Abstract
Arc (aka Arg 3.1) is induced by neural activity and learning experience. Arc mRNA is rapidly exported into dendrites where it localizes near activated synapses. By imaging green fluorescent protein (GFP)-tagged mRNA in living neurons in culture, we show that fusion transcripts containing the Arc 3′UTR (untranslated region) localize with remarkable precision in a microdomain at the base of dendritic spines. Transcripts with the Arc 3′UTR that encode a reporter protein rather than Arc show precise localization. Localization persists in the presence of translation inhibitors, indicating that localization does not require ongoing translation. Similarly, polyribosome complexes remained stably positioned at spine bases in brain tissue treated with the translation inhibitor (puromycin) that releases ribosomes from mRNA. Single particle tracking revealed that Arc mRNA particles positioned at spine bases exhibited highly constrained submicron movements. These observations imply the existence of a microdomain at the spine base where Arc mRNA docks in association with a previously unknown mRNA-binding structural element.
INDEXING TERMS: Arc mRNA, microdomain, dendritic spines
In neurons, many proteins are thought to contribute to time-dependent consolidation of changes in synaptic efficacy (Sanes and Lichtman, 1999; Guzowski et al., 2000; Bramham, 2008; Miyashita et al., 2008; Richter and Klann, 2009). Of the proteins implicated in consolidation, Arc, also known as Arg 3.1, is unique because of the way it is induced by learning experiences and delivered to synapses (Dynes and Steward, 2008). Arc transcription is strongly induced in selected neuronal populations as a result of learning experiences (Guzowski et al., 1999, 2005; Miyashita et al., 2008). Newly transcribed Arc mRNA is rapidly delivered throughout dendrites (Link et al., 1995; Lyford et al., 1995; Miyashita et al., 2008), and Arc mRNA and protein rapidly accumulate near activated synapses (Steward et al., 1998; Steward and Worley, 2001). This pattern of expression and activity-dependent synaptic targeting suggests that Arc could serve as a transcription- and translation-dependent link between synaptic activity and subsequent synaptic modifications, including the ones that underlie memory consolidation.
Recent studies have provided evidence that Arc protein is a critical but transient component of mechanisms that control synaptic efficacy. One line of evidence suggests that Arc protein contributes to the stabilization of changes in the subsynaptic actin network induced by long-term potentiation (LTP) (Messaoudi et al., 2007). Other evidence indicates that Arc protein contributes to AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) receptor endocytosis leading to decreased synaptic strength (Plath et al., 2006; Rial Verde et al., 2006; Shepherd et al., 2006). In both proposed functions, the distribution of Arc protein in dendrites would constrain, and in part determine, the specificity of plastic changes at synapses. An unresolved question, however, is the precision of Arc targeting to synapses, that is, whether Arc mRNA and protein target precisely to individual synapses or more generally to local dendritic domains.
Recently, we used exogenous expression of mRNA and live cell imaging to assess characteristics of Arc mRNA transport (Dynes and Steward, 2007). As this system utilizes high affinity binding of MS2 phage coat protein to short RNA sequence tags, we refer to the mRNA expressed from chimeric Arc constructs as “Arc/MS2 mRNA.” Messenger RNAs containing the 3′ untranslated region (UTR) of Arc assemble into particles that are transported bi-directionally at a variety of rates in dendrites of rat cortical neurons in culture (Dynes and Steward, 2007). Slow, short distance movements were often seen as rapidly moving mRNA comes to a stop, suggesting a final precise targeting of mRNA at the subcellular level. Whether this final targeting occurred at synaptic sites was not determined, however.
In the course of our studies of Arc mRNA transport in dendrites, we noticed that apparently stationary Arc/MS2 mRNA particles were often precisely localized beneath the base of dendritic spines. Here we use the Arc/MS2 system and confocal microscopy to assess the selectivity of this subcellular localization and to determine if the spine base possesses a translation-independent docking site for mRNA. Our results show that Arc/MS2 mRNA particles are positioned with a very high degree of precision in a small domain at the spine base. Because the Arc/MS2 transcript does not encode Arc protein, localization is dependent on the mRNA rather than Arc protein. This positioning persists in the presence of inhibitors of mRNA translation. Together, our results suggest the existence of a previously unknown structure at the spine base upon which mRNA is bound and translated.
MATERIALS AND METHODS
Cell culture and chemicals
Cultures of embryonic day (E)18 rat cortical neurons (Sprague–Dawley) were derived from tissue provided by BrainBits (care of Dr. Gregory J. Brewer, Southern Illinois University, Springfield) and prepared and cultured in Neurobasal + B27 medium (both Invitrogen, Carlsbad, CA) according to the technique of Brewer et al. (1993). Cultures were plated at 1.3 × 105/cm2 and grown from 16 to 20 days at 37°C in 5% CO2. Neurons were cultured on 35-mm plates in which the integrated coverslip was first coated with poly-D-lysine by the manufacturer (Mattek, Ashland, MA) and then coated with 0.5 mg/ml poly-L-lysine (Sigma-Aldrich, St. Louis, MO). Puromycin (Sigma-Aldrich) was used at 1 mM; Cycloheximide (Sigma-Aldrich) was used at 500 μM and D-AP5 (D-(-)-2-Amino-5-phospho-nopentanoic acid; Tocris Biochemicals, St. Louis, MO) was used at 20 μM. Puromycin, cycloheximide, and D-AP5 treated neurons were incubated at 37°C and imaged at room temperature in the presence of the drug.
DNA constructs and transfection conditions
Details of the constructs GFP-MS2-NLS, CMV-DsRE-6xBS-rArc3′UTR, and CMV-DsRE-6xBS-no3′UTR are in Dynes and Steward (2007). In short: in GFP-MS2-NLS, a cytomegalovirus (CMV) promoter, drives expression of an open reading frame encoding a fusion of a bipartite nuclear localization sequence, enhanced green fluorescent protein (eGFP), and the MS2 coat protein. This protein binds to mRNA made from CMV-DsRE-6xBS-rArc3′UTR, which contains a CMV promoter driving expression of the red fluorescent protein DsRE and followed by a 3′UTR containing a 6× multimerized MS2 coat protein binding sequence and the 3′UTR from the rat Arc gene. CMV-DsRE-6xBS-no3′UTR is similar but lacks the rat Arc 3′UTR sequence; the transcript is terminated from a simian virus 40 polyadenylation sequence. The rat Dendrin 3′UTR sequence was amplified from total hippocampal mRNA by polymerase chain reaction and cloned into CMV-DsRE-6xBS-rArc3′UTR SalI/XhoI-SacII, replacing the Arc 3′UTR and creating CMV-DsRE-6xBS-rDendrin3′UTR. Neurons were transfected biolistically with a Bio-Rad (Hercules, CA) Helios Gene Gun 16 to 26 hours before imaging as in Dynes and Steward (2007). The 1.6 μm gold beads were used for transfection; these were coated with 7.5 μg of GFP-MS2-NLS and 30 μg of either CMV-DsRE-6xBS-rArc3′UTR, CMV-DsRE-6xBS-no3′UTR, or CMV-DsRE-6xBS-rDendrin3′UTR.
Microscopy
Images were acquired using a Bio-Rad Radiance 2000 laser scanning confocal microscope and 60× PlanApo 1.4 N.A. objective as previously reported (Dynes and Steward, 2007). Neurons were maintained in the low fluorescence version of the ambient CO2 medium Hibernate (BrainBits, Springfield, IL) at room temperature. Time-lapse images were acquired from rotated regions of interest at a rate of one every 10 seconds with a pixel size of 0.1 μm. Red and green fluorescence channels were acquired simultaneously. The confocal aperture was increased for the experiments to increase the depth of field.
Neuronal morphology
Imaged neurons were selected for inclusion in the dataset according to the criteria established in Dynes and Steward (2007) with the following modifications. Neurons coexpressing the transcripts were identified by the presence of green fluorescence in the primary dendrite(s) and accumulation at the initial dendritic branch points, similar to the pattern of localization of endogenous dendritic mRNA (Knowles et al., 1996).
Dendritic spines were identified as protrusions from the dendritic shaft at least 0.2 μm in length and were distinguished from small dendritic branches on the basis of confocal optical sectioning of the imaged dendritic segment at a 0.5-μm interval.
