareham NJ, McCarthy MI, Holdstock J, Kalloo-Hosein H, Krook A, Swinn RA, O’Rahilly S. Homozygosity for a common polymorphism in the islet-specific promoter of the glucokinase gene is associated with a reduced early insulin response to oral glucose in pregnant women. Diabet Med. 1997;14(3):228–234. [DOI] [PubMed] [Google Scholar]
- 15.Stone LM, Kahn SE, Fujimoto WY, Deeb SS, Porte D Jr. A variation at position -30 of the β-cell glucokinase gene promoter is associated with reduced β-cell function in middle-aged Japanese-American men. Diabetes. 1996;45(4):422–428. [DOI] [PubMed] [Google Scholar]
- 16.Elbein SC, Sun J, Scroggin E, Teng K, Hasstedt SJ. Role of common sequence variants in insulin secretion in familial type 2 diabetic kindreds: the sulfonylurea receptor, glucokinase, and hepatocyte nuclear factor 1α genes. Diabetes Care. 2001;24(3):472–478. [DOI] [PubMed] [Google Scholar]
- 17.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Langenberg C, Hofmann OM, Dupuis J, Qi L, Segrè AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bengtsson Boström K, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Couper DJ, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jørgensen T, Kao WH, Klopp N, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Perry JR, Petersen AK, Platou C, Proença C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparsø T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet-Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Mohlke KL, Morris AD, Palmer CN, Pramstaller PP, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Wareham NJ, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Hu FB, Meigs JB, Pankow JS, Pedersen O, Wichmann HE, Barroso I, Florez JC, Frayling TM, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI; MAGIC investigators; GIANT Consortium . Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra EJ, Below JE, Krithika S, Valladares A, Barta JL, Cox NJ, Hanis CL, Wacher N, Garcia-Mena J, Hu P, Shriver MD, Kumate J, McKeigue PM, Escobedo J, Cruz M, Escobedo J, Cruz M; Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Genome-wide association study of type 2 diabetes in a sample from Mexico City and a meta-analysis of a Mexican-American sample from Starr County, Texas. Diabetologia. 2011;54(8):2038–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P, Been LF, Chia KS, Dimas AS, Hassanali N, Jafar T, Jowett JB, Li X, Radha V, Rees SD, Takeuchi F, Young R, Aung T, Basit A, Chidambaram M, Das D, Grundberg E, Hedman AK, Hydrie ZI, Islam M, Khor CC, Kowlessur S, Kristensen MM, Liju S, Lim WY, Matthews DR, Liu J, Morris AP, Nica AC, Pinidiyapathirage JM, Prokopenko I, Rasheed A, Samuel M, Shah N, Shera AS, Small KS, Suo C, Wickremasinghe AR, Wong TY, Yang M, Zhang F, Abecasis GR, Barnett AH, Caulfield M, Deloukas P, Frayling TM, Froguel P, Kato N, Katulanda P, Kelly MA, Liang J, Mohan V, Sanghera DK, Scott J, Seielstad M, Zimmet PZ, Elliott P, Teo YY, McCarthy MI, Danesh J, Tai ES, Chambers JC; DIAGRAM Consortium; MuTHER Consortium . Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43(10):984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim X, Takeuchi F, Wu Y, Go MJ, Yamauchi T, Chang YC, Kwak SH, Ma RC, Yamamoto K, Adair LS, Aung T, Cai Q, Chang LC, Chen YT, Gao Y, Hu FB, Kim HL, Kim S, Kim YJ, Lee JJ, Lee NR, Li Y, Liu JJ, Lu W, Nakamura J, Nakashima E, Ng DP, Tay WT, Tsai FJ, Wong TY, Yokota M, Zheng W, Zhang R, Wang C, So WY, Ohnaka K, Ikegami H, Hara K, Cho YM, Cho NH, Chang TJ, Bao Y, Hedman AK, Morris AP, McCarthy MI, Takayanagi R, Park KS, Jia W, Chuang LM, Chan JC, Maeda S, Kadowaki T, Lee JY, Wu JY, Teo YY, Tai ES, Shu XO, Mohlke KL, Kato N, Han BG, Seielstad M, Han BG, Seielstad M; DIAGRAM Consortium; MuTHER Consortium . Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2011;44(1):67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estrada K, Aukrust I, Bjørkhaug L, Burtt NP, Mercader JM, García-Ortiz H, Huerta-Chagoya A, Moreno-Macías H, Walford G, Flannick J, Williams AL, Gómez-Vázquez MJ, Fernandez-Lopez JC, Martínez-Hernández A, Jiménez-Morales S, Centeno-Cruz F, Mendoza-Caamal E, Revilla-Monsalve C, Islas-Andrade S, Córdova EJ, Soberón X, González-Villalpando ME, Henderson E, Wilkens LR, Le Marchand L, Arellano-Campos O, Ordóñez-Sánchez ML, Rodríguez-Torres M, Rodríguez-Guillén R, Riba L, Najmi LA, Jacobs SB, Fennell T, Gabriel S, Fontanillas P, Hanis CL, Lehman DM, Jenkinson CP, Abboud HE, Bell GI, Cortes ML, Boehnke M, González-Villalpando C, Orozco L, Haiman CA, Tusié-Luna T, Aguilar-Salinas CA, Altshuler D, Njølstad PR, Florez JC, MacArthur DG; SIGMA Type 2 Diabetes Consortium . Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311(22):2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39(8):977–983. [DOI] [PubMed] [Google Scholar]
- 23.Manning AK, LaValley M, Liu CT, Rice K, An P, Liu Y, Miljkovic I, Rasmussen-Torvik L, Harris TB, Province MA, Borecki IB, Florez JC, Meigs JB, Cupples LA, Dupuis J. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP × environment regression coefficients. Genet Epidemiol. 2011;35(1):11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, Potter SC, Erdos MR, Sanna S, Hottenga JJ, Wheeler E, Kaakinen M, Lyssenko V, Chen WM, Ahmadi K, Beckmann JS, Bergman RN, Bochud M, Bonnycastle LL, Buchanan TA, Cao A, Cervino A, Coin L, Collins FS, Crisponi L, de Geus EJ, Dehghan A, Deloukas P, Doney AS, Elliott P, Freimer N, Gateva V, Herder C, Hofman A, Hughes TE, Hunt S, Illig T, Inouye M, Isomaa B, Johnson T, Kong A, Krestyaninova M, Kuusisto J, Laakso M, Lim N, Lindblad U, Lindgren CM, McCann OT, Mohlke KL, Morris AD, Naitza S, Orrù M, Palmer CN, Pouta A, Randall J, Rathmann W, Saramies J, Scheet P, Scott LJ, Scuteri A, Sharp S, Sijbrands E, Smit JH, Song K, Steinthorsdottir V, Stringham HM, Tuomi T, Tuomilehto J, Uitterlinden AG, Voight BF, Waterworth D, Wichmann HE, Willemsen G, Witteman JC, Yuan X, Zhao JH, Zeggini E, Schlessinger D, Sandhu M, Boomsma DI, Uda M, Spector TD, Penninx BW, Altshuler D, Vollenweider P, Jarvelin MR, Lakatta E, Waeber G, Fox CS, Peltonen L, Groop LC, Mooser V, Cupples LA, Thorsteinsdottir U, Boehnke M, Barroso I, Van Duijn C, Dupuis J, Watanabe RM, Stefansson K, McCarthy MI, Wareham NJ, Meigs JB, Abecasis GR. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41(1):77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparsø T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proença C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tönjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paré G, Chasman DI, Parker AN, Nathan DM, Miletich JP, Zee RY, Ridker PM. Novel association of HK1 with glycated hemoglobin in a non-diabetic population: a genome-wide evaluation of 14,618 participants in the Women’s Genome Health Study. PLoS Genet. 2008;4(12):e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jablonski KA, McAteer JB, de Bakker PI, Franks PW, Pollin TI, Hanson RL, Saxena R, Fowler S, Shuldiner AR, Knowler WC, Altshuler D, Florez JC; Diabetes Prevention Program Research Group . Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59(10):2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM; Diabetes Prevention Program Research Group . Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346(6):393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunham LR, Singaraja RR, Pape TD, Kejariwal A, Thomas PD, Hayden MR. Accurate prediction of the functional significance of single nucleotide polymorphisms and mutations in the ABCA1 gene. PLoS Genet. 2005;1(6):e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–1223. [DOI] [PubMed] [Google Scholar]
