Abstract
Context:
Genome-wide association studies identified >50 type 1 diabetes (T1D) associated non-human leukocyte antigens (non-HLA) loci.
Objective:
The purpose of this study was to assess the contribution of non-HLA single nucleotide polymorphisms (SNPs) to risk of disease progression.
Design and Setting:
The TrialNet Pathway to Prevention Study follows relatives of T1D patients for development of autoantibodies (Abs) and T1D.
Participants:
Using the Immunochip, we analyzed 53 diabetes-associated, non-HLA SNPs in 1016 Ab-positive, at-risk non-Hispanic white relatives.
Main Outcome Measure:
Effect of SNPs on the development of multiple Abs and T1D.
Results:
Cox proportional analyses included all substantial non-HLA SNPs, HLA genotypes, relationship to proband, sex, age at initial screening, initial Ab type, and number. Factors involved in progression from single to multiple Abs included age at screening, relationship to proband, HLA genotypes, and rs3087243 (cytotoxic T lymphocyte antigen-4). Significant factors for diabetes progression included age at screening, Ab number, HLA genotypes, rs6476839 [GLIS family zinc finger 3 (GLIS3)], and rs3184504 [SH2B adaptor protein 3 (SH2B3)]. When glucose area under the curve (AUC) was included, factors involved in disease progression included glucose AUC, age at screening, Ab number, relationship to proband, HLA genotypes, rs6476839 (GLIS3), and rs7221109 (CCR7). In stratified analyses by age, glucose AUC, age at screening, sibling, HLA genotypes, rs6476839 (GLIS3), and rs4900384 (C14orf64) were significantly associated with progression to diabetes in participants <12 years old, whereas glucose AUC, sibling, rs3184504 (SH2B3), and rs4900384 (C14orf64) were significant in those ≥12.
Conclusions:
In conclusion, we identified five non-HLA SNPs associated with increased risk of progression from Ab positivity to disease that may improve risk stratification for prevention trials.
In Ab-positive relatives, non-HLA SNPs contribute to disease progression in addition to known factors such as age, relationship to proband, HLA genotypes, and number of Abs.
Type 1 diabetes (T1D) is a chronic autoimmune disease resulting from the interaction of genes and environmental factors (1). T1D has a strong genetic component. Concordance rates for disease development exceed 50% in monozygotic twins with long-term follow-up, compared with 6% to 10% in dizygotic twins, which is similar to what is found in nontwin siblings (2, 3). T1D prevalence by age 30 is higher in siblings of patients (6%) than in the general population (0.4%). Several genomic loci have been confirmed as T1D susceptibility regions by linkage and association studies. The human histocompatibility complex on chromosome 6p21 contains genes involved in innate and adaptive immune functions; genes coding for variants of human leukocyte antigen (HLA), especially selected allele variants at the class II HLA DR and DQ loci are considered the major susceptibility locus for T1D: an estimated 30% to 50% of the overall genetic risk is attributed to this region (4, 5). With the advent of genome-wide association studies, more than 50 non-HLA susceptibility gene markers have been associated with diabetes risk (6–9). Most appear to have effects in the immune system and have recently been shown to localize to enhancer sequences active in thymus, T and B cells, and CD34+ stem cells (10); however, there is increasing appreciation that many are also expressed in islet cells and modulate beta cell function and responses to various stimuli (11). In the Diabetes Autoimmunity Study in the Young (DAISY), PTPN22, UBASH3A, INS, and IFIH1 were significantly associated with islet autoimmunity and/or T1D, after adjusting for family history of T1D and HLA-DR3/4-DQ2/8 genotype (12, 13). In The Environmental Determinants of Diabetes in the Young (TEDDY) study, eight single nucleotide polymorphisms (SNPs) achieved significant association to development of islet autoimmunity (IA) using time-to-event analysis, including rs2476601 in PTPN22, rs2292239 in ERBB3, rs3184504 in SH2B adaptor protein 3 (SH2B3), rs1004446 in INS, rs2816316 in RGS1, rs10517086 on chromosome 4p15.2, rs4948088 in COBL, and rs12708716 in CLEC16A (14).
It is becoming increasingly evident that various phenotypes and genotypes are likely to contribute to the different presentations of diabetes (variable age of onset, severity of presentation, etc.) and that understanding disease pathophysiology and staging of diabetes will help facilitate individualized treatment (15). The goal of this study was to evaluate if non-HLA SNPs can improve risk stratification for prevention trials in relatives enrolled in the TrialNet Pathway to Prevention (PTP) study.