Image processing
Image processing was performed with the public-domain program ImageJ (NIH, Bethesda, MD). Images were subpixel aligned using the StackReg plugin for ImageJ (Sage et al., 2005). Alignment was restricted to translation along the X- and Y-axis. Fluorescence from biolistic gold beads was manually removed to avoid errors in automated alignment. Twenty aligned images were used to generate the averaged images of dendrites. For display purposes, green channel images were adjusted for brightness and contrast only, while the red channel images were also gamma-adjusted to enhance the visibility of spine necks.
The positions of spine bases were determined to the nearest pixel from the aligned, averaged red channel fluorescence images through the use of a topographic lookup table. The positions of Arc/MS2 mRNA particles were derived from aligned and averaged green fluorescent images after local background subtraction using a Log of Gaussian filter (radius of 1 pixel). Identified particles were limited to masked regions of dendrites, which were produced by thresholding and then eroding (radius of 1 pixel) a Gaussian blur (radius of 1 pixel) of the red fluorescence channel image. Arc/MS2 mRNA particles were defined as encompassing at least two adjacent pixels whose intensity was at least 4 standard deviations from the mean of regions of dendritic shaft devoid of particles. Restricting particles to at least two adjacent pixels minimized the inclusion of spurious puncta due to noise. Arc/MS2 particle position was calculated using the center of mass of the identified particles.
To assess the average position of Arc/MS2 particles, images of multiple spines were aligned by first subtracting background fluorescence occurring outside the imaged dendrite. Regions of interest centered on the spine base were then selected, flipped vertically if needed, and rotated until the region of the dendritic shaft around the spine base was horizontal. Orientation of the spine with respect to the proximal-distal axis of the dendrite was preserved. Aligned regions of images surrounding spine bases were averaged to generate composite images. Ratiometric images were generated by dividing the green fluorescent images by the red fluorescent images and masking the result using a standard template. Ratiometric images were normalized by fluorescence intensity measured in the dendritic shaft using a standard, nearly rectangular template beneath the spine base.
Images of the accumulation of fluorescence localized near spine bases were derived from composite, averaged green fluorescence images by subtracting an estimate of the dendritic shaft associated green fluorescence. For this analysis, dendritic shaft associated green fluorescence was assumed to be proportional to the red fluorescent protein that fills the neuronal processes. A spine base region of interest (ROI) was defined as a roughly circular region 0.35 μm in radius centered on the identified spine base. This ROI was moved if needed to center it on fluorescent puncta near the spine base. The ratio of green fluorescence to red fluorescence was calculated for two 0.3 × 0.7 μm regions positioned in the dendritic shaft flanking the ROI. This value was multiplied by the composite, averaged red fluorescence image to produce the dendritic shaft associated green fluorescence image. Background subtracted images were normalized to the level of green fluorescence in the 0.3 × 0.7 μm regions flanking the ROI. Green fluorescence accumulation near the spine base was measured as the mean fluorescence value within the circular spine base ROI.
Automated subpixel particle tracking of restricted regions (typically 3–5 μ on a side) was performed with the SpotTracker plugin for ImageJ (Sage et al., 2005). For each frame the search area was limited to 9 to 11 pixels square (0.81–1.11 μm2 per 5 sec) centered on the particle’s previous position, which corresponds to the area predicted for free diffusion of a 920 nm long mRNA (0.95 μm2 per 5 sec) calculated as in Fusco et al. (2003). High intensity green channel pixels not associated with the labeled dendrite were removed manually to avoid tracking errors. The precision of particle tracking was calculated based on values in Sage et al. (2005) in which the root mean square localization error is graphed as a function of peak signal-to-noise ratio (PSNR). Because particle movement reduces the apparent peak particle intensity in averaged images, the average of subsets of images containing at least 18 consecutive frames were used to measure peak particle intensity, after subtraction of a scaled estimate of the background. Noise was calculated by measuring the standard deviation of fluorescent intensity values in the dendritic shaft adjacent to the spine base in each frame and averaging. PSNR values were calculated by dividing the peak intensity by the noise, taking the log base 10 of this value, and multiplying by 20. The red fluorescently labeled dendritic shaft and spine neck were tracked using small rectangular regions (0.5 μm across for spine necks, 1 μm across for dendritic shaft) and calculating their center of mass. The resulting positional offsets were applied to the particle trajectories. Typical errors in dendritic shaft position were determined by measuring the center of mass at three positions within 10 μm and calculating the closest two of the three positions (mean value of 7 nm).
Neurophysiological techniques
All procedures involving living animals were approved by the Institutional Animal Care and Use Committee of the University of California Irvine. Adult male Sprague–Dawley rats were anesthetized by intraperitoneal injection of 20% urethane (500 mg/kg body weight). Supplemental injections of urethane were given approximately every 10 minutes until the animal was completely unresponsive to tail pinch. The animal was then positioned in a stereotaxic apparatus, a craniotomy was performed, and stimulating and recording electrodes were positioned stereotaxically so as to selectively activate medial perforant path projections while recording in the dentate gyrus. The stimulating electrode (an insulated tungsten microelectrode) was positioned at 4.0 lateral to the mid-line and 1.0 mm anterior to the transverse sinus. The depth of the stimulating electrode was adjusted so as to obtain a maximal evoked response in the dentate gyrus at minimal stimulus intensity. The recording electrode was a glass micropipette filled with puromycin in 0.9% saline (10 mg/ml). This was lowered into the brain at 3.5 posterior to bregma and 1.5–2.0 mm lateral to the mid-line and then positioned in the cell body layer based on responses evoked by stimulation of the entorhinal cortex. Stimulus intensity was set so as to evoke an ≈3 mV population spike; single test pulses were then delivered at a rate of 1/10 seconds for 10–15 minutes. Then three bouts of high-frequency stimulation (10 trains of 8 pulses at 400 Hz) were delivered at a rate of 1/10 seconds. Between each bout of 10 trains, 10 test responses were collected to determine the extent of synaptic potentiation. After the third bout, high-frequency trains were delivered at a rate of 1/10 seconds for 30 minutes. At the end of the period of high-frequency stimulation, animals received an anesthetic overdose and were perfused for electron microscopy with 2% paraformaldehyde / 2% glutaraldehyde in cacodylate buffer.
Electron microscopy
The brain was sectioned at 40 μm on a vibratome. Sections through the hippocampus that contained the microelectrode tract were immunostained for c-fos to document the extent of protein synthesis inhibition due to puromycin. The primary antibody was from Santa Cruz Biologicals (Santa Cruz, CA; Cat. no. SC52) diluted 1:500 in phosphate-buffered saline (PBS) with 5% normal goat serum. This antibody was raised in rabbit using a peptide containing amino acids 3–16 from human c-fos. In western blots the antibody stains a 62-Kd band corresponding to the expected molecular weight of c-fos (manufacturer’s data sheet). Additional evidence for specificity is the staining pattern in the hippocampus following activation, in which the nuclei of activated neurons are selectively stained (see Results). Prior to incubation in the primary antibody, sections in buffer were placed in boiling water for 5 minutes for antigen retrieval. Adjacent sections were postfixed with 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 hour, rinsed in ddH20 and en bloc-stained with 0.1% uranyl acetate in ddH20, rinsed in ddH20 for 2 × 10 minutes, dehydrated in increasing serial dilutions of ethanol (70%, 85%, 95%, 100% × 2) for 10 minutes each, immersed in propylene oxide for 2 × 10 minutes, incubated in propylene oxide/Spurr’s resin (1:1 mix) for 1 hour, and in Spurr’s resin overnight. The vibratome sections were flat-embedded between two sheets of “Aclar” film and polymerized overnight at 60°C.
One micrometer sections were cut using a Leica Ultra-cut UCT ultramicrotome and stained in toluidine blue (1% toluidine blue and 2% sodium borate in ddH20) at 60°C for 3 minutes, mounted on slides, and coverslipped. Ultra-thin sections of 60 nm thickness were cut, mounted on copper grids, and stained with uranyl acetate and lead citrate. Using the toluidine blue-stained sections as a guide, dendrites within the area of effective protein synthesis inhibition were viewed using a JEOL 1400 electron microscope.
Statistical analysis
Statistical tests were performed using GraphPad Prism v. 4.0 (GraphPad Software, San Diego, CA). Random points for Monte Carlo analysis were generated and the distance to the nearest spine calculated using a custom macro program written for ImageJ. Spine base-Arc/MS2 mRNA particle distances were calculated and scatter-plots generated using Microsoft Excel (Redmond, WA).