- 33.The Diabetes Prevention Program Research Group The Diabetes Prevention Program: baseline characteristics of the randomized cohort. Diabetes Care. 2000;23(11):1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Utzschneider KM, Prigeon RL, Faulenbach MV, Tong J, Carr DB, Boyko EJ, Leonetti DL, McNeely MJ, Fujimoto WY, Kahn SE. Oral disposition index predicts the development of future diabetes above and beyond fasting and 2-h glucose levels. Diabetes Care. 2009;32(2):335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitabchi AE, Temprosa M, Knowler WC, Kahn SE, Fowler SE, Haffner SM, Andres R, Saudek C, Edelstein SL, Arakaki R, Murphy MB, Shamoon H; Diabetes Prevention Program Research Group . Role of insulin secretion and sensitivity in the evolution of type 2 diabetes in the diabetes prevention program: effects of lifestyle intervention and metformin. Diabetes. 2005;54(8):2404–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Diabetes Prevention Program The Diabetes Prevention Program. Design and methods for a clinical trial in the prevention of type 2 diabetes. Diabetes Care. 1999;22(4):623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb). 2005;95(3):221–227. [DOI] [PubMed] [Google Scholar]
- 40.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye L, Xu Y, Zhu Y, Fan Y, Deng H, Zhang J. Association of polymorphism in neurogenic differentiation factor 1 gene with type 2 diabetes. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19(6):484–487. [PubMed] [Google Scholar]
- 42.Kanatsuka A, Tokuyama Y, Nozaki O, Matsui K, Egashira T. Β-cell dysfunction in late-onset diabetic subjects carrying homozygous mutation in transcription factors NeuroD1 and Pax4. Metabolism. 2002;51(9):1161–1165. [DOI] [PubMed] [Google Scholar]
- 43.Mochizuki M, Amemiya S, Kobayashi K, Kobayashi K, Ishihara T, Aya M, Kato K, Kasuga A, Nakazawa S. The association of Ala45Thr polymorphism in NeuroD with child-onset type 1a diabetes in Japanese. Diabetes Res Clin Pract. 2002;55(1):11–17. [DOI] [PubMed] [Google Scholar]
- 44.Chiu KC, Chuang LM, Chu A, Wang M. Transcription factor 1 and β-cell function in glucose-tolerant subjects. Diabet Med. 2003;20(3):225–230. [DOI] [PubMed] [Google Scholar]
- 45.Urhammer SA, Hansen T, Ekstrøm CT, Eiberg H, Pedersen O. The Ala/Val98 polymorphism of the hepatocyte nuclear factor-1α gene contributes to the interindividual variation in serum C-peptide response during an oral glucose tolerance test: evidence from studies of 231 glucose-tolerant first degree relatives of type 2 diabetic probands. J Clin Endocrinol Metab. 1998;83(12):4506–4509. [DOI] [PubMed] [Google Scholar]
- 46.Rissanen J, Wang H, Miettinen R, Kärkkäinen P, Kekäläinen P, Mykkänen L, Kuusisto J, Karhapää P, Niskanen L, Uusitupa M, Laakso M. Variants in the hepatocyte nuclear factor-1α and -4α genes in Finnish and Chinese subjects with late-onset type 2 diabetes. Diabetes Care. 2000;23(10):1533–1538. [DOI] [PubMed] [Google Scholar]
- 47.Lee HJ, Ahn CW, Kim SJ, Song YD, Lim SK, Kim KR, Lee HC, Huh KB. Mutation in hepatocyte nuclear factor-1α is not a common cause of MODY and early-onset type 2 diabetes in Korea. Acta Diabetol. 2001;38(3):123–127. [DOI] [PubMed] [Google Scholar]
- 48.Chiu KC, Chuang LM, Ryu JM, Tsai GP, Saad MF. The I27L amino acid polymorphism of hepatic nuclear factor-1α is associated with insulin resistance. J Clin Endocrinol Metab. 2000;85(6):2178–2183. [DOI] [PubMed] [Google Scholar]
- 49.Ihara A, Yamagata K, Nammo T, Miura A, Yuan M, Tanaka T, Sladek FM, Matsuzawa Y, Miyagawa J, Shimomura I. Functional characterization of the HNF4α isoform (HNF4α8) expressed in pancreatic β-cells. Biochem Biophys Res Commun. 2005;329(3):984–990. [DOI] [PubMed] [Google Scholar]
- 50.AMP-T2D Program. T2D-GENES Consortium STDC. Available at: www.type2diabetesgenetics.org/rs11086926. Accessed 31 December 2014.