Materials and Methods
Study population and design
The TrialNet PTP study screens for the presence of autoantibodies (Abs) and follows relatives of T1D patients at increased risk of diabetes as previously described (16). All subjects are screened for Abs to glutamic acid decarboxylase Ab (GADA), insulin Ab (IAA), and insulinoma-associated antigen 2 Ab (IA-2A); if any of these are detected, Abs to zinc transporter 8 Ab (ZnT8A) and islet cell antibodies (ICAs) are tested as well. Islet autoimmunity was defined as confirmed positive for at least one positive Ab, including IAA, GADA, IA-2A, ZnT8A, and ICA. Since ZnT8A and ICA measurements were not consistently performed, only GADA, IAA, and IA-2A were considered for analyses of progression from single to multiple (≥2) Ab. Relatives positive for at least one Ab are followed with 2-hour oral glucose tolerance test (OGTT) surveillance for the development of diabetes. In this study, we analyzed 1016 non-Hispanic white relatives with Ab positivity and for whom we had available Immunochip (Illumina) data; only one subject per family was included for these genetic analyses (Supplemental Fig. 1 (265.5KB, tif) ). On follow-up, 284 relatives were diagnosed with diabetes according to American Diabetes Association guidelines (17). All study participants gave informed consent and the study was approved by the responsible ethics committee at each study site.
HLA typing
Relatives were typed for HLA class II DRB1, DQA1, and DQB1 alleles at full resolution using DNA-based typing with oligonucleotide probes, as previously reported (18). HLA genotypes were categorized as HLA-DR3/4*0302, HLA-DR4*0302/4*0302, HLA-DR3/3, HLA-DR4*0302/X, HLA-DR3/X, and HLA-DRX/X, where X equals not HLA-DR3, not HLA-DR4.
SNPs
SNPs were genotyped using the Illumina Immunochip at the Center for Public Health Genomics at the University of Virginia, Charlottesville, VA. The Immunochip is a custom array of 186,000 SNPs selected from regions of the genome robustly associated with autoimmune diseases. For the purpose of this study, we analyzed a total of 53 non-HLA SNPs in 44 gene regions previously associated with T1D risk (10, 14) (Supplemental Table 1 (19.4KB, docx) ). The insulin rs689 SNP failed quality control and was therefore excluded from analyses, but another insulin SNP rs7111341 was included in the analyses.
Statistical analysis
Statistical analyses were performed using SAS software (version 9.4, SAS Institute, Cary, NC). Student t tests and Pearson χ2 tests were used to compare groups. For each SNP, the number of minor alleles was used and analyzed as a continuous variable in the univariate Cox proportional hazards model. Because our analyses were based on a priori hypotheses, P values were not corrected for multiple testing. Multivariate Cox proportional hazards models with backward selection were performed to obtain hazard ratios (HRs) with 95% confidence intervals (CIs). Models included all SNPs univariately substantial with P value < 0.1 and the following variables: age at the initial screening, sex, relationship to proband and HLA DR-DQ risk group, and type of first Ab for progression to multiple Abs and number of positive Abs for progression to T1D. An additional model included glucose area under the curve (AUC) from 2-hour initial OGTT. Univariate and multivariate Cox proportional hazards models were also performed after stratification by age; a cut-off of 12 years of age was chosen to be consistent with typical age inclusion criteria into prevention trials in children. Further stratification in younger children (<6 years old) was not possible because of small numbers. Kaplan–Meier curves were generated to examine the development of T1D or multiple Abs in those initially identified with a single Ab; differences between the curves were assessed with log-rank tests. P values < 0.05 were considered statistically significant.
Results
Characteristics of the PTP participants who were included in the analysis are shown in Table 1. Subjects who progressed to diabetes were more likely to be males, younger at initial positive visit, sibling to the proband, and more often carried the high-risk HLA DR3/4 genotype. Among the 1016 Ab-positive TrialNet participants, a total of 239 were single confirmed Ab positive. The 254 excluded subjects had similar characteristics to the 1016 subjects included in the analyses, except for relationship to proband with slightly less parent and offspring in the excluded group (Supplemental Table 2 (14.8KB, docx) ).
Table 1.
Characteristics of Ab-Positive (Ab+) TrialNet Participants
| Ab+ Subjects Without T1D (N = 732) | Ab+ Subjects Who Progressed to T1D (N = 284) | P Valuea | |
|---|---|---|---|
| Age at initial Ab+, median (IQR) | 12.4 (7.2 to 31.7) | 9.7 (6.0 to 14.7) | <0.001 |
| Follow-up, median (IQR)b | 5.1 (3.1 to 7.2) | 2.6 (1.5 to 4.5) | <0.001 |
| Sex: male, N (%) | 351 (48) | 152 (54) | 0.02 |
| AUC glucose at first OGTT (mg/dL), mean (SD) | 2353 (298) | 2648 (408) | <0.001 |
| Relationship to proband, N (%) | <0.001 | ||
| Sibling | 376 (51) | 166 (59) | |
| Parent | 150 (21) | 28 (10) | |
| Offspring | 149 (20) | 69 (24) | |
| Other/unknown | 57 (8) | 21 (7) | |
| HLA genotypes, N (%) | <0.001 | ||
| HLA-DR3/3 | 35 (5) | 14 (5) | |
| HLA-DR3/4 | 107 (15) | 78 (27) | |
| HLA-DR3/Xc | 116 (16) | 33 (12) | |
| HLA-DR4/4 | 61 (8) | 28 (10) | |
| HLA-DR4/Xc | 221 (30) | 84 (29) | |
| Other/unavailable | 192 (26) | 47 (17) |
Abbreviation: IQR, interquartile range.