RESULTS
Arc/MS2 mRNA localizes precisely at the base of dendritic spines
We used the same combination of exogenous expression of mRNA, GFP-labeling, and live imaging of neurons in culture that we used previously to identify patterns of movement (Dynes and Steward, 2007). Here, however, we averaged consecutive images to sensitively identify Arc/MS2 mRNA particles that remained stationary over the recording interval.
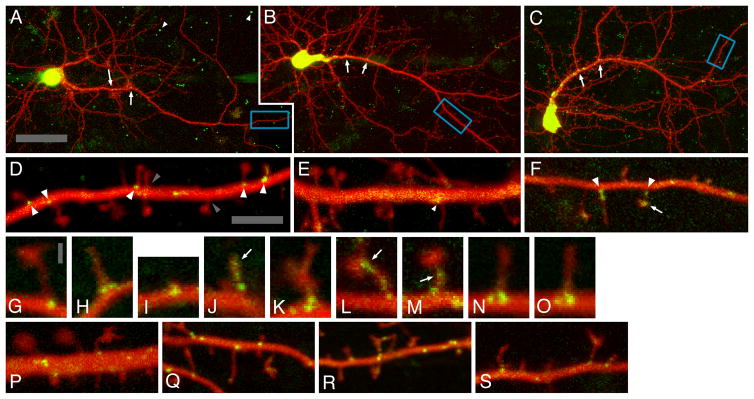
A total of seven neurons (seven independent transfections; average of 18 days in vitro [DIV]) were imaged 16–26 hours (average of 21 hours) after cotransfection with a construct encoding a GFP fusion protein (GFP-MS2-NLS) that links to a tagged mRNA containing the Arc 3′UTR, expressed from a second construct (CMV-DsRE-6xBS-rArc3-UTR). The morphology of transfected neurons was similar to those described in Dynes and Steward (2007), except that spines were visible in the more mature neurons in the present study (17–20 DIV here vs. 10–16 DIV in Dynes and Steward, 2007). Red fluorescence was present throughout the neuron, including distal dendrites and dendritic spines. GFP fluorescence was high in the cell body (Fig. 1A), and discrete GFP fluorescent particles were present throughout dendrites. A magenta/green version of Figure 1 is available online as Supporting Figure 1.
Figure 1.
Arc/MS2 and Den/MS2 mRNA particles are found near the base of dendritic spines and Arc/MS2 mRNA particles persist with protein synthesis inhibition. E18 rat cortical neurons that were cotransfected with GFP-MS2-NLS and either CMV-DsRE-6xBS-rArc3′UTR (A,D), CMV-DsRE-6xBS-no3′UTR (B,E), or CMV-DsRE-6xBS-rDendrin3′UTR (C,F). Neurons were imaged as a series of optical section that were projected brightest point (A–C) or as single optical sections which were imaged 20 times, subpixel aligned, and averaged (D–S). Merged green and red channels are shown. Red channel images were gamma-adjusted to enhance the visibility of spine necks. Arrows in (A–C) indicate accumulation of tagged mRNA at proximal branch points. Blue boxed regions in A–C correspond to the dendritic segments shown in D–F, respectively. Arrowheads indicate Arc/MS2 (D) or Den/MS2 (F) or fluorescent puncta in neurons cotransfected with GFP-MS2-NLS and CMV-DsRE-6xBS-no3′UTR (E). Gray arrowheads (D) indicate spine bases that lack Arc/MS2 mRNA particles. (G–K) Spines with associated fluorescent mRNA particles from neurons cotransfected with GFP-MS2-NLS and CMV-DsRE-6xBS-rArc3′UTR. Arrow in (J) indicates Arc/MS2 mRNA in the spine head. (L,M) Spines with associated particles from neurons cotransfected with GFP-MS2-NLS and CMV-DsRE-6xBS-rDendrin3′UTR. Arrows in (L,M) indicate Den/MS2 mRNA in the lower spine head/upper neck (L) and upper neck (M). Dendrites were imaged either 25 minutes (P) or 6 hours (Q) after or both 1 minute before (N) and 25 minutes after (O) 1 mM puromycin treatment. (R) Dendrite from Arc/MS2 transfected neuron treated for 6 hours with 500 μM cycloheximide. (S) Dendrite from Arc/MS2 transfected neuron treated for 4 hours with 20 μM D-AP5. A magenta-green version of this figure has been provided as Supporting Figure 1 for the assistance of colorblind readers. Scale bars = 40 μm in A (applies to B,C); 5 μm in D (applies to E,F,P–S); 1 μm in G (applies to H–O).
Spine-bearing dendritic segments selected for imaging and analysis ranged from 24–207 μm from the soma (average of 100 μm), with a combined length of 479 μm. Twenty consecutive images (one every 10 seconds) of a dendritic segment were acquired from a single focal plane, aligned and averaged (Fig. 1D). With this averaging approach, Arc/MS2 particles that are stationary during the sampling interval appear as distinct well-defined spots; particles that move rapidly during the recording period are not visible.
A total of 235 Arc/MS2 mRNA particles were identified by thresholding of averaged images (average of one particle per 2.0 μm of dendrite) and 216 dendritic spines were identified (average of one spine per 2.2 μm of dendrite). In these images, many stationary Arc/MS2 mRNA particles were localized at or near the base of dendritic spines and many spines had underlying Arc/MS2 particles (Fig. 1D,G–K). Importantly, only the spines that extend in the horizontal plane are visible in the images; spines that extend perpendicular to the surface would be invisible. Thus, some stationary Arc/MS2 particles may be associated with spines that are not visible in these images. However, not all spine bases had a visible Arc/MS2 mRNA particle nearby (Fig. 1D).
In control experiments, constructs lacking the Arc 3′UTR (see Materials and Methods) were transfected and neurons were imaged using similar procedures as above; these control constructs were not localized at spines (Fig. 1B,E). These analyses involved a total of six neurons and dendrites (four independent transfections; average of 18 DIV) similarly cotransfected 16–25 hours before imaging (average of 20 hours). The dendritic segments imaged ranged from 33–269 μm from the soma (average of 145 μm), with a combined length of 444 μm; 200 dendritic spines were identified (an average of one spine per 2.2 μm of dendrite). GFP fluorescence was found in the cell body (Fig. 1B). Fluorescence in dendrites was at lower levels than in the case of neurons transfected with Arc/MS2 constructs and was diffuse. In neurons with high levels of fluorescence, there were higher levels of fluorescence at dendritic branch points (Fig. 1B) and some indication of higher levels of fluorescence near the base of some dendritic spines (Fig. 1E). However, discrete fluorescent particles were much less common than in neurons labeled with Arc/MS2 constructs, consistent with previous observations (Dynes and Steward, 2007). Only 13 fluorescent accumulations were classified as particles, at a density 17-fold less than for Arc/MS2 mRNA particles (0.29 vs. 4.9 per 10 μm of dendrite).
Arc/MS2 mRNA particles were found at the base of different types of dendritic spines, including mushroom, thin, and stubby spines (Fig. 1G–I, respectively). Arc/MS2 mRNA particles were also localized at the base of other dendritic protuberances with more variable shapes that may or may not be spines (Fig. 1K). Arc/MS2 mRNA particles were found at several sites at or near spine bases, either directly beneath the site at which the spine neck and dendritic shaft adjoin (Fig. 1G) or displaced ≈200–300 nm from the spine base (for example, Fig. 1H). Displacement was more evident when two particles were present at the base of the same spine (Fig. 1H,N). Less frequently, Arc/MS2 mRNA particles were found in spine heads and necks (Fig. 1J). Many of these particles were found in the lower spine neck (Fig. 1J; also see Fig. 3F–J). In some cases less discrete Arc/MS2 fluorescence extended into the upper portion of the spine neck and spine head.
Figure 3.
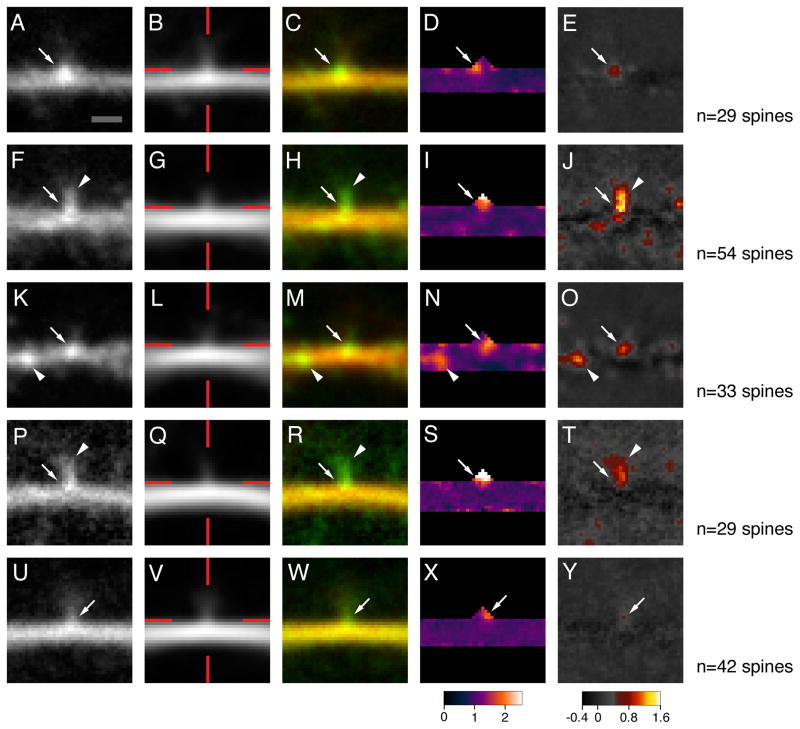
Arc/MS2 mRNA accumulates at a distinct and reproducibly positioned site at the base of dendritic spines. Regions of images surrounding the bases of dendritic spines were aligned by the spine base and averaged. Green fluorescence, red fluorescence, green-red composite, ratiometric (pseudocolored), and dendritic shaft fluorescence subtracted green fluorescence (pseudocolored) images are shown for three neurons transfected with both GFP-MS2-NLS and CMV-DsRE-6xBS-rArc3′UTR (A–E,F–J,K–O) and one each with GFP-MS2-NLS and CMV-DsRE-6xBS-rDendrin3′UTR (P–T) and GFP-MS2-NLS and CMV-DsRE-6xBS-no3′UTR (U–Y). Red lines in B,G,L,Q,V indicate the X-and Y-axis positions of the aligned spine bases. Arrows indicate accumulation of green fluorescence at the bases of dendritic spines; arrowheads indicate accumulation of green fluorescence in spine necks (F,H,J,P,R,T) and in the dendritic shaft (K,M,N,O). Scale bar = 1 μm; 1 pixel = 0.1 μm.
In order to quantify this association, we determined the position of every Arc/MS2 mRNA particle and spine base in the imaged dendrites. The position of spine bases was determined visually, while the position of Arc/MS2 mRNA particles was defined by the particle’s center of mass. Fourteen percent (3/21) to 52% (15/29; average of 32%, 69/216) of spines on a dendritic segment had an Arc/MS2 mRNA particle at or near the spine base (defined as within 0.35 μm), while 17% (5/29) to 71% (15/21) had no visible particle within 1 μm (average 33%; 71/216). In addition, between 23% (5/22) and 44% (15/34) of particles in a dendritic segment lie within 0.35 μm of a spine base (average 30% 71/235). Association with the spine base was not dependent on fluorescence intensity of the particle, as both bright and dim particles were found within 0.35 μm of a spine base (Fig. 1D, right two small arrowheads).
Monte Carlo analysis
A close association of Arc/MS2 particles with spine bases is evident in the confocal images and distance measurements, but we wanted to obtain a more general measure of the degree of association to assess its significance. In formulating an analysis, one issue to consider is that spines are not uniformly distributed along dendrites of neurons in culture. Instead, on some dendrites there are clusters of spines that are closely spaced separated by stretches of several μm or more in length that were relatively spine free (see, for example, Fig. 1D–F). About 30% of particles lie within 0.35 μm of a spine base, but this excess of small Arc/MS2 mRNA particle-spine base distances might result, in part, from the clustering of spines. Indeed, clustering would lead to an apparent excess of small Arc/MS2 mRNA particle-spine base distances even if mRNA particle and spine base positions were independent. Therefore we sought an assessment method that would be independent of local variation in spine density.
Accordingly, we used Monte Carlo analysis to assess the selectivity of the localization. Here we compared measurements of the distance to the nearest spine base for each Arc/MS2 mRNA particle with the same number of measurements for randomly positioned points within the same dendritic segment. In this analysis spine clustering will have equal effects on the distances of mRNA particles and random points to spine bases. The set of spine base distances for each dendrite (n = 7) were combined for both Arc/MS2 mRNA particles and randomly spaced points (235 total distances each). The distribution of Arc/MS2 mRNA particles was significantly different from a random distribution (P = 0.019, Mann–Whitney U, two-tailed), with a higher proportion of shorter distances to dendritic spine bases than predicted by chance.
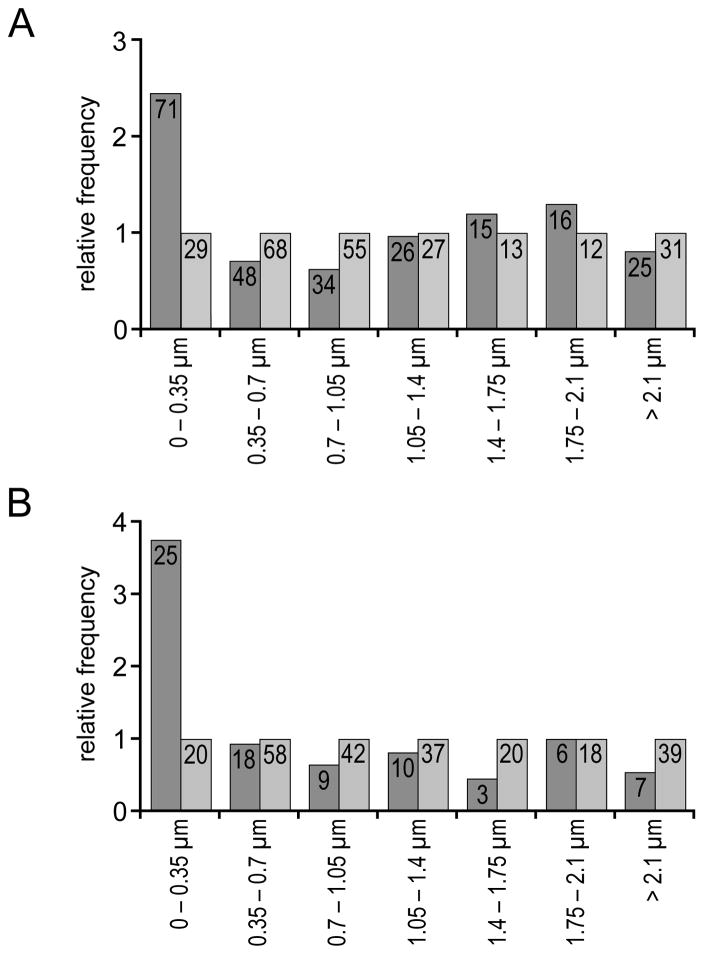
How precise is the localization of Arc/MS2 mRNA at spine bases? Uncertainty in the position of the spine base, the size of the localization site, and the size of the Arc/MS2 mRNA particle limits the precision with which this can be assessed. If a spine base-particle distance of 0.35 μm is used as an upper limit for colocalization, then Arc/MS2 mRNA particles are found to colocalize with spine bases significantly more frequently than random particles (P < 0.0001; χ2, Fisher’s exact test). Significant localization of Arc/MS2 mRNA particles at spine bases is found when shorter (0.2 μm, P = 0.0023; 0.25 and 0.3 μm, P < 0.001) or longer (0.4 and 0.45 μm, P < 0.001) distance criteria are used as well. The selectivity of Arc/MS2 localization is illustrated in the histogram in Figure 2A, which has been normalized to the number of random distances in each distance class. Enrichment of Arc/MS2 mRNA particles was limited to the immediate vicinity of the spine base, as fewer Arc/MS2 mRNA particles than random points were found at distances between 0.35 and 1.05 μm from the spine base.
Figure 2.
Arc/MS2 and Den/MS2 mRNA particles are nonrandomly positioned near the base of dendritic spines. A: The relative frequency of distances to the nearest spine base for Arc/MS2 mRNA particles (dark gray bars) and randomly placed points (light gray bars) was plotted with a 0.35-μm interval. The relative frequency was normalized to the number of random distances in each distance class. The number of observation in each class is listed at the top of each bar. Note that fewer Arc/MS2 mRNA particles than random points are found at distances between 0.35 and 1.05 μm from the spine base. B: The relative frequency of distances to the nearest spine base for Den/MS2 mRNA particles (dark gray bars) and three times as many randomly placed points (light gray bars).