Wilcoxon test for comparing medians; χ2 for proportions between two groups.
Follow-up: time to diabetes or last visit for those who did not develop diabetes.
Where X equals not DR3, not DR4. HLA DR X/X (i.e., not 3/3, 3/4, 3/X, 4/4, or 4/X) is used as reference group.
Among relatives initially identified with a single Ab (n = 239), three SNPs were associated with progression to multiple Abs in univariate analyses: rs3087243 [cytotoxic T lymphocyte antigen-4 (CTLA4); HR = 0.61, P = 0.03], rs75793288 (IL2/IL21; HR = 1.50, P = 0.048), and rs62447205 (IKZF1; HR = 1.62, P = 0.04); two other SNPs showed a trend toward association for progression to multiple Abs, rs4505848 (IL2/IL21; HR = 1.42, P = 0.093) and rs72928038 (BACH2 ; HR = 1.50, P = 0.07), and were included in multivariate analyses. Among all Ab-positive relatives, the following SNPs were associated with progression to diabetes in univariate analyses: rs6476839 [GLIS family zinc finger 3 (GLIS3); HR = 1.35, P = 0.001], rs3184504 (SH2B3; HR = 1.22, P = 0.02), and rs7221109 (CCR7; HR = 0.82, P = 0.03). Another five SNPs showed a trend toward association with progression to T1D, and were also included in multivariate analyses: rs113010081 (CCR5; HR = 0.80, P = 0.098), rs7020673 (GLIS3; HR = 0.86, P = 0.063), rs2290400 (ORMDL; HR = 0.86, P = 0.079), rs1893217 (PTPN2; HR = 0.81, P = 0.05), and rs11203203 (UBASH3A; HR = 0.85, P = 0.052).
Multivariate Cox proportional backward selection analyses were performed including all substantial non-HLA SNPs, HLA genotypes, relationship to proband (sibling, offspring, parent), sex, age at initial screening and type of first Ab for progression to multiple Abs and number of positive Ab for progression to diabetes T1D. Factors involved in progression to multiple Abs in those single Ab-positive subjects included age at initial screening, being parent of a patient, HLA genotypes, and rs3087243 (CTLA4) (Table 2). Factors that stayed significant for disease progression included age at initial screening, number of Ab, HLA-DR3/4, HLA-DR4/4, rs6476839 (GLIS3), and rs3184504 (SH2B3) (Table 3).
Table 2.
Influence of SNPs on Progression to Multiple Abs in Single Ab-Positive (Ab+) Subjects (N = 239)
| Variable | HR (95% CI) | P Value |
|---|---|---|
| Age at initial screen | 0.89 (0.83 to 0.95) | 0.001 |
| Sex: male | 1.83 (0.82 to 4.09) | 0.141 |
| Offspring | 8.13 (1.00 to 66.43) | 0.051 |
| Parent | 47.59 (2.54 to 890.06) | 0.010 |
| Sibling | 4.66 (0.61 to 35.86) | 0.139 |
| HLA-DR3/3 | 3.98 (0.54 to 29.30) | 0.175 |
| HLA-DR3/4 | 8.26 (1.28 to 53.18) | 0.026 |
| HLA-DR3/Xa | 4.74 (0.88 to 25.57) | 0.071 |
| HLA-DR4/4 | 14.19 (2.24 to 90.04) | 0.005 |
| HLA-DR4/Xa | 4.58 (0.92 to 22.87) | 0.064 |
| IA-2A first | 0.80 (0.24 to 2.68) | 0.722 |
| IAA first | 0.63 (0.25 to 1.58) | 0.321 |
| rs3087243 (CTLA4) | 0.47 (0.28 to 0.80) | 0.005 |
Multivariate Cox proportional hazard model including the SNPs that were univariately substantial (P ≤ 0.10), age, sex, HLA-DR, relationship to proband, and type of first Ab. GADA is the reference group.
Where X equals not DR3, not DR4. HLA DR X/X (i.e., not 3/3, 3/4, 3/X, 4/4, or 4/X) is used as reference group.
Table 3.