Because of the paucity of discrete particles in neurons that were cotransfected with control constructs that lacked the Arc 3′UTR, Monte Carlo analysis was of questionable reliability. However, there was a difference between the distributions of fluorescent particles (13 total) and random points (260 total), and this difference was significant (P = 0.029, Mann–Whitney U, two-tailed), with a higher proportion of shorter distances to dendritic spine bases than predicted by chance. Only 2 of 13 particles were found within 0.35 μm of a spine base.
Averaging across dendritic segments
It is possible that the mRNA that lacks the Arc 3′UTR might accumulate at many or even most spine bases, but this might not be apparent because the mRNA did not aggregate into discrete mRNA particles. To address this question, we combined images of multiple spines from a given dendrite to look for higher levels of fluorescence in the region of the spine base. Images that include the area surrounding the base of each dendritic spine on a given dendrite (between 21 and 54 spines) were aligned by the base and averaged. Three examples are shown in Figure 3A–C,F–H,K–M. Relative concentration of Arc/MS2 mRNA at the spine was calculated by dividing the green fluorescence image by the red fluorescence image (Fig. 3D,I,N) and relative accumulation at the spine base was calculated by subtracting out the average fluorescence in the dendritic shaft (Fig. 3E,J,O). By each of these three measures—simple average, relative concentration, and relative accumulation—Arc/MS2 fluorescence was enriched at a discrete site at the base of dendritic spines in six out of seven dendrites. The existence of discrete distributions found in averaged images of dendritic spines implies that the position of Arc/MS2 mRNA particles relative to the spine base does not vary substantially between spine bases in a given dendritic segment. In one dendrite (Fig. 3A–E), the average accumulation of fluorescence for Arc/MS2 mRNA was consistently displaced to one side of the spine base, whereas in another (Fig. 3F–J), fluorescence was nearly coincident with the spine base. Higher levels of fluorescence were also found in the spine neck (Fig. 3F–J) and ≈1.5 μm cell proximal to the spine base (Fig. 3K–O).
Green fluorescence from neurons cotransfected with constructs lacking the Arc 3′UTR was also concentrated at the spine base, but the degree of enrichment was less than in the case of transcripts containing the 3′UTR of Arc (Fig. 3U–Y). One possible explanation is that ribosomes are also concentrated beneath spines, which could capture any transcripts capable of translation (Steward and Levy, 1982; Ostroff et al., 2002).
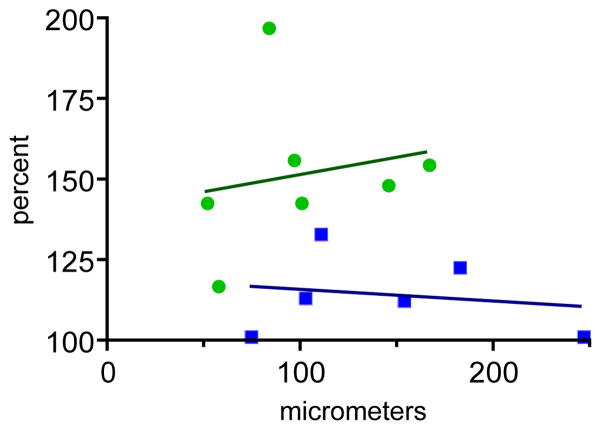
Does inclusion of the Arc 3′UTR enhance accumulation of mRNA at the spine base? We measured relative accumulation in composite, averaged green fluorescence images in which fluorescence associated with the dendritic shaft was subtracted out (Fig. 3E,J,O,Y). Mean values of fluorescent puncta located near the averaged spine base were compared for neurons cotransfected with constructs that contain or lack the Arc 3′UTR. One potential complication of the analysis is that the dendritic segments imaged were at different distances from the soma, and on average the segments imaged from neurons transfected with constructs containing the Arc 3′UTR were closer to the cell body than those transfected with constructs lacking the Arc 3′UTR. To assess whether this was a significant variable, mean fluorescence at spine bases was plotted as a function of the distance of the imaged dendritic segment from the cell body. Figure 4 shows a 2D scatterplot of values derived from neurons cotransfected with constructs that contain or lack the Arc 3′UTR. In both cases the degree of fluorescence accumulation was not related to the distance of the dendritic segment from the cell body. Accordingly, we compared the mean accumulation for all neurons in each population. Subtraction-adjusted fluorescence at the spine base was significantly higher in neurons cotransfected with constructs containing the Arc 3′UTR than with those lacking the Arc 3′UTR (51% and 14% of dendritic shaft associated fluorescence, respectively; P = 0.0029, unpaired t-test, one-tailed). Thus, enrichment of mRNA at the base of dendritic spines can be conferred by sequences in the Arc 3′UTR.
Figure 4.
Inclusion of the Arc 3′UTR leads to enrichment of tagged mRNA at the spine base. Two-dimensional scatterplot of mean accumulation of fluorescence at the spine base (as a percentage of dendritic shaft green fluorescence) versus distance from the cell body. Neurons cotransfected with GFP-MS2-NLS and CMV-DsRE-6xBS-rArc3′UTR are plotted as circles (regression is indicated by the solid line), and those cotransfected with GFP-MS2-NLS and CMV-DsRE-6xBS-no3′UTR are plotted as squares (regression is indicated by the dotted line). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
mRNA containing the 3′UTR of Dendrin also localizes to the base of dendritic spines
It was of interest to assess whether other known dendritic mRNAs would show the same high degree of localization at spine bases. For this, neurons were transfected with constructs in which the Arc 3′UTR was replaced with the 3′UTR from Dendrin, another mRNA that is localized in the dendrites of many forebrain neurons (Herb et al., 1997; Kremerskothen et al., 2006). Analyses involved a total of 10 neurons and dendrites (seven independent transfections; average of 18 DIV). The dendritic segments imaged ranged from 10–225 μm from the soma (average of 141 μm), with a combined length of 719 μm. A total of 243 dendritic spines were identified (an average of one spine per 2.9 μm of dendrite). Den/MS2 mRNA assembled into particles that were delivered into dendrites by 16–26 hours posttransfection (Fig. 1C,F). A total of 78 Den/MS2 mRNA particles were identified by thresholding of averaged images (an average of one particle per 9.3 μm of dendrite). These were localized in a way that was largely similar to Arc/MS2 mRNA particles (Fig. 1F, also see Figs. 2B, 3P–T). Observed distances of Den/MS2 from spine bases differed significantly from distances of random particles (P < 0.0001 Mann–Whitney U, two-tailed). The fraction of particles within 0.35 μm of the spine base also differed significantly from random particles, χ2, Fisher’s exact test (P < 0.0001). However, the density of Den/MS2 mRNA particles was 4–5-fold lower than Arc/MS2 mRNA particles (1.1 particles per 10 μm for Den/MS2 vs. 4.9 particles per 10 μm for Arc/MS2) and fluorescence from Den/MS2 mRNA was more frequently found in the upper spine neck and lower spine head (Fig. 1L,M). Thus, other mRNAs are capable of localizing to the spine base, and the pattern of localization depends in part on the sequence of the mRNA.
Localization is independent of active translation
Localization of Arc/MS2 mRNA at spine bases might occur because the mRNA is engaged with ribosomes located at spine bases, the mRNA is docked independent of translation, or both. We sought to assess the requirement for active translation by dissociating ribosome-mRNA complexes within living neurons by applying the chain terminating translational inhibitor puromycin and the peptidyl transferase translational inhibitor cycloheximide. Twenty-five minutes after treatment of transfected neurons (n = 5; five independent transfections) with puromycin (1 mM), Arc/MS2 mRNA particles were found at the base of dendritic spines (n = 36) in a manner similar to untreated neurons (Fig. 1P). Moreover, particles at spine bases were found to persist after puromycin application in cases where the same spine base could be clearly identified before and after treatment (Fig. 1N,O). This was true for as many as 77% (10/13) of spine bases from a given neuron. The size and relative position of spine-associated particles were largely unaffected by puromycin treatment. Arc/MS2 mRNA particles were also found at the base of dendritic spines after 6 hours in 1 mM puromycin (Fig. 1Q; n = 5 neurons), although fewer particles were typically seen and dendritic varicosities were formed (Fig. 1Q). Arc/MS2 mRNA particles were also found at the base of dendritic spines after 6 hours in 500 μM cycloheximide (Fig. 1R; n = 5 neurons). Thus, localization of Arc/MS2 mRNA at the base of dendritic spines does not depend on ongoing translation.