Influence of SNPs on Progression to T1D in Ab-Positive (Ab+) Participants (N = 1016)
| Variable | HR (95% CI) | P Value |
|---|---|---|
| Age at initial screen | 0.98 (0.96 to 0.99) | 0.012 |
| Number Ab+ | 1.51 (1.04 to 2.19) | 0.030 |
| Sex: male | 1.24 (0.94 to 1.64) | 0.133 |
| Offspring | 1.34 (0.69 to 2.58) | 0.383 |
| Parent | 0.73 (0.29 to 1.82) | 0.449 |
| Sibling | 1.37 (0.74 to 2.55) | 0.321 |
| HLA-DR3/3 | 2.04 (0.85 to 4.89) | 0.109 |
| HLA-DR3/4 | 3.12 (1.55 to 6.28) | 0.001 |
| HLA-DR3/Xa | 1.60 (0.76 to 3.39) | 0.219 |
| HLA-DR4/4 | 2.78 (1.28 to 6.02) | 0.010 |
| HLA-DR4/Xa | 2.00 (1.00 to 4.01) | 0.051 |
| rs6476839 (GLIS3) | 1.29 (1.05 to 1.57) | 0.013 |
| rs3184504 (SH2B3) | 1.23 (1.01 to 1.48) | 0.035 |
Multivariate Cox proportional hazard model including the SNPs that were univariately substantial (P ≤ 0.10), age, sex, HLA-DR, relationship to proband, and number of Abs.
Where X equals not DR3, not DR4. HLA DR X/X (i.e., not 3/3, 3/4, 3/X, 4/4, or 4/X) is used as reference group.
When adding glucose AUC to the model, factors involved in progression to diabetes included glucose AUC, age at screening, number of Ab, being an offspring or sibling, HLA DR3/4, HLA DR4/X, and rs6476839 (GLIS3); rs3184504 (SH2B3) was no longer significant, while rs7221109 (CCR7) became newly significant in this model (Table 4). For progression from single to multiple Abs in participants with glucose AUC data, results were similar overall, but only age at initial screening, HLA-DR4/4 and rs3087243 (CTLA4) stayed significant in this model (Table 5).
Table 4.
Influence of SNPs on Progression to T1D in Ab-Positive (Ab+) Subjects With AUC Glucose (N = 1016)
| Variable | HR (95% CI) | P Value |
|---|---|---|
| Age at initial screen | 0.97 (0.95 to 0.99) | <0.001 |
| Glucose AUC (first OGTT) | 1.003 (1.002 to 1.003) | <0.001 |
| Number Ab+ | 1.48 (1.01 to 2.16) | 0.046 |
| Sex: male | 1.12 (0.84 to 1.49) | 0.438 |
| Offspring | 2.43 (1.14 to 5.19) | 0.022 |
| Parent | 1.66 (0.59 to 4.66) | 0.339 |
| Sibling | 2.71 (1.31 to 5.62) | 0.007 |
| HLA-DR3/3 | 2.49 (0.96 to 6.46) | 0.061 |
| HLA-DR3/4 | 2.78 (1.27 to 6.06) | 0.010 |
| HLA-DR3/Xa | 1.79 (0.78 to 4.14) | 0.169 |
| HLA-DR4/4 | 2.27 (0.96 to 5.35) | 0.062 |
| HLA-DR4/Xa | 2.44 (1.12 to 5.34) | 0.025 |
| rs6476839 (GLIS3) | 1.27 (1.04 to 1.56) | 0.022 |
| rs7221109 (CCR7) | 0.70 (0.57 to 0.87) | 0.001 |
Multivariate Cox proportional hazard model including the SNPs that were univariately substantial (P ≤ 0.10), age, sex, HLA-DR, relationship to proband, number of Ab+, and glucose AUC.
Where X equals not DR3, not DR4. HLA DR X/X (i.e., not 3/3, 3/4, 3/X, 4/4, or 4/X) is used as reference group.
Table 5.
Influence of SNPs on Progression to Multiple Abs in Single Ab-Positive (Ab+) Subjects With AUC Glucose (N = 239)
| Variable | HR (95% CI) | P Value |
|---|---|---|
| Age at initial screen | 0.94 (0.91 to 0.97) | <0.001 |
| AUC glucose (first OGTT) | 1.00 (0.99 to 1.01) | 0.841 |
| Sex: male | 1.48 (0.68 to 3.22) | 0.324 |
| HLA-DR3/3 | 2.62 (0.39 to 17.84) | 0.325 |
| HLA-DR3/4 | 5.42 (0.94 to 31.44) | 0.059 |
| HLA-DR3/Xa | 3.03 (0.56 to 16.30) | 0.198 |
| HLA-DR4/4 | 9.94 (1.60 to 61.80) | 0.014 |
| HLA-DR4/Xa | 3.85 (0.81 to 18.36) | 0.091 |
| IA-2A first | 0.91 (0.25 to 3.40) | 0.891 |
| IAA first | 0.72 (0.30 to 1.74) | 0.458 |
| rs3087243 (CTLA4) | 0.43 (0.25 to 0.76) | 0.003 |
Multivariate Cox proportional hazard model including the SNPs that were univariately substantial (P ≤ 0.10), age, sex, HLA-DR, type of first Ab, and AUC glucose. GADA is the reference group. The number of subjects with nonfirst degree proband was too small, thus the relationship to proband variable was removed from this model.