Polysome rosettes remain intact and associated with dentate granule spines after treatment with puromycin in vivo
As Arc/MS2 mRNA associates with the base of dendritic spines independently of active translation, we wondered if ribosomes might associate with the base of dendritic spines independently of active translation as well. To test this, we assessed whether the spatial organization of ribosomes at spines would persist following blockade of protein synthesis with puromycin, which dissociates mRNA from ribosomes by serving as a polypeptide chain terminating tRNA analog. If ribosomes are anchored, dissociation of ribosomes from mRNA should not affect polyribosome clusters. Alternatively, if ribosome clusters are held together by the mRNA, ribosomes should disperse from the rosettes with puromycin treatment. For this experiment we turned to an in vivo model where we could directly assess efficacy of puromycin in blocking protein synthesis.
High-frequency stimulation (HFS) of perforant path projections to the dentate gyrus induces expression of a number of immediate early genes (IEGs), which can be measured using immunocytochemistry. Figure 5A illustrates this by immunostaining for the IEG c-fos 30 minutes after HFS. Local infusion of puromycin via micropipette recording electrodes blocks protein synthesis in a small region, documented by a blockade of activity-induced c-fos synthesis around the recording micropipette (Fig. 5A). Electron microscopic examination of dendrites in the area with confirmed blockade of protein synthesis revealed a continued presence of polyribosome rosettes at the base of dendritic spines (Fig. 5B–D). Thus, polyribosome positioning at spine bases does not depend on ongoing translation.
Figure 5.
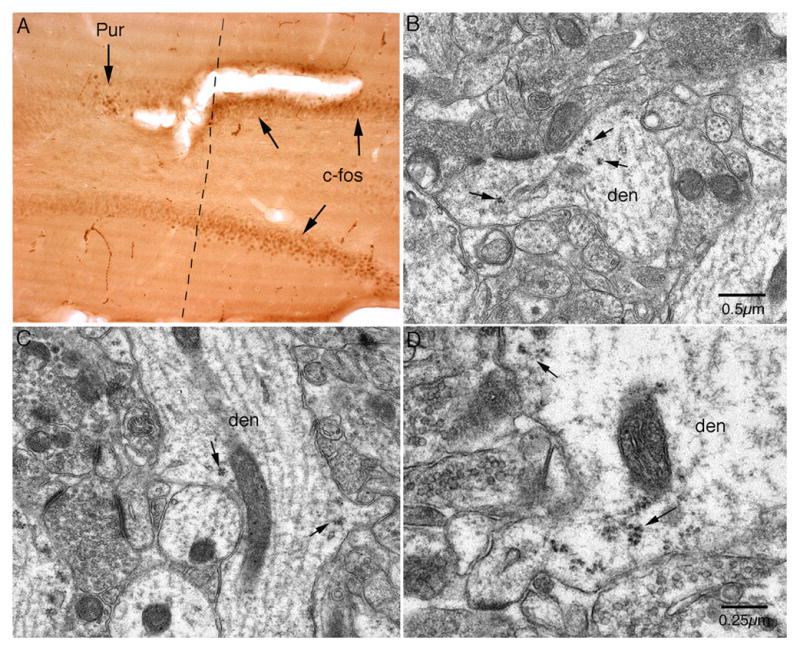
Polyribosome complexes remain intact and associated with spine bases after puromycin treatment. Puromycin (10 mg/ml in saline) was delivered into the dentate gyrus through the recording micropipette for 1 hour while the perforant path was stimulated via an electrode in the entorhinal cortex. A: Section of the dentate gyrus immunostained with an anti-c-fos antibody. The dotted line indicates the region of protein synthesis inhibition, defined by the absence of c-fos immunostaining. B–D: Electron micrographs of puromycin-treated dendrites. Spines with associated polyribosome clusters or “rosettes” are indicated by small arrows.
Tracking Arc/MS2 mRNA at the spine base
Images of Arc/MS2 mRNA derived from averaging reveal localization patterns that are a few hundred nanometers across, discrete, stable, and reproducibly positioned across dendrites. This is surprising because the projected length of Arc/MS2 mRNA (2,627 nucleotides) is over 900 nm when extended (at 0.35 nm/nucleotide), and it suggests that Arc/MS2 mRNA docks at a discrete site at the spine base. Therefore, it became of interest to assess the microscale positioning of the GFP tag using single particle tracking to identify time periods in which particle position is highly stable, consistent with physical docking of mRNA. Movement of the GFP tag during tracking could be due to several different causes, such as structural rearrangement of the mRNA, movement of the mRNA as a whole, movement of any structure to which the mRNA might be bound, or any combination of these. Nevertheless, if the GFP tag exhibits periods of positional stability, this jointly constrains mRNA structure and position.
To assess the degree to which GFP tag position was stable over time, we tracked the movements of the most intensely labeled Arc/MS2 particles with subpixel resolution (Sage et al., 2005; Dynes and Steward, 2007). An example is illustrated in Figure 6. In these cases the particle could be clearly tracked, as illustrated in the kymographs of particle position over time in the X and Y dimensions (Fig. 6C,D).
Figure 6.
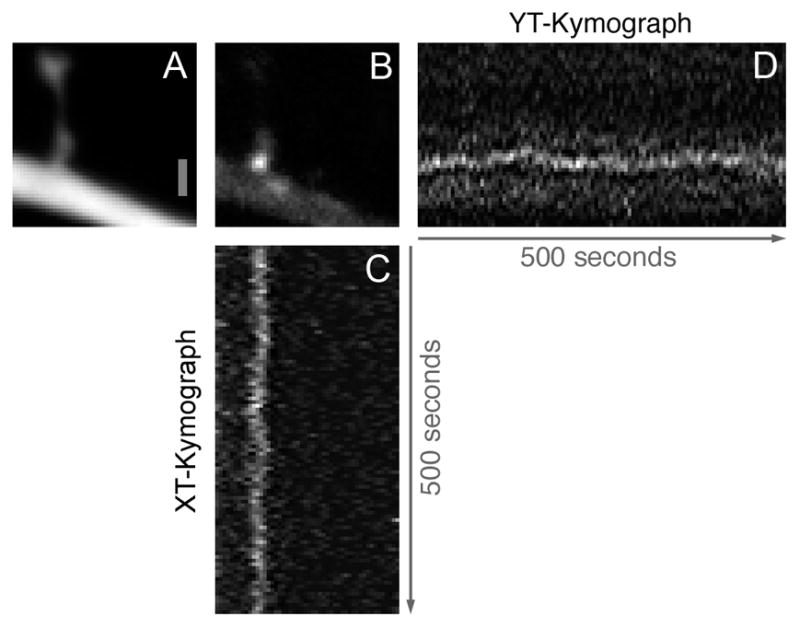
Bright single Arc/MS2 mRNA particles can be consistently tracked from single images of time-lapse image series. A,B: Average of 100 consecutive images of a dendritic spine labeled with RFP (A) with associated Arc/MS2 mRNA particle (B). C,D: XT and YT kymographs of Arc/MS2 mRNA particles, respectively. Scale bar = 1 μm in A (applies to B–D).
The peak signal to noise ratio for all tracked particles averaged 7.6, resulting in an average root mean square localization error of 40 nm calculated as in Sage et al. (2005). To compensate for movement of the preparation, regions from the red fluorescently labeled dendritic shaft and spine neck were also tracked using their center of mass and the resulting positional offsets were applied to the measured particle positions. In this way, the position of the dendrite itself provided the control for any drift in overall position or fluctuations in data capture.