Where X equals not DR3, not DR4. HLA DR X/X (i.e., not 3/3, 3/4, 3/X, 4/4, or 4/X) is used as reference group.
Multivariate Cox proportional hazards analyses stratified by age including glucose AUC are shown in Supplemental Table 3 (14.3KB, docx) . For subjects <12 years old at the initial screening visit, glucose AUC, age at screening, being a sibling, HLA genotypes, rs6476839 (GLIS3), and rs4900384 (C14orf64) were significant factors involved in diabetes progression; glucose AUC, being a sibling, rs3184504 (SH2B3), and rs4900384 (C14orf64) were associated with disease progression in those older than 12 years of age. Interestingly, rs4900384 (C14orf64) had opposite effects in these age groups as it was associated with disease progression in those aged 12 or older (HR = 1.85, 95% CI, 1.24 to 2.74) but showed a protective effect in those younger than 12 (HR = 0.74,95% CI, 0.57 to 0.97).
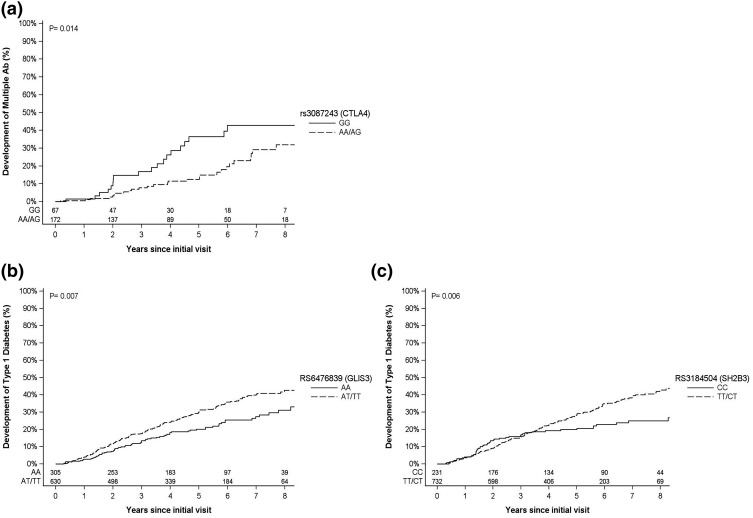
Survival curves were generated for the significant SNPs described previously. Risk for development of multiple Abs by 6 years was 43% for those with rs3087243 GG genotype (CTLA4) compared with 20% for those with AA/AG genotypes [P = 0.014; Fig. 1(a)]. By year 6 after the initial visit, subjects with rs6476839 AT/TT genotype (GLIS3) had a higher risk for diabetes development (36%) compared with those with the AA (25%) genotypes [P < 0.007; Fig. 1(b)]. Participants with rs3184504 TT/CT genotypes (SH2B3) had a higher risk of diabetes development compared with those with the CC genotype [35% vs 23% respectively, P = 0.006; Fig. 1(c)].
Figure 1.
Kaplan-Meier curves for the significant SNPs in Cox proportional hazards models. (a) Progression to multiple Abs by rs3087243 (CTLA4). (b) Progression to T1D by rs6476839 (GLIS3). (c) Progression to T1D by rs3184504 (SH2B3).
In addition, we explored whether there was an interaction between CTLA4 and HLA-DR3 or HLA-DR4 haplotype. There was no interaction effect between HLA-DR3 and CTLA4. However, there was a significant interaction between HLA-DR4 and CTLA4. Subjects with rs3087243 GG genotype and HLA-DR4 had a higher risk of developing multiple Abs by 6 years than those with AA/AG genotypes and HLA DR4 (51% vs 17% respectively, P = 0.003).
Discussion
Several prospective studies such as DAISY (12, 13), BABYDIAB (19, 20), and TEDDY (14) have looked at the additional effects of known susceptibility non-HLA genes on progression from islet autoimmunity to diabetes. All of these studies involve the prospective follow-up of very young children identified to be at genetic risk at birth. The TrialNet PTP cohort examined here differs in that relatives of T1D patients are screened and enrolled in the study at an older age, between the ages of 1 and 45 years [median age at screening for the participants in this study was 11.3 years (interquartile range = 6.8 to 23.7)]. Given the well-known influence of age on disease progression, it is possible that genetic associations may differ according to age. To our knowledge, this the largest cohort of Ab-positive subjects studied to date, whereas DAISY, TEDDY, and BABYDIAB studies primarily analyzed the risk of developing Ab in genetically at-risk populations. This is also the first study, to our knowledge, to combine genetics and dysglycemia assessment (glucose AUC) to improve prediction of disease progression. This study in relatives at high-risk for development of diabetes shows that non-HLA SNPs contribute to progression to diabetes or progression from single to multiple Abs in addition to known factors such as age, relationship to proband (sibling, offspring), HLA genotypes, and number of Ab. Whereas both rs6476839 (GLIS3) and rs3184504 (SH2B3) are important factors involved in progression to diabetes among Ab-positive subjects, rs3087243 (CTLA4) was involved in progression to multiple Abs in those single Ab-positive participants.
GLIS3 is a transcription factor important for the regulation of pancreatic β-cell development and insulin gene expression (21). GLIS3 has been associated with diabetes (7, 19) and permanent neonatal diabetes (22). In DAISY, GLIS3 showed borderline association with development of islet autoimmunity in multivariate analyses adjusting for family history and HLA-DR3/4 genotype (13). Both CTLA4 and SH2B3 have been associated with diabetes and celiac disease (9). Eight SNPs, including SH2B3, were associated with time to diabetes in the study of children with high-risk HLA alleles participating in TEDDY (14). The encoded protein (SH2B) is a key negative regulator of cytokine signaling. CTLA4 plays a role in the inhibition of T-cell activation. A clinical trial in patients with new onset T1D showed that costimulation modulation with abatacept (CTLA4-Ig) slows the decline of β-cell function and improves HbA1c (23, 24). This has led to the currently ongoing TrialNet prevention trial with abatacept in relatives with multiple Abs.
When adding glucose AUC to the model, rs6476839 (GLIS3) but not rs3184504 (SH2B3) stayed significantly associated with disease progression besides glucose AUC, age at screening, number of Ab, relation to proband and HLA genotypes. Interestingly, rs7221109 (CCR7) became significant in the overall model with AUC glucose. However, rs7221109 (CCR7) was not significant in the base model without glucose AUC nor in the stratified model by age with AUC glucose. C-C chemokine receptor type 7 (CCR7) has been shown to be involved in the recruitment of T cells into inflamed islets and thus plays a role in the pathogenesis of T1D (25). Only age at initial screening, HLA-DR4/4 and rs3087243 (CTLA4) stayed significant for progression to multiple Abs; this suggests that dysglycemia is not associated with progression from single to multiple Abs. Prevention trials are currently only available for relatives with multiple Abs. However, single Ab-positive subjects with CTLA4 rs3087243_A and HLA-DR4 might benefit from early prevention trials to prevent spreading of Abs and preservation of β-cell mass at the early stages of disease.
In stratified analyses by age, glucose AUC, age at screening, being a sibling, HLA genotypes, rs6476839 (GLIS3), and rs4900384 (C14orf64) were significantly associated with progression to diabetes in subjects <12 years old at initial visit, whereas glucose AUC, being a sibling, rs3184504 (SH2B3), and rs4900384 (C14orf64) were important factors in those ≥12 years. Interestingly, rs4900384 (C14orf64) had an opposite (protective) effect in the younger group, similar to what has been shown in TEDDY, a cohort of young children at increased risk for T1D (14). The differences in genetic associations may imply a different mechanism of disease in younger and older subjects. Identifying specific risk factors according to age could lead to personalized prevention trials which might have a better chance in succeeding to prevent T1D.
Besides the effects of age, variance in results between this study and other prospective studies such as DAISY, BABYDIAB, and TEDDY could be due to differences in ascertainment (TEDDY cohort has mainly young general population children, whereas BABYDIAB follows offsprings of T1D parents) and in the SNPs tested. The strengths of this study include well-characterized population of relatives at increased risk for diabetes representing an older age group than in the prospective studies from birth. Among the single Ab-positive confirmed group (N = 239), a total of 45 subjects (19%) developed T1D (including 14 with multiple Abs) and another 36 subjects (15%) developed multiple Abs but not T1D. The progression to T1D among single Ab-positive confirmed subjects has typically been around 15% in PTP, so these numbers might be slightly higher. The limitations of this study include that participants are not followed since birth and, therefore, the time of seroconversion is often unknown. Analyses were limited to participants with at least one positive Ab. The factors that we identified as contributors to progression from single to multiple Abs and from Ab positivity to overt disease may differ from those that influence the triggering of islet autoimmunity. Some of the analyses were limited by a relatively small number of subjects. To avoid confounding effects of genetic heterogeneity across populations, we only analyzed non-Hispanic whites, who represent the major ethnic group in the PTP study; whether these findings apply to other races/ethnicities warrants further study.