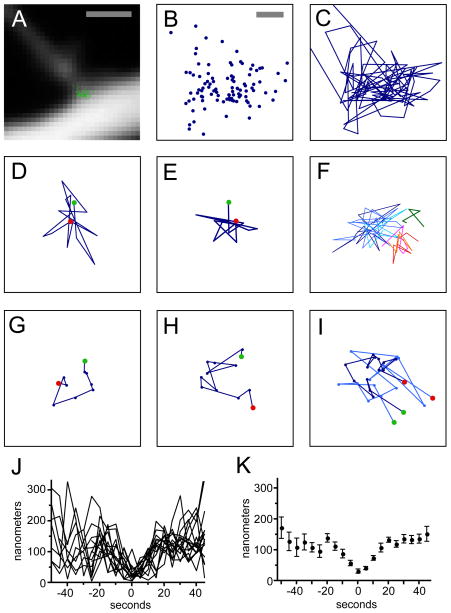
Plots of the position of GFP puncta over the course of 500-second imaging sessions (1 per 5 seconds; 12 spines from 5 neurons) revealed that particles largely kept to a circumscribed area roughly 200–300 nm diameter in the region of the spine base (Fig. 7A). GFP puncta positions were not uniformly distributed, instead exhibiting regions of high and low density (Fig. 7B). Tracing the trajectory of the GFP puncta revealed extensive movement within this highly restricted spatial domain (Fig. 7C); mean displacement was 135 nm per 5 seconds. GFP puncta trajectories were examined for bouts in which the mean displacement between successive positions was small. To our surprise these bouts corresponded to two small, circumscribed, but clearly defined patterns of movement. The first class, termed stellate trajectories, exhibited a central region ≈50 nm in diameter and extended ≈50–150 nm from the center (Fig. 7D,E). These trajectories lasted as long as 105 seconds. Movements of the GFP puncta within stellate trajectories were typically short and back-and-forth. Twenty-three stellate trajectories were identified lasting at least 30 seconds. These trajectories constituted 20% of the 5940 second total imaging time.
Figure 7.
Arc/MS2 mRNA trajectories at the spine base. A: Subpixel Arc/MS2 mRNA particle positions (green circles) from 100 consecutive images superimposed upon an averaged image of the spine base. B: Subpixel Arc/MS2 mRNA particle positions (blue circles) from the same image set at higher magnification. C: Trajectory of Arc/MS2 mRNA particle from the same image set. D,E: Stellate trajectories of Arc/MS2 mRNA particles lasting 70 and 55 seconds, respectively. The green and red circles in (D,E,G,H,I) indicate the beginning and ending of particle trajectories, respectively. F: Stellate trajectories from a neuron treated with D-AP5. Three colors of blue show three overlapping trajectories covering 225 of 400 seconds of imaging. Three colors of red-orange show another three overlapping trajectories. A seventh spatially restricted trajectory is shown in green. G,H: Crescent trajectories lasting 45 (G) and 55 (H) seconds. I: Two overlapping crescent trajectories (dark and light blue) separated by 115 seconds. J,H: Plots of the distance to the crescent trajectory starting point. J: All distances plotted; H: mean and SEM, respectively. Some values are missing between −50 and −5 seconds because they are part of earlier crescent trajectories (n = 4) or crescent trajectories that began less than 50 seconds after the start of the time-lapse imaging session (n = 4). Bounding squares are 3 μm (A) and 500 nm (B–I). Scale bars = 1 μm in A; 100 nm in B–I.
The GFP puncta revisited the site of previous stellate trajectories over the course of 500 seconds. The stellate trajectories shown in Figure 7F (treated with D-AP5; see below) are largely restricted to one centrally located site, but additional, adjacent, and largely nonoverlapping trajectories were also seen (Fig. 7F). However, it cannot be excluded that the presence of these additional sites was due to errors in tracking the dendrite over time. Nevertheless, given that the core of these movement patterns is only 50–100 nm in diameter, these observations are consistent with the physical docking of Arc/MS2 mRNA at a fixed site accompanied by small shifts in the structure or orientation of the mRNA.
To assess whether these micromovements were dependent on synaptic activity, neurons were treated with the NMDA receptor antagonist D-AP5 (20 μM) for 4 hours. Stellate trajectories were still evident after 4 hours of NMDA receptor blockade (Fig. 7F; n = 4 spines from 2 neurons; also see Fig. 1S).
The second class of trajectories were ones in which movements occurred in rough arcs with a radius from 40–100 nm (Fig. 7G,H). To avoid confusion with the name of the Arc gene, we have termed these “crescent trajectories.” Movements in crescent trajectories were on the whole progressive but typically exhibited short kinks or larger back-and-forth movements, which were commonly found at the beginning and ending of the trajectories. Crescent trajectories persisted for 30–60 seconds, and one crescent trajectory was seen on average every 400 seconds (15 total at 12 spines from 5 neurons; 5,940 seconds of total imaging time). Over the course of a 500-second imaging session multiple crescent trajectories were seen in the same relative position at the spine base, extending in the same direction (Fig. 7I). However, some back-and-forth trajectories that occurred at the site of previous or subsequent crescent trajectory did not progress (n = 6); in two of these cases particle positions when plotted formed an arc (data not shown).
As individual crescent trajectories were progressive and extended over similar periods of time, we aligned them so they could be examined collectively (Fig. 7J). We estimated the probable starting position of the crescent trajectory and measured the distance of the GFP puncta to this point over time. Distance functions were aligned so their minima were at time 0 seconds. Alignment of trajectories revealed that movements were stereotyped. Strikingly, the pattern of movement preceding the crescent trajectory was also stereotyped, with the position of GFP puncta being progressively restricted as it moves toward the start of the crescent trajectory. The rate of movement of the GFP puncta away from the starting point (t = 5 to 20 seconds), as well as rate of approach of the GFP puncta toward the start (t = −20 to 0 seconds), were calculated from the slope of lines derived from GFP puncta distance plots via linear regression (Fig. 7K). The GFP puncta approached the start at a mean rate of 5.4 ± 0.7 nm/sec (95% confidence limits 4.0 and 6.8 nm/second; r2 = 0.45) and moved through the crescent trajectory at 6.1 ± 0.8 nm/sec (95% confidence limits 4.5 and 7.8 nm/sec; r2 = 0.49). These rates strongly constrain possible mechanisms underlying this pattern of movement.
DISCUSSION
Arc mRNA is unique among known dendritic mRNAs because it accumulates in activated dendritic domains in response to synaptic activity strong enough to activate NMDA receptors (Steward et al., 1998; Steward and Worley, 2001). This activity-dependent localization inspired the hypothesis that Arc may act selectively at activated synapses, thereby creating a synapse-specific link between synaptic activation and subsequent protein synthesis-dependent consolidation (Steward et al., 1998; Steward and Worley, 2001). Unresolved, however, was whether Arc mRNA localized at individual synapses or rather in more broad dendritic domains. Here we show that mRNA containing the Arc 3′UTR localizes with precision at or very near the base of dendritic spines of neurons in culture. Thus, a mechanism exists to target Arc mRNA not only to particular regions of a dendrite, but also to a microdomain at the base of individual spine synapses. This highly precise localization means that mRNA targeting could in principle restrict gene expression induced by activity to individual synapses.
Previous studies have reported that mRNA can be found throughout dendrites of neurons in culture, including near spine bases (Martone et al., 1996; Tiruchinapalli et al., 2003; Kao et al., 2010). The very precise positioning at spine bases that we document here has not been previously reported for other dendritic mRNAs, however. One possibility is that Arc mRNA is targeted in a more precise way to individual spines than the other mRNAs whose localization has been examined at the submicrometer level (Martone et al., 1996; Tiruchinapalli et al., 2003; Kao et al., 2010). There may also be a technical explanation; for example, Arc/MS2 mRNA may be present at lower levels than the endogenous mRNAs that have been previously examined, and the lower abundance might make a selective localization more apparent. We also cannot exclude the possibility that Arc/MS2 mRNA localizes more precisely than endogenous Arc mRNA. So far, the localization of endogenous Arc mRNA has not been assessed at high resolution as done here, either in intact neural tissue or in neurons in culture. There is no obvious reason, however, why Arc/MS2 mRNA would localize more precisely than endogenous Arc mRNA.
Stable and selective positioning of mRNAs and ribosomes at subcellular sites has been widely reported, as has the existence of translation “hot spots” (Cervera et al., 1981; Bassell et al., 1994; Aakalu et al., 2001; Ju et al., 2004; Dieterich et al., 2010). The patterns of mRNA localization we observe here are far more precise and stereotyped than would be expected for simple anchoring. The mRNA localization site at the spine base is small, discrete, stable, and reproducibly positioned across dendrites. Single particle tracking indicates that the core of the mRNA docking subdomain is on the order of 100 nm or less. It has long been known that translational machinery including polyribosomes and associated membranous cisterns is preferentially localized at spine bases in mature neurons in vivo (Steward and Levy, 1982) and we show here that the positioning persists even after inhibition of translation with puromycin. Taken together, these observations provide compelling evidence for the existence of a previously unknown structure, or structures, at the spine base that mediates docking of mRNA and ribosomes upon which mRNA is bound and translated.