In conclusion, we suggest that non-HLA genes are active during disease progression after the initial triggering of autoimmunity. Non-HLA SNPs help to refine risk for Ab-positive relatives and may improve risk stratification for prevention trials.
Acknowledgments
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health (NIH) or the Juvenile Diabetes Research Foundation International (JDRF). A.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Acknowledgments
This study was sponsored by the Type 1 Diabetes TrialNet Pathway to Prevention Study Group, which is a clinical trials network funded by the NIH through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and The Eunice Kennedy Shriver National Institute of Child Health and Human Development, through the cooperative agreements U01 DK061010, U01 DK061034, U01 DK061042, U01 DK061058, U01 DK085465, U01 DK085453, U01 DK085461, U01 DK085463, U01 DK085466, U01 DK085499, U01 DK085504, U01 DK085505, U01 DK085509, U01 DK103180, U01-DK103153, U01-DK085476, U01-DK103266, and the JDRF. A.K.S. was supported by the American Diabetes Association Grant 1-14-CD-17. J.M.W. was supported by a Mentored Clinical Researcher Fellowship from JDRF Australia.
Acknowledgments
Author contributions: A.K.S. and A.P. conducted the study, designed the analyses, analyzed data, and wrote the manuscript. P.X. and S.G. provided statistical support, analyzed data, and reviewed the manuscript. M.J.R., P.A., J.M.W., and J.S. reviewed data, discussed findings and analysis plans, and edited the manuscript. S.O.-G., W.-M.C., and S.S.R. performed Immunochip typing, reviewed data, and edited the manuscript.
Acknowledgments
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- Ab
- autoantibody
- AUC
- area under the curve
- CI
- confidence interval
- CTLA4
- cytotoxic T lymphocyte antigen-4
- DAISY
- Diabetes Autoimmunity Study in the Young
- GADA
- glutamic acid decarboxylase autoantibody
- GLIS3
- GLIS family zinc finger 3
- HLA
- human leukocyte antigen
- HR
- hazard ratio
- IA-2A
- insulinoma-associated antigen 2 autoantibody
- IAA
- insulin autoantibody
- ICA
- islet cell antibody
- OGTT
- oral glucose tolerance test
- PTP
- Pathway to Prevention
- SH2B3
- SH2B adaptor protein 3
- SNP
- single nucleotide polymorphism
- T1D
- type 1 diabetes
- TEDDY
- The Environmental Determinants of Diabetes in the Young
- ZnT8A
- zinc transporter 8 autoantibody.
References
- 1.Bluestone JA, Herold K, Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464(7293):1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS, Orban T. Concordance for islet autoimmunity among monozygotic twins. N Engl J Med. 2008;359(26):2849–2850. [DOI] [PubMed] [Google Scholar]
- 3.Hyttinen V, Kaprio J, Kinnunen L, Koskenvuo M, Tuomilehto J. Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: a nationwide follow-up study. Diabetes. 2003;52(4):1052–1055. [DOI] [PubMed] [Google Scholar]
- 4.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59(5):1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 5.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14(9):661–673. [DOI] [PubMed] [Google Scholar]
- 6.Burren OS, Adlem EC, Achuthan P, Christensen M, Coulson RM, Todd JA. T1DBase: update 2011, organization and presentation of large-scale data sets for type 1 diabetes research. Nucleic Acids Res. 2011;39(Database issue):D997–D1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS; Type 1 Diabetes Genetics Consortium . Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, Warram JH, Todd JA. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40(12):1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, Howson JM, Stevens H, McManus R, Wijmenga C, Heap GA, Dubois PC, Clayton DG, Hunt KA, van Heel DA, Todd JA. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359(26):2767–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, Farber E, Bonnie JK, Szpak M, Schofield E, Achuthan P, Guo H, Fortune MD, Stevens H, Walker NM, Ward LD, Kundaje A, Kellis M, Daly MJ, Barrett JC, Cooper JD, Deloukas P, Todd JA, Wallace C, Concannon P, Rich SS; Type 1 Diabetes Genetics Consortium . Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47(4):381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fløyel T, Kaur S, Pociot F. Genes affecting β-cell function in type 1 diabetes. Curr Diab Rep. 2015;15(11):97. [DOI] [PubMed] [Google Scholar]
- 12.Steck AK, Wong R, Wagner B, Johnson K, Liu E, Romanos J, Wijmenga C, Norris JM, Eisenbarth GS, Rewers MJ. Effects of non-HLA gene polymorphisms on development of islet autoimmunity and type 1 diabetes in a population with high-risk HLA-DR,DQ genotypes. Diabetes. 2012;61(3):753–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steck AK, Dong F, Wong R, Fouts A, Liu E, Romanos J, Wijmenga C, Norris JM, Rewers MJ. Improving prediction of type 1 diabetes by testing non-HLA genetic variants in addition to HLA markers. Pediatr Diabetes. 2014;15(5):355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Törn C, Hadley D, Lee HS, Hagopian W, Lernmark Å, Simell O, Rewers M, Ziegler A, Schatz D, Akolkar B, Onengut-Gumuscu S, Chen WM, Toppari J, Mykkänen J, Ilonen J, Rich SS, She JX, Steck AK, Krischer J, Group TS; TEDDY Study Group . Role of type 1 diabetes-associated SNPs on risk of autoantibody positivity in the TEDDY Study. Diabetes. 2015;64(5):1818–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skyler JS, Bakris GL, Bonifacio E, Darsow T, Eckel RH, Groop L, Groop PH, Handelsman Y, Insel RA, Mathieu C, McElvaine AT, Palmer JP, Pugliese A, Schatz DA, Sosenko JM, Wilding JP, Ratner RE. Differentiation of diabetes by pathophysiology, natural history, and prognosis. Diabetes. 2017;66(2):241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahon JL, Sosenko JM, Rafkin-Mervis L, Krause-Steinrauf H, Lachin JM, Thompson C, Bingley PJ, Bonifacio E, Palmer JP, Eisenbarth GS, Wolfsdorf J, Skyler JS; TrialNet Natural History Committee; Type 1 Diabetes TrialNet Study Group . The TrialNet Natural History Study of the Development of Type 1 Diabetes: objectives, design, and initial results. Pediatr Diabetes. 2009;10(2):97–104. [DOI] [PubMed] [Google Scholar]
- 17.American Diabetes Association (2) Classification and diagnosis of diabetes. Diabetes Care. 2015;38(Suppl):S8–S16. [DOI] [PubMed] [Google Scholar]
- 18.Mychaleckyj JC, Noble JA, Moonsamy PV, Carlson JA, Varney MD, Post J, Helmberg W, Pierce JJ, Bonella P, Fear AL, Lavant E, Louey A, Boyle S, Lane JA, Sali P, Kim S, Rappner R, Williams DT, Perdue LH, Reboussin DM, Tait BD, Akolkar B, Hilner JE, Steffes MW, Erlich HA; T1DGC . HLA genotyping in the international Type 1 Diabetes Genetics Consortium. Clin Trials. 2010; 7(Suppl 1):S75–S87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkler C, Krumsiek J, Buettner F, Angermüller C, Giannopoulou EZ, Theis FJ, Ziegler AG, Bonifacio E. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia. 2014;57(12):2521–2529. [DOI] [PubMed] [Google Scholar]
- 20.Bonifacio E, Krumsiek J, Winkler C, Theis FJ, Ziegler AG. A strategy to find gene combinations that identify children who progress rapidly to type 1 diabetes after islet autoantibody seroconversion. Acta Diabetol. 2014;51(3):403–411. [DOI] [PubMed] [Google Scholar]
- 21.Kang HS, Kim YS, ZeRuth G, Beak JY, Gerrish K, Kilic G, Sosa-Pineda B, Jensen J, Pierreux CE, Lemaigre FP, Foley J, Jetten AM. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol. 2009;29(24):6366–6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senée V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec JC, Charon C, Nicolino M, Boileau P, Cavener DR, Bougnères P, Taha D, Julier C. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet. 2006;38(6):682–687. [DOI] [PubMed] [Google Scholar]
- 23.Orban T, Bundy B, Becker DJ, DiMeglio LA, Gitelman SE, Goland R, Gottlieb PA, Greenbaum CJ, Marks JB, Monzavi R, Moran A, Raskin P, Rodriguez H, Russell WE, Schatz D, Wherrett D, Wilson DM, Krischer JP, Skyler JS; Type 1 Diabetes TrialNet Abatacept Study Group . Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet. 2011;378(9789):412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orban T, Bundy B, Becker DJ, Dimeglio LA, Gitelman SE, Goland R, Gottlieb PA, Greenbaum CJ, Marks JB, Monzavi R, Moran A, Peakman M, Raskin P, Russell WE, Schatz D, Wherrett DK, Wilson DM, Krischer JP, Skyler JS; Type 1 Diabetes TrialNet Abatacept Study Group . Costimulation modulation with abatacept in patients with recent-onset type 1 diabetes: follow-up 1 year after cessation of treatment. Diabetes Care. 2014;37(4):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan Z, Xu B, Mikulowska-Mennis A, Michie SA. CCR7 directs the recruitment of T cells into inflamed pancreatic islets of nonobese diabetic (NOD) mice. Immunol Res. 2014;58(2-3):351–357. [DOI] [PubMed] [Google Scholar]