We have no evidence as to this structure’s molecular composition, although molecules that are concentrated at spine bases are candidates. For example, members of the septin protein family are selectively positioned at the spine base (Tada et al., 2007; Xie et al., 2007). Very recently, the RNA binding protein Fxr1p, a member of the Fragile-X Mental Retardation protein family, was shown to share this localization pattern (Cook et al., 2011). Like Arc/MS2 mRNA, Fxr1p is found only at a subset of spines in neurons in culture, and it is more frequently found at the base and lower spine neck than in the spine head. The close correspondence between these two localization patterns invites the speculation that Fxr1p might bind to Arc mRNA in dendrites at spines.
At some spines, Arc/MS2 mRNA particles were also stably positioned to the side of the spine base or in the lower spine neck in addition to its more central position at the spine base. It is unclear whether the structure mediating localization at the base extends or moves to these locations, or whether there are additional structures surrounding the spine base.
Immobilization of ribosomes during translation might account for an apparently puzzling submicron mRNA movement pattern we observe. At the spine base, GFP tags on mRNA move slowly and progressively in patterns we have termed crescent trajectories. The size, shape, position, and velocity of crescent trajectories are consistent with the movement of single mRNAs through anchored polyribosome complexes. This model also accounts for the striking pattern of progressively restricted and stereotyped movements of the GFP tag preceding the start of crescent trajectories. In the DNA construct we use, the GFP tag follows the stop codon of the red fluorescent protein open reading frame. In this case, movements preceding crescent trajectories would correspond to the approach of the stop codon to the first ribosome of the polyribosome complex as the mRNA is translocated during elongation. Movement through the crescent trajectory would correspond to movement of the stop codon as it passes in turn each ribosome of the polyribosome complex. Further study is necessary to fully establish the nature of these movements, as to our knowledge translocation of individual mRNAs during translation has never been documented in living cells (Marshall et al., 2008; Aitken et al., 2010). If the movement does reflect translation of a single mRNA, this implies that individual Arc/MS2 particles contain only a single Arc/MS2 mRNA molecule.
Highly specific patterns of subcellular localization are indicators of specialized cellular functions. Docking of Arc mRNA might enable a rapid translational response to changing patterns of synaptic stimulation (Park et al., 2008; Waung et al., 2008), or allow for rapid translocation of Arc mRNA into the spine head, as occurs with Homer 1a/Vesl 1S (Okada et al., 2009), in response to particular synaptic signals. Docking of Arc mRNA might allow for local or cotranslational delivery of Arc protein to specific subsynaptic sites. Arc protein interacts with the endocytotic machinery that internalizes AMPA receptors (Chowdhury et al., 2006; Rial Verde et al., 2006) and it is noteworthy that endosomes are positioned at the base of dendritic spines (Wang et al., 2008). The proximity between the sites of Arc synthesis and Arc function could allow for the spatially linked and synapse specific expression of Arc protein as may occur in mGluR1-dependent synaptic depression (Park et al., 2008; Waung et al., 2008).
The existence of a specialized microdomain for mRNA docking and translation at the entrance to the spine supports the view that newly synthesized protein in dendrites may act preferentially or exclusively at a single, immediately adjacent synapse (Wang et al., 2008). The critical question for understanding Arc’s role in synaptic modification is whether Arc protein made at the spine base or in the spine proper is targeted exclusively to that synapse or is available to other nearby synapses. The answer to this question will constrain models of Arc protein function. For example, if Arc protein is involved in synaptic rescaling (Shepherd et al., 2006), and is targeted only to the synapse at which it is made, then Arc-dependent homeostasis only occurs in dendritic regions and at individual synapses where Arc mRNA is targeted. Further studies will be required to determine the destination of Arc protein that is made at the synapse.
Supplementary Material
Acknowledgments
We thank Ilse Sears-Kraxberger for performing the electron microscopy, Kelly Matsudaira Yee for performing the immunohistochemistry, and Jason Shepard for assistance in cloning the rat Dendrin 3′UTR. We thank also Roderick Hori, Alison Barth, and Gail Lewandowski for helpful discussion as well as comments on a previous version of the article. Author contributions: J.L.D. performed all the experiments except for electron microscopy and immunohistochemistry. Electron microscopy was performed by Ilse Sears-Kraxberger and O.S. Immunohistochemistry was performed by Kelly Matsudaira Yee. Image analysis and statistical testing was performed by J.L.D. The article was written by J.L.D and O.S.
Grant sponsor: National Institutes of Health (NIH); Grant number: NIH-NS12333 (to O.S.).
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Aakalu G, Smith WB, Jiang C, Nguyen N, Schuman EM. Dynamic visualization of dendritic protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Aitken CE, Petrov A, Puglisi JD. Single ribosome dynamics and the mechanism of translation. Annu Rev Biophys. 2010;39:491–513. doi: 10.1146/annurev.biophys.093008.131427. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Singer RH, Kosik KS. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- Bramham CR. Local protein synthesis, actin dynamics, and LTP consolidation. Curr Opin Neurobiol. 2008;18:524–531. doi: 10.1016/j.conb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Cervera M, Dreyfuss G, Penman S. Messenger RNA is translated when associated with the cytoskeletal framework in normal and VSV-infected HeLa cells. Cell. 1981;23:113–120. doi: 10.1016/0092-8674(81)90276-2. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Sanchez-Carbente MDR, LaChance C, Radzioch D, Tremblay S, Khandjian EW, DesGroseillers L, Murai KK. Fragile X related protein 1 clusters with ribosomes and messenger RNAs at a subset of dendritic spines in the mouse hippocampus. PLOS One. 2011;6:e26120. doi: 10.1371/journal.pone.0026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes JL, Steward O. Dynamics of bi-directional transport of Arc mRNA in neuronal dendrites. J Comp Neurol. 2007;500:433–447. doi: 10.1002/cne.21189. [DOI] [PubMed] [Google Scholar]
- Dynes JL, Steward O, editors. Dendritic transport of mRNA, the IEG Arc, and synaptic modifications involved in memory consolidation. Vol. 4. Oxford: Elsevier; 2008. [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific induction of the immediate early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent Arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr Opin Neurobiol. 2005;15:599–606. doi: 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Herb A, Wisden W, Catania dMV, Marechal D, Dresse A, Seeberg PH. Prominent dendritic localization in fore-brain neurons of a novel mRNA and its product, dendrin. Mol Cell Neurosci. 1997;8:367–374. doi: 10.1006/mcne.1996.0594. [DOI] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- Kao DI, Aldridge GM, Weiler IJ, Greenough WT. Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci U S A. 2010;107:15601–15606. doi: 10.1073/pnas.1010564107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremerskothen J, Kindler S, Finger I, Veltel S, Barnekow A. Postsynaptic recruitment of Dendrin depends on both dendritic mRNA transport and synaptic anchoring. J Neurochem. 2006;96:1659–1666. doi: 10.1111/j.1471-4159.2006.03679.x. [DOI] [PubMed] [Google Scholar]
- Link W, Konietzko G, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford G, Yamagata K, Kaufmann W, Barnes C, Sanders L, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu Rev Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- Martone ME, Pollock JA, Jones YZ, Ellisman MH. Ultra-structural localization of dendritic messenger RNA in adult rat hippocampus. J Neurosci. 1996;16:7437–7446. doi: 10.1523/JNEUROSCI.16-23-07437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Kubik S, Lewandowski G, Guzowski JF. Networks of neurons, networks of genes: an integrated view of memory consolidation. Neurobiol Learn Mem. 2008;89:269–284. doi: 10.1016/j.nlm.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada D, Ozawa F, Inokuchi K. Input-specific spine entry of soma-derived Vesl-1S protein conforms to synaptic tagging. Science. 2009;324:904–909. doi: 10.1126/science.1171498. [DOI] [PubMed] [Google Scholar]
- Ostroff LE, Fiala JC, Allwardt B, Harris KM. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35:535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bosl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Can molecules explain long-term potentiation? Nat Neurosci. 1999;2:597–604. doi: 10.1038/10154. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, Sheng M. Role of septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Edwards JG, Riley N, Provance DW, Jr, Karcher R, Li XD, Davison IG, Ikebe M, Mercer JA, Kauer JA, Ehlers MD. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Vessey JP, Konecna A, Dahm R, Macchi P, Kiebler MA. The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr Biol. 2007;17:1746–1751. doi: 10.1016/j.cub.2007.08.